Abstract
One of the most critical infrastructures during the COVID-19 pandemic are intensive care units (ICU). ICU’s crucial task is to preserve the lives of patients and mitigate the pandemic’s impact on the population. However, most ICUs plan only one day ahead. This short-term planning becomes an obstacle during disaster situations since physicians need to decide efficiently and ensure the timely treatment of high-risk patients. Integrating machine learning (ML) systems for clinical decision support could improve this process by predicting the mortality risk of critically ill patients. Several ML approaches tackling this problem have already shown promising results. However, these systems mostly neglect the integration of explicit domain knowledge, which is crucial to ensure prediction quality and adaptability. Otherwise, black-box systems might base their decision on confounding variables and improper relationships. Following design science research, we utilize a unique dataset of patients diagnosed with SARS-CoV-2 in ICU care to design a clinical decision support system by combining ML and expert knowledge in the form of a severity score. We show that by augmenting the system with expert knowledge, its overall performance improves compared to the baseline approach.
Similar content being viewed by others
1 Introduction
The healthcare sector faces tremendous challenges: Costs continue to rise, and physicians have less time available for their patients compared to recent years [5, 36]. Global crises such as the COVID-19 pandemic increase the tremendous time pressure on physicians even further. To cope with such limited resources, using technologies like Artificial Intelligence (AI) seems to be a promising approach to relieve the burden on physicians. Recent literature in the COVID-19 pandemic has shown the success of clinical decision support systems (CDSS) which yield high predictive performance [3, 10, 14]. However, focusing solely on the predictive performance of respective systems bears the danger of overfitting and the lack of adaptability to other data sources [39]. The COVID-19 pandemic demonstrated the need for more flexible approaches. Early datasets available only included a limited and likely skewed study population. In addition, rising infections in the population as well as changing virus variants and mutations require fast adaptability for potential predictive approaches. Therefore, we propose including explicit expert domain knowledge in the prediction process [22] to overcome this limitation. Experts could identify relevant information and help remove biases in the datasets and improve the generalizability of such CDSS.
One of the most critical infrastructures during the COVID-19 pandemic are intensive care units (ICU). Their crucial task is to preserve the lives of patients and mitigate the pandemic’s impact on the population. However, most ICUs plan only one day ahead [23]. In this regard, CDSS could help physicians to make better and more efficient decisions for each patient by predicting the critical state of a patient. When physicians know the severity of a disease in advance, they may be able to treat the patient in time. Therefore, we present the design of a data-centric clinical decision support system that integrates expert knowledge and predicts the mortality risk on the patient level to increase the quality of care in ICUs. With our CDSS design, we aim to support the decision-making process and reduce staff workload in ICUs.
We use a unique dataset of patients diagnosed with SARS-CoV-2 in ICU care of a German university hospital for the evaluation of our system design. The dataset consists of time-variant data during the entire stay of patients including monitoring data, laboratory values and treatments such as artificial ventilation. Additionally, we augment the dataset with expert knowledge from on-site physicians. Relying on this expert knowledge enables us to integrate decision processes of ICUs into our system and prevent the integration of misleading confounders [15].
The majority of the mortality risk prediction systems in the literature lack the integration of explicit domain expert knowledge into clinical decision support systems. Therefore, we want to answer the following research question: How to design clinical decision support systems predicting the mortality risk of COVID-19 patients that benefit from explicit expert knowledge?
To answer this RQ, we follow a design science research approach to first define our problem and derive generic design requirements (DR) for a CDSS based on a literature review. Second, we suggest and develop our solution as an IT artifact, and third, evaluate the performance of the artifact in comparison to a baseline system without explicit expert knowledge to finally draw our conclusion [19, 29, 31]. Design science aims to create and evaluate IT artifacts to solve a problem [19]. Vast research methodologies define iterative design processes built on a commonly understood framework [29]. We will elaborate on the problem formulation based on the theoretical background in more detail and explain our design as a suggestion to cope with the problem in the following section. We describe our methodological approach in the third section and evaluate it based on our use case of mortality risk prediction of patients in the ICU in light of the COVID-19 pandemic. Finally, we conclude the design science process of our artifact.
2 Background
2.1 Clinical Decision Support Systems
Integrating data science methods, such as machine learning (ML), into the clinical decision process can support physicians in multiple applications and tasks [5]. Since physicians cannot manually inspect and process large quantities of data, they require algorithms to handle big data [36]. ML algorithms can extract knowledge from large datasets and find patterns in the data hidden from human experts [22]. These algorithms often outperform humans regarding predictive performance and have been adopted for healthcare already in the late 1980s for clinical reasoning [13, 24]. Nowadays, improved algorithms and reduced costs for computational resources foster the applicability and predictive performance of such algorithms. The application of ML in the healthcare sector covers a wide range of use cases, e.g., cost prediction [32], risk profiling [30], or even the identification of suicidal people in online social media [7]. Additionally, there is a wide range of approaches for supporting clinical decision-making for critically ill patients, such as patients in the ICU. Current research shows promising performances by using different ML algorithms to make decisions. Thus, we propose the first DR 1: Ensure high predictive performance.
The literature on ML adoption in the ICU range from the prediction of a patient’s mortality risk [10, 16, 34], length of stay [23, 34], and subgroup identification [37]. These systems often rely on decision trees [10, 16, 23, 40], Bayesian inference [30], neural networks [23, 34], or support vector regressions [23]. For the COVID-19 pandemic, there has been enormous interest in applying ML on patient-level prognostics. For instance, Gao et al. [14] built an early warning system for COVID-19 mortality prediction based on the data of the first COVID-19 patients. Assaf et al. [3] utilize ML to predict the risk for the critical state of 162 hospitalized patients based on data available at hospital admission. Similarly, Cheng et al. [9] predict the ICU transfer of hospitalized patients. A few publications provide easy-to-use risk scores for the prediction task that can handle missing data without requiring complex computations (e.g., [28]). However, most approaches assume that all variables included in their models are available to other hospitals, a shortcoming addressed in our work. Consequently, we propose DR 2: Ensure the integration of adaptable and easily interpretable features.
2.2 The Role of Human Experts
Physicians using ML-based decision support systems should be aware of their development and limitations; otherwise, their usage may lead to ethical problems and potential medical errors [6]. Since humans are the decision-makers in the end, they need to take responsibility and compensate for technological errors [26]. Incorporating a physician in the system development process and integrating expert knowledge into an ML-based system may be beneficial, especially for complex tasks [22]. Because humans and computers have complementary capabilities that can augment each other [11], an efficient combination of humans and ML systems could result in better predictions compared to the performance of the individual parts [13]. A human can interact with the ML component in different ways, e.g., by providing input data to the ML system, preprocessing the data, or checking the ML system’s results [21]. This “human in the loop” (HITL) setting enables human experts to constantly audit and alter ML systems [17]. Accordingly, a human can learn from the ML system, and the ML system can learn from a human [1, 11]. This complementarity is especially beneficial to overcome limitations in the available data for ML training [38]. Human experts can help reduce the search space [21] and decrease the data required for ML models while increasing the reliability and robustness of the models [12]. However, most approaches in the healthcare context do not integrate features that rely on expert knowledge and hence, miss complementary information outside the dataset. Thus, we will focus on the initial altering phase of HITL and propose our last DR 3: Integrate domain expert knowledge in CDSS.
To the best of our knowledge, there is no COVID-19 related mortality risk CDSS demonstrating the benefit of expert-based risk scores as additional input for CDSS. In this paper, we will therefore focus on the additional value of integrating expert knowledge while satisfying our design requirements. We present our clinical decision support system design for mortality risk prediction based on expert knowledge in the following.
3 Methodology
3.1 Data
Our datasets consist of time-variant data of 439 patients diagnosed with SARS-CoV-2 in ICU care from 2020 to 2021 of a German university hospital.Footnote 1 The three datasets, namely patient meta, monitoring, and laboratory data, were retrieved from the hospital’s electronic health record (EHR). To be precise, we have minute-wise monitoring data of patients’ stay in the ICU with approximately 9.17 million observations and laboratory data with 82.788 observations for our set of 439 patients. For each patient, the patient meta dataset contains information about gender, age, admission and discharge date (of both the hospital and the ICU), detailed ventilation information, and whether they survived or passed away during their ICU stay. The monitoring dataset contains the minute-wise values of each patient’s vital condition, including – among others – heart rate, blood pressure (arterial and/or non-invasive), oxygen saturation (SpO\(_{2}\)), and temperature. Notably, not every value is available for each patient and each timespan (e.g., all patients have either an entry for the systolic arterial blood pressure or the systolic non-invasive blood pressure, but only half of the patients have an entry for both). This incompleteness is a common issue with EHR data, and we will address it in our preprocessing. Furthermore, not all entries are subject to high data quality. For instance, we had to omit temperature since 88% of the values available were below 30\(^{\circ }\)C, including negative values. The laboratory dataset captures a wide range of laboratory values for the patients measured at different irregular points in time, such as IL6, albumin, or pCO\(_{2}\).
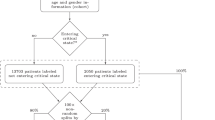
Figure 1 shows the distribution of age in our dataset. Around 76% of the patients are male and the average age is 62 years (around 3% of the patients are less than 30 years old, 82% are 50 years or older, and around 11% are 80 years or older). Around half of the patients received invasive ventilation via endotracheal intubation, 14% received an ECMO, and around 39% passed away.
3.2 Preprocessing
While each patient is identifiable through a unique ID, the monitoring and laboratory datasets additionally contained a time attribute. Therefore, we merged the different datasets by first, merging monitoring and laboratory datasets on the unique ID and time values, and second, merging this dataset with the meta dataset. As a result, our new dataset contains the entire monitoring history of each patient recorded in the meta dataset. After merging the data, we checked for missing values in the laboratory attributes. Thus, variables were excluded from further processing steps if they were not measured once for at least 20% of patients. Figure 2 summarizes the datasets and the workflow of our preprocessing and evaluation method.
Afterwards, we calculated the length of stay for each patient in the ICU with the available admission date and discharge date or date of death. As Fig. 1 illustrates, many patients only stayed for a few days in the ICU (on average 14 days). The figure also shows that there are a few outliers which we excluded in our analysis. In total, we removed 15 patients in this preprocessing step, which left us with 424 patients.
To deal with potential outlier values originating from monitoring and laboratory data due to measurement errors, we acquired additional information from the on-site physicians regarding ranges of physiological possible values. If a value was either above or below the respective maximum or minimum range value, we replaced it with the respective number. We imputed missing data by forward filling available values on the patient-level. We repeated this step with backward filling if necessary. Finally, we applied a rolling mean with a ten-minute window for each numerical value to smooth the data and mitigate the impact of outliers which potentially occur through measurement errors since we are interested in the forecast of a more extended period. Afterwards, we reduced our dataset to the tenth record of every patient (i.e., a data record every 10 min per patient).
Furthermore, we selected attributes to evaluate our system and removed patients who were missing these attributes. This additional step reduced the number of patients that we included in our dataset to 323. By utilizing expert guidance and statistical correlation analysis, we selected the following features: age, arterial blood pressure medium, heart rate, FiO\(_{2}\), SpO\(_{2}\), positive end-expiratory pressure (PEEP), C-reactive protein, albumin, pCO\(_{2}\), thrombocytes, IL-6, aspartate aminotransferase (GOT), and HCO\(_{3-}\). In addition, we defined two binary variables indicating when a patient received invasive ventilation via endotracheal intubation or was treated with ECMO, respectively. We used the intubation and ECMO time frame provided through the meta dataset. As we have time-variant data, we did not include any complications that occurred during the later stages of the patients’ ICU stay as features that could potentially leak information to a previous time period. Finally, we created lags for our set of features. To be precise, we created lags of six hours, i.e., 36 lags for each observation and variable.
For the outcome variable of our prediction task, we defined the mortality risk as low if a patient survived their stay in the ICU and high if a patient did not. Our dataset indicates the mortality with the binary attribute “death” for each patient. We assume that the (last) two days before the death of a patient are the most indicative of their mortality risk. Therefore, each patient in the final dataset had to stay at least two days in the ICU. Since most ICUs only plan one day ahead [23] and in order to predict the mortality risk at least one day ahead of a patient’s death, we also removed the last day from the dataset. Thus, the dataset only contains the penultimate day of each patient. Our final cohort of patients that we utilized for evaluation consisted of 306 patients, of which 168 (55%) survived their stay in ICU care and 138 (45%) did not.
We enabled the integration of explicit domain knowledge into the system of our IT artifact with our implementation of the severity score. Concretely, we incorporated expert knowledge in the form of severity thresholds (see appendix). For this purpose, the on-site physicians provided information on the severity threshold of multiple variables. We encoded this information as a binary variable, only for the respective longitudinal variable at each point in time to prevent temporal leakage. The severity of a variable equals one if the current value of the patient exceeds or goes below the threshold and zero otherwise. The severity score is the sum of these binary severity features. In total, we encoded nine severity features to produce the severity score. Importantly, we note that the severity score only includes severity features, whose (continuous) features were available in evaluating both expert-augmented and baseline systems.
3.3 Evaluation
We evaluated two clinical decision support system designs: One baseline approach without additional information by medical domain experts as a benchmark and another expert-augmented system with encoded domain knowledge. In order to predict the mortality risk of each patient, we used a long short-term memory (LSTM) network [20]. LSTM is a class of recurrent neural networks that performs particularly well on sequential and multidimensional time-series data [34] and is commonly used in healthcare (e.g., [8, 33,34,35]). We created the following network architecture for the LSTM model: One LSTM input layer with 32 units, one hidden dense layer with 128 units, and a single unit output layer. We added one dropout layer with a 0.75 rate after each layer except for the output layer for regularization purposes and to prevent overfitting.
Moreover, we used the ReLU activation function for our hidden layer and the sigmoid function for our output layer, as our prediction problem is a binary classification task. To compile our LSTM model, we chose the recommended binary cross-entropy loss function, Adam optimizer, and the metric accuracy. Additionally, we used a batch size of 32 and five training epochs. These hyperparameters are commonly used in previous literature and we did not optimize them in this case [8].
To get a more reliable estimate of the performance of both systems, we performed repeated cross-validation. Specifically, we repeated cross-validation by using the same five random seeds for both systems and stratified group k-fold with five splits for both sets of features – one including the severity score and one excluding it. In total, we conducted 25 different evaluations of both system performances. First, we utilized stratified group k-fold to ensure that each patient’s data is either in the training or the validation set to prevent data leakage. Second, each set contains approximately the same percentage of samples of both outcome classes. Finally, we scaled each fold using min-max normalization to fit on and transform the training set and transform the validation set with the fitted scaler. In the following section, we compare the mean performance of both systems by reporting binary classification metrics.
In addition, we evaluated the severity score separately without utilizing a ML model in two ways. First, we set different values of the severity score as thresholds to compute the same set of metrics that we used to compare our system designs. The thresholds indicate whether a patient will survive their stay or not. We evaluate these thresholds to show the added benefit of our system design in comparison to standalone expert knowledge. Second, we plotted the mean severity score of the patients that we used for our system and added a 95% confidence interval to the scores to compare patients that survived their stay in the ICU and those that did not.
3.4 Results
Overall, the performance of the expert-augmented system is better than the baseline system based on the chosen classification metrics. Figure 3 shows the area under the receiver operating characteristic (AUROC) for the expert-augmented system. In Table 1, we observe that the AUROC, accuracy, recall and F1 metric show improved results compared to the baseline approach. Notably, the highest increase is in recall (bold).Footnote 2 The results are comparable to the high predictive performance of other CDSS in the literature, which used cross-sectional data of COVID-19 patients to predict the mortality risk (e.g. [2, 4, 14, 27]).
With respect to the standalone severity score, we show the evaluation in Table 2. This table presents classification metrics for different values of severity score thresholds ranging from the minimum zero to the maximum nine. For each threshold value, we implemented the binary decision rule that a patient will survive their stay in the ICU if their severity score is below the threshold value. Afterwards, we computed different metrics to compare them with our system performance. With increasing threshold value, the precision of the severity score increases, while the recall decreases indicating the trade-off between precision and recall for different values of the threshold. Notably, for the threshold equaling 5, we find the most balanced performance of the severity score across the specified metrics. In particular, F1 score and accuracy have the highest values, while precision and recall have similar values in comparison to other thresholds. When we compare these values with the metrics of our expert-augmented CDSS design, our results show that the system design has a better performance overall. Figure 4 shows the mean severity score with a confidence interval of 95% of the last 48 h of all patients we have evaluated (48 on the x-axis represents the last point in time of patients in ICU care). Moreover, the two graphs represent the two classes of patients that we are interested in for our expert-augmented system, i.e., surviving and deceased patients. The figure illustrates that patients who survived their stay in the ICU have a much lower score than the patients who deceased. Distinctively, in the group of deceased patients, we observe that the mean severity score increases over time, whereas the score decreases for the surviving patients. This result suggests that the severity score can serve as an indicator for the vulnerable state of a patient in the ICU.
4 Discussion
The results of our evaluation show the potential of integrating expert knowledge into clinical decision support systems. Following the integration of an iterative search process from the design science methodology [29], we considered the performance difference and interplay of the different metrics. We evaluated our three design requirements by comparing (DR 1) the predictive performance of the baseline system and the expert-augmented system based on (DR 2) adaptable and easily interpretable features derived from (DR 3) domain experts.
Since the baseline system already performs quite well, one can expect minor overall improvements in the expert-augmented system. However, we argue that these improvements can still contribute substantially to better decision-making. To be precise, we looked at designing CDSS to predict the mortality risk of COVID-19 patients in the ICU. A CDSS built for this task is, in fact, used for a matter of life or death. Hence, any performance increase can potentially lead to better treatment of patients by the attending physicians, possibly detecting their deteriorating condition earlier and preserving their lives.
Most importantly, we emphasize that the highest performance increase of the expert-augmented system compared to the baseline system was recall (for a tradeoff in precision performance). We argue that recall is the most relevant metric for our use case. Specifically, it is more important to avoid false negatives, i.e., systems falsely predicting patients to have a low mortality risk than false positives. Failing to recognize patients with high mortality risk has drastic consequences, as they are actually at risk of dying. Moreover, we compared our system design with the standalone severity score at different threshold values. Our results showed that the expert-augmented system performs better than the severity score by itself which demonstrates the added benefit of using the expert-augmented system instead of expert knowledge alone. In sum, the expert-augmented system better predicts the relevant group of high-risk patients, satisfying DR 1.
As the results showed, the mean severity score increases over time for deceased patients, while it decreases for surviving patients. Intuitively, this makes sense, as the health condition of the deceased patients worsens until their death. In contrast, the health condition of patients that survive improves as they are discharged, and thus, their severity score decreases. Therefore, we argue that the severity score derived from expert knowledge has utility as an IT artifact in and of itself because it clearly differentiates the health condition of patients at high risk of mortality from those that are not. Furthermore, the severity score can be a general indicator of a patient’s health condition. Therefore, any medical personnel can quickly and efficiently interpret the sum of severity features as a decision support tool. This ease of use becomes increasingly important when time is a crucial factor as physicians have to make many decisions efficiently and need to avoid information overload. The score can also prompt physicians to investigate the relevant variables further and make appropriate decisions based on their value, e.g., a high severity score can urge personnel to act more quickly. Additionally, researchers and practitioners can adapt the score to different hospitals’ needs based on the available variables of each hospital and the thresholds that they choose to utilize. This adaptability is essential because not every hospital has the same resources available to them.Footnote 3 Thereby, we offer an adaptable and easily interpretable feature based on domain expertise, satisfying DR 2 and DR 3.
Moreover, physicians may feel more comfortable using expert-augmented systems due to the inclusion of domain experts in the ML model development process. The literature suggests that trust is the main hindrance to ML adoption in healthcare [15]. However, the inclusion of medical professionals in ML system development and implementation as part of the “human in the loop” process might increase their adoption rates [18].
Finally, we may even further extend the use case of the severity score if we assume that a variable that was not measured during a specific point in time (the observation being N/A) is not severe. This idea follows the assumption that hospitals often only measure different variables for a patient’s health condition if they deem the condition to be harmful or severe. Using this assumption allows system designers to use multiple variables that have many missing values and, as a result, more patients’ data to potentially improve the system’s performance. Evaluating this performance is crucial in the design process to ensure the artifact’s utility, quality, and efficacy [19].
5 Limitations and Conclusion
In this paper, we designed a clinical decision support system to predict the mortality risk of ICU patients diagnosed with SARS-CoV-2 by integrating domain expert knowledge into the system. We utilized design science research to formulate our problem definition based on the theoretical background and identified as well as satisfied the three design requirements: high predictive performance (DR 1), use of adaptable and easily interpretable features (DR 2), and integration of domain expert knowledge (DR 3). Furthermore, we followed design science to implement an IT artifact and evaluate its performance compared to the baseline approach through a design search process [19]. The results indicate that domain expert knowledge has the potential to improve the performance of systems. It also enables a more adaptable and transparent use of CDSS. We note that although the performance of our baseline system is comparable to the literature, we were able to show that our expert-augmented approach outperformed this system and thus enabled a better forecasting of the relevant group of people: patients at high risk of deceasing. Consequently, we argue that other CDSS applying our adaptable expert-augmented approach can benefit from the increased performance and improved interpretability of features.
We implemented our design by imputing N/A values with the rolling average of each value. We acknowledge that other promising imputation strategies are also feasible. However, the results of the baseline approach indicate that using this imputation strategy performs sufficiently well to predict our target variable. We have also used the assumption that the severity score is additive with a limited set of binary severity threshold-based features. Future research could explore other scoring methods such as different weighting approaches or test other severity feature combinations and more sensitive (multiclass) thresholds to improve the scoring mechanism.
Additionally, we note that there is a potential selection bias in the variables integrated into the CDSS. We selected the variables based on their availability in our dataset, the current state of research and expert guidance. We also only used a single dataset relying on the patients of one hospital only. This limitation stems from the fact that large-scale time-variant datasets of COVID-19 patients were almost nonexistent at the time of analysis, and data privacy regulations hamper critical health-related data aggregation. Future work can follow a multicenter approach to integrate multiple datasets from different hospitals.
With this paper, we contribute to the literature on clinical decision support systems and the integration of expert knowledge from the healthcare domain into the system of our IT artifact following design science research methodology [19]. We defined design requirements for expert-augmented CDSS and showed with our evaluation that by augmenting CDSS with expert knowledge, the system’s performance increases compared to the baseline approach and standalone expert knowledge. In addition, we created the severity score as an adaptable measure of a patient’s health condition. The severity score serves two purposes: First, it improves the general performance of the ML-based CDSS, and second, physicians can use it as an adjustable standalone decision support tool by combining different available attributes and thresholds if machine learning know-how is not available to the user. Incorporating domain expert knowledge in CDSS can aid medical decision-makers in a very critical environment.
Notes
We obtained the data retrospectively. Thus, our CDSS results did not influence any medical decisions.
Since some researchers expressed concerns about learning weights from past values with LSTMs, we repeated our evaluation with a temporal convolutional network [25]. As we show in the appendix, our evaluation produced the same trends in all performance metrics. However, our LSTM model performs better for most evaluation metrics.
To assess adaptability and generalizability, we conducted an ablation study to evaluate the predictive performance with or without each feature such that only one feature was removed during each evaluation. If the feature that we removed is part of the severity score, then we also removed its occurrence in the score. In the end, the expert-augmented system yielded a similar predictive performance on average compared to the full set of features suggesting adaptability and generalizability.
References
Abdel-Karim BM, Pfeuffer N, Rohde G, Hinz O (2020) How and what can humans learn from being in the loop? KI-Künstliche Intell 34(2):199–207 (Publisher: Springer)
Abdulaal A, Patel A, Charani E, Denny S, Mughal N, Moore L (2020) Prognostic modeling of COVID-19 using artificial intelligence in the United Kingdom: model development and validation. J Med Internet Res 22(8):e20259 (Publisher: JMIR Publications Inc., Toronto, Canada)
Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S, Shilo N, Epstein A, Mor-Cohen R, Biber A (2020) Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern Emerg Med 15(8):1435–1443 (Publisher: Springer)
Bertsimas D, Lukin G, Mingardi L, Nohadani O, Orfanoudaki A, Stellato B, Wiberg H, Gonzalez-Garcia S, Parra-Calderón CL, Robinson K, Schneider M, Stein B, Estirado A, Beccara L, Canino R, Dal Bello M, Pezzetti F, Pan A, Group THCS (2020) Covid-19 mortality risk assessment: an international multi-center study. PLOS ONE 15(12):1–13. https://doi.org/10.1371/journal.pone.0243262
Bhardwaj R, Nambiar AR, Dutta D (2017) A study of machine learning in healthcare. In: 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC), vol. 2, pp 236–241. IEEE
Char Danton S, Shah Nigam H (2018) Magnus David. Implementing machine learning in health care-addressing ethical challenges. N Engl J Med 378:981–983
Chau M, Li TM, Wong PW, Xu JJ, Yip PS, Chen H (2020) Finding people with emotional distress in online social media: a design combining machine learning and rule-based classification. MIS Q 44(2):933–955
Che Z, Purushotham S, Cho K, Sontag D, Liu Y (2018) Recurrent neural networks for multivariate time series with missing values. Sci Rep 8(1):1–12 (Publisher: Nature Publishing Group)
Cheng FY, Joshi H, Tandon P, Freeman R, Reich DL, Mazumdar M, Kohli-Seth R, Levin MA, Timsina P, Kia A (2020) Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J Clin Med 9(6):1668 (Publisher: MDPI)
Darabi HR, Tsinis D, Zecchini K, Whitcomb WF, Liss A (2018) Forecasting mortality risk for patients admitted to intensive care units using machine learning. Proc Computer Sci 140:306–313 (Publisher: Elsevier)
Dellermann D, Ebel P, Söllner M, Leimeister JM (2019) Hybrid intelligence. Bus Inf Syst Eng 61(5):637–643 (Publisher: Springer)
Deng C, Ji X, Rainey C, Zhang J, Lu W (2020) Integrating machine learning with human knowledge. iScience 23(11):101656. https://doi.org/10.1016/j.isci.2020.101656
Fuegener A, Grahl J, Gupta A, Ketter W (2022) Cognitive challenges in human-artificial intelligence collaboration: investigating the path toward productive delegation. Inf Syst Res 33(2):678–696 (Publisher: INFORMS)
Gao Y, Cai GY, Fang W, Li HY, Wang SY, Chen L, Yu Y, Liu D, Xu S, Cui PF (2020) Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat Commun 11(1):1–10 (Publisher: Nature Publishing Group)
Gennatas ED, Friedman JH, Ungar LH, Pirracchio R, Eaton E, Reichmann LG, Interian Y, Luna JM, Simone CB, Auerbach A (2020) Expert-augmented machine learning. Proc Natl Acad Sci 117(9):4571–4577 (Publisher: National Acad Sciences)
Ghose S, Mitra J, Khanna S, Dowling J (2015) An improved patient-specific mortality risk prediction in ICU in a random forest classification framework. Stud Health Technol Inform 214:56–61
Grønsund T, Aanestad M (2020) Augmenting the algorithm: emerging human-in-the-loop work configurations. J Strateg Inf Syst 29(2):101614
Habehh H, Gohel S (2021) Machine learning in healthcare. Curr Genom 22(4):291
Hevner AR, March ST, Park J, Ram S (2004) Design science in information systems research. MIS Q 28(1):75–105
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780. https://doi.org/10.1162/neco.1997.9.8.1735
Holzinger A (2016) Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inf 3(2):119–131 (Publisher: Springer)
Holzinger A, Plass M, Holzinger K, Crişan GC, Pintea CM, Palade V (2016) Towards interactive Machine Learning (iML): applying ant colony algorithms to solve the traveling salesman problem with the human-in-the-loop approach. In: International Conference on Availability, Reliability, and Security, pp 81–95. Springer
Houthooft R, Ruyssinck J, van der Herten J, Stijven S, Couckuyt I, Gadeyne B, Ongenae F, Colpaert K, Decruyenaere J, Dhaene T (2015) Predictive modelling of survival and length of stay in critically ill patients using sequential organ failure scores. Artif Intell Med 63(3):191–207 (Publisher: Elsevier)
Hu PJH (2003) E-diagnosis support systems: a web-based decision support system to aid complex lower back pain diagnosis & management. E-Health
Ismail AR, Jovanovic S, Ramzan N, Rabah H (2023) Ecg classification using an optimal temporal convolutional network for remote health monitoring. Sensors 23(3):1697
Jussupow E, Spohrer K, Heinzl A, Gawlitza J (2021) Augmenting medical diagnosis decisions? An investigation into physicians’ decision-making process with artificial intelligence. Inf Syst Res 32(3):713–735 (Publisher: INFORMS)
Kar S, Chawla R, Haranath SP, Ramasubban S, Ramakrishnan N, Vaishya R, Sibal A, Reddy S (2021) Multivariable mortality risk prediction using machine learning for COVID-19 patients at admission (AICOVID). Sci Rep 11(1):1–11 (Publisher: Nature Publishing Group)
Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, Dunning J, Fairfield CJ, Gamble C, Green CA (2020) Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. bmj 370. Publisher: British Medical Journal Publishing Group
Kuechler B, Vaishnavi V (2008) On theory development in design science research: anatomy of a research project. Eur J Inf Syst 17(5):489–504
Lin YK, Chen H, Brown RA, Li SH, Yang HJ (2017) Healthcare predictive analytics for risk profiling in chronic care: a Bayesian multitask learning approach. Mis Q 41(2):473–495
Meth H, Mueller B, Maedche A (2015) Designing a requirement mining system. J Assoc Inf Syst 16(9):2
Morid MA, Sheng ORL, Del Fiol G, Facelli JC, Bray BE, Abdelrahman S (2020) Temporal pattern detection to predict adverse events in critical care: case study with acute kidney injury. JMIR Med inf 8(3):e14272 (Publisher: JMIR Publications Inc., Toronto, Canada)
Morid MA, Sheng ORL, Dunbar J (2023) Time series prediction using deep learning methods in healthcare. ACM Trans Manag Inf Syst 14(1):1–29
Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M (2018) Scalable and accurate deep learning with electronic health records. NPJ Digit Med 1(1):1–10 (Publisher: Nature Publishing Group)
Shastri S, Singh K, Kumar S, Kour P, Mansotra V (2020) Time series forecasting of covid-19 using deep learning models: India-usa comparative case study. Chaos Solitons Fract 140:110227
Topol EJ (2019) High-performance medicine: the convergence of human and artificial intelligence. Nat Med 25(1):44–56. https://doi.org/10.1038/s41591-018-0300-7. (Number: 1 Publisher: Nature Publishing Group)
Vranas KC, Jopling JK, Sweeney TE, Ramsey MC, Milstein AS, Slatore CG, Escobar GJ, Liu VX (2017) Identifying distinct subgroups of intensive care unit patients: a machine learning approach. Crit Care Med 45(10):1607–1615. https://doi.org/10.1097/CCM.0000000000002548
Wiethof C, Bittner E (2021) Hybrid Intelligence – Combining the Human in the Loop with the Computer in the Loop: A Systematic Literature Review. ICIS 2021 Proceedings. https://aisel.aisnet.org/icis2021/ai_business/ai_business/11
...Wynants L, Calster BV, Collins GS, Heinze G, Schuit E, Bonten MMJ, Damen JAA, Debray TPA, Vos MD, Dhiman P, Haller MC, Harhay MO, Henckaerts L, Kreuzberger N, Lohman A, Luijken K, Ma J, Martin G, Andaur CL, Reitsma JB, Sergeant JC, Shi C, Skoetz N, Smits LJM, Snell KIE, Sperrin M, Spijker R, Steyerberg EW, Takada T, Kuijk SMJV, Royen FSV, Wallisch C, Wilkinson J, Hooft L, Moons KGM, Smeden MV (2020) Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ (Clin Res Edn). https://doi.org/10.1136/bmj.m1328
Yan L, Zhang HT, Goncalves J, Xiao Y, Wang M, Guo Y, Sun C, Tang X, Jing L, Zhang M, Huang X, Xiao Y, Cao H, Chen Y, Ren T, Wang F, Xiao Y, Huang S, Tan X, Huang N, Jiao B, Cheng C, Zhang Y, Luo A, Mombaerts L, Jin J, Cao Z, Li S, Xu H, Yuan Y (2020) An interpretable mortality prediction model for COVID-19 patients. Nat Mach Intell 2(5):283–288. https://doi.org/10.1038/s42256-020-0180-7. (Number: 5 Publisher: Nature Publishing Group)
Funding
Open Access funding enabled and organized by Projekt DEAL. Johannes Chen was funded by the Alfons und Gertrud Kassel-Stiftung. Maximilian Lowin and Domenic Kellner were funded by the German Federal Ministry of Education and Research as part of the egePan project (Grant 01KX2021). We thank the University Hospital Frankfurt for the kind support during the research investigation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Appendix
Appendix
1.1 List of Severity Thresholds
We used the following severity thresholds to calculate the severity score of each patient:
-
pCO\(_2\) > 50 mmHg
-
SpO\(_2\) \(\ge\) 93%
-
Thrombocytes < 100/nl
-
C-reactive protein \(\ge\) 10 mg/l
-
IL-6 \(\ge\) 10 pg/ml
-
GOT > 40 U/l
-
Albumin < 3.5 g/dL
-
ARDS*: PO\(_{2}\)/FiO\(_{2}\) \(\le\) 100 mmHg with PEEP \(\ge\) 10 cm H\(_{2}\)O
-
Severe metabolic acidemia*: pH\(_t\) < 7.20, HCO\(_{3-}\) < 22 mmol/L, aBE \(\le\) 5 mmol/L, pCO\(_{2}\) \(=\) 1.5 \(*\) (HCO\(_{3-}\)) \(+\) 8 ± 2 mmHg
*) We simplified these rules in an iterative manner to ensure higher predictive performance.
1.2 Classification Metrics of Temporal Convolutional Network (TCN)
Table 3 shows the classification metrics of baseline and expert-augmented designs based on temporal convolutional network [25] with the same set of metrics as our LSTM model. While the direction of the performance differences is the same between LSTM and TCN model for each metric when comparing baseline and expert-augmented designs with recall being the highest increase (bold), our LSTM model yields greater overall performance in all metrics but precision.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, J., Lowin, M., Kellner, D. et al. Designing Expert-Augmented Clinical Decision Support Systems to Predict Mortality Risk in ICUs. Künstl Intell (2023). https://doi.org/10.1007/s13218-023-00808-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13218-023-00808-7