Abstract
Chitinase is an enzyme that catalyzes the degradation of chitin, commonly induced upon the attack of pathogens and other stresses. A cDNA (MsChi1) was isolated from Metroxylon sagu and expressed predominantly in the inflorescence tissue of M. sagu, suggesting its role in developmental processes. The chitinase cDNA was detected and isolated via differential display and rapid amplification of cDNA ends (RACE). Primers specific to M. saguchitinase were used as probes to amplify the 3′-end and 5′-end regions of chitinase cDNA. Transcript analysis showed that chitinase is expressed in inflorescence and meristem tissues but was not detected in the leaf tissue. Sequence analysis of amplified cDNA fragments of 3′-end and 5′-end regions indicated that the chitinase cDNA was successfully amplified. The M. saguchitinase cDNA isolated was approximately 1,143 bp long and corresponds to 312 predicted amino acids. Alignments of nucleotide and amino acid have grouped this chitinase to family 19 class I chitinase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metroxylon sagu or locally known as sago palm, belongs to the Lepidocaryoid subfamily of Arecaceae (Palmae). It is a once-flowering (hapaxanthic), tillering or suckering perennial that thrives in swamp areas. The trunk of the M. sagu is used to obtain starch for human consumption (Flach 1984) and it is an important plant contributing to the economy of Sarawak. The advantages of M. sagu cultivation are that it requires no fertilization, has few natural pests or diseases and can be grown in swamp areas where it is impossible for other crops to thrive (Abd-Aziz 2002).
Chitinases are proteins that catalyze the hydrolysis of β-1,4-linkages of N-acetyl-d-glucosamine polymer of chitin; a major component of the exoskeleton of insects, crustacean shells and cell wall of many fungi (Bishop et al. 2000; El-Sayed et al. 2000; Passarinho and de Vries 2002). Chitinases are present in many higher plant species, although higher plants themselves do not contain chitin, chitosan or chitin-like substrate (Boller et al. 1983; Hirano et al. 1988). Chitinases are often described as pathogenesis-related proteins because they are constitutively expressed at low levels and increase dramatically in response to fungal, bacteria or viral infections (Graham and Sticklen 1994; van Loon 1999). Chitinases also play a role in plant defense mechanism by damaging chitin structures of parasites (Bishop et al. 2000; Odjakova and Hadjiivanova 2001). Apart from that, chitinase can also be induced by stress or elicitors such as wounding, salicylic acid and ethylene (Graham and Sticklen 1994; Leon et al. 2001).
Plant chitinases are classified in to classes I–V depending on their sequences and primary structures (Collinge et al. 1993; Neuhaus et al. 1996). Several studies have revealed that some chitinase are expressed at higher levels in healthy floral and flower-predominant organs such as potato (Wemmer et al. 1994) and tomato (Harikrishna et al. 1996). The expression of chitinase in flowers have also been detected in Arabidopsis thaliana (Samac et al. 1990; Passarinho et al. 2001), petunia (Leung 1992), parsley (Ponath et al. 2000), rice (Takakura et al. 2000) and tobacco (Lotan et al. 1989; Trudel and Asselin 1989; Neale et al. 1990). Chitinase expressions were also found in other tissues such as the roots of Arabidopsis thaliana (Samac and Shah 1991), rice (Lamb et al. 1991) and tobacco (Memelink et al. 1990; Neale et al. 1990); as well as in embryogenic cultures of carrot (van Hengel et al. 1998) and spruce (Egertsdotter 1996; Dong and Dunstan 1997). In other plants such as barley (Leah et al. 1994), carrot (van Hengel et al. 1998), pea (Petruzzelli et al. 1999) and soybean (Yeboah et al. 1998) the chitinase gene was detected in the seeds. Expression of chitinase due to infection by pathogen has also been shown in several plants such as pineapples (Taira et al. 2005) and grapes (Vasanthaiah et al. 2008).
Here we report the differential expression of chitinase in the leaf, meristem and inflorescence of M. sagu, the isolation and characterization of a near complete chitinase cDNA from inflorescence tissue.
Materials and methods
Plant materials
Three tissue types were selected in this study: leaf, meristem and inflorescence. The leaves of M. sagu (Fig. 1a, b) were collected from the UNIMAS plant house. The meristem and inflorescence (Fig. 1c–e) were collected from Sri Aman areas. All the samples were stored at −80 °C,
The leaves, meristem and inflorescence samples of M. sagu. a The leaf samples. b The vegetative growth of the palm. c The meristem samples. d The inflorescence sample. e The inflorescence developing palm (photo copyright CRAUN Research Sdn. Bhd, taken from Tie 2004)
Ribonucleic acids (RNA) isolation and purification
Isolation of total RNA was carried out using the method described by Gasic et al. (2004), with modifications for small scale extraction. The quality and quantity of the isolated RNA were verified by agarose gel electrophoresis and spectrophotometry (Ultrospec® 1100 pro, Amersham Pharmacia Biotech). The total RNA samples were treated with RQ1 RNase-Free DNase (Promega), to ensure the samples were completely free of genomic DNA contaminants (Tan and Roslan 2008).
First-strand cDNA synthesis
First-strand cDNA was synthesized using RevertAidTM Moloney Murine Leukemia Virus Reverse Transcriptase (M-MuLV RT) (Fermentas) from purified total RNA of leaf, meristem and inflorescence according to the method described by Tan and Roslan (2008). The oligo(dt)15ACP was used as a cDNA synthesis primer.
Differential display reverse transcription PCR
Differential display reverse transcription PCR (DDRT-PCR) was undertaken according to the method described by Kim et al. (2004) with modifications in the PCR steps. Two annealing control primers (ACP2 and ACP3) were used to screen for differentially expressed genes in the selected tissues. PCR was performed using two combinations of ACP primers; oligo(dt)15ACP/AP2 and oligo(dt)15ACP/AP3. PCR was carried out in a final volume of 25 μl. The reaction mixture included: 2.5 μl of 10× PCR buffer, 0.25 μl of Taq DNA polymerase (5 U/μl) (Fermentas), 1.0 μl of each forward and reverse primers (10 μM), 1.5 μl of 25 mM MgCl2, 0.5 μl of 10 mM dNTPs, sterile distilled water and 3.0 μl of 10× diluted RT product. PCR was performed using a Mastercycler Personal (Eppendorf) with thermal cycling conditions of one cycle of 94 °C for 4 min followed by five cycles at 94 °C for 1 min, 36 °C for 1 min, and 72 °C for 2 min. A further 35 cycles was undertaken at 94 °C for 1 min, 65 °C for 1 min, 72 °C for 2 min, and a 5-min final extension at 72 °C. The amplified PCR products were separated in 2.5% agarose gel stained with ethidium bromide.
Isolation of the 3′-end of chitinase cDNA
The 3′ and 5′RACE were conducted according to the method described by Frohman et al. (1988). The strategy employed to isolate the chitinase cDNA is given in the diagram below.

The first strand cDNA was generated using oligo(dt)17 primer (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′). A combination of emChi-f (5′-GGTGTCATCACCAACATCATCAA-3′) and oligo(dt)17 was used to amplify the 3′-end of the chitinase cDNA in meristem, inflorescence and leaf tissues of M. sagu. PCR amplification was carried out in a final volume of 25 μl. The reaction mixture includes: 2.5 μl of 10× PCR buffer, 0.25 μl of Taq DNA polymerase (5 U/μl), 0.25 μl of each primer (10 μM), 1.0 μl of 25 mM MgCl2, 0.5 μl of 10 mM dNTPs, sterile water and 3.0 μl of diluted template. Thermal cycling condition was one cycle of 94 °C for 4 min; 35 cycles of 94 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min; and a final extension at 72 °C for 7 min.
Isolation of the 5′-end of chitinase cDNA
The first strand of cDNA from purified inflorescence total RNA was generated using gene specific primer, Chi-sp1 (5′-GCCTCTGGTTGTAGCAGTCCA-3′). The cDNA was purified and a terminal deoxynucleotidyl transferase (Fermentas) was used to tail the 3′-end of the cDNA with dATP prior to PCR amplification using Chi-sp2 (5′-GCCCTCCATTGATGATGTTG-3′) and oligo(dt)17 primer combination. Amplification was carried out in a final volume of 25 μl. The reaction mixture includes: 2.5 μl of 10× PCR buffer, 0.25 μl of Taq DNA polymerase (5 U/μl), 1.0 μl of each primer (10 μM), 1.5 μl of 25 mM MgCl2, 0.5 μl of 10 mM dNTPs, sterile water and 1.0 μl of diluted template. Thermal cycling condition was one cycle of 94 °C for 4 min; 30 cycles of 94 °C for 30 s, 55 °C for 45 s, 72 °C for 1.5 min; and a final extension at 72 °C for 7 min.
Cloning of PCR products
The purified PCR products were cloned into the pGEM-T Vector (Promega). The positive clones were screened via PCR using universal primer set T7 (5′-TAATACGACTCACTATAGGG-3′) and SP6 (5′-TATTTAGGTGACACTATAG-3′). Clones corresponding to the expected size were selected for plasmid extraction using the GeneJET™ Plasmid Miniprep Kit (Fermentas) and sequenced.
DNA sequencing and bioinformatics analysis
Direct sequencing on plasmid DNA was performed by 1st BASE Laboratories Sdn Bhd (Malaysia) using ABI PRISM® 377 DNA sequencer. The sequences were analyzed using ChromasPro version 1.34 software. Alignment of nucleotide sequences was conducted using European Bioinformatics Institute’s (EBI) Clustal W multiple alignment software and similarity sequence searches were carried out using programs at the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLASTx and BLASTn).
Results and discussion
DDRT-PCR of M. sagu tissues
The DDRT-PCR that was performed using oligo(dt)15ACP/AP2 primers combination generated several amplicons (indicated by arrows in Fig. 2a). Several amplicons were selected (a, b and c in Fig. 2a), cloned into a cloning vector and sequenced. From the BLAST analysis, the amplicons were determined to be derived from chitinase (msAP21, msAP22, and msAP23).
A 2.5% (w/v) agarose gel-electrophoresis of fragments amplified using (a) combination of oligo(dt)15ACP/AP2 primers. The fragments labelled as a, b and c represent fragments msAP21, msAP22 and msAP23, respectively. b Combination of oligo(dt)15ACP/AP3 primers. The arrow represents fragment msAP33. The M, I and L represent meristem, inflorescence and leaf cDNA samples, respectively. S represents a 100 bp Forever Ladder (Seegene)
The DDRT-PCR method using oligo(dt)15ACP/AP3 primer combination generated several amplicons from which a few were selected, sequenced and one was identified to have high similarity to chitinase (msAP33) (indicated by arrow in Fig. 2b). The results showed chitinase is expressed in meristem and inflorescence tissues with higher expression in the inflorescence compared to meristem. However, no chitinase expression was detected in the leaves of M. sagu. These results were also in accordance with several researches in which they showed that chitinase was highly expressed in healthy floral organs and developing flowers, and either not expressed or at an extremely low level in vegetative organs (Neale et al. 1990; Wemmer et al. 1994; Hamel and Bellemare 1995; Harikrishna et al. 1996; Takakura et al. 2000).
Isolation and analysis of chitinase cDNA from M. sagu
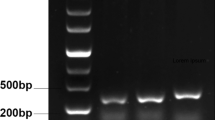
Several steps of RACE were undertaken to isolate chitinase cDNA from M. sagu tissues. The primers; emChi-f, Chi-sp1 and Chi-sp2, were designed for chitinase cDNA 3′RACE and 5′RACE. The 3′RACE of the meristem, inflorescence and leaf cDNA samples generated two bands of approximately 400 and 300 bp in all tissues (Fig. 3). Subsequent 5′RACE of the cDNA managed to produce an amplicon of approximately 900 bp (not shown). Amplification of 5′-end region was carried out only in the cDNA derived from inflorescence tissue of M. sagu because chitinase had been found to be expressed in higher levels in inflorescence tissue when compared to meristem and leaf tissues (Fig. 2).
A 2.0% (x/v) agarose gel-electrophoresis of amplified 3′-end region of chitinase in M. sagu meristem, inflorescence and leaf tissues. LaneS represents a 100 bp Forever Ladder (Seegene). LanesM, I and L represent amplified product from meristem, inflorescence and leaf tissue, respectively. Lane C represents the negative control. Arrow indicates the 400 bp fragment and dashed arrow shows the 300 bp fragment
The 3′-end and 5′-end fragments of chitinase cDNA of M. sagu were combined and analysed using NCBI’s BLASTn software. Sequence analysis showed that amplified chitinase cDNA sequence of M. sagu (1,143 bp) share between 70 and 74% identity with chitinase mRNA of Ananas comosus, Bambusa oldhamii, Bromus inermis, Citrus unshiu, Festuca arundinacea, Fragaria x ananassa, Hordeum vulgare, Medicago sativa, Leucaena leucocephala, Musa x paradisiaca, Nepenthes khasiana, Oryza sativa, Petroselinum crispum, Pinus halepensis, Secale cereale rscc, Triticum aestivum and Vitis vinifera (Table 1). From the nucleotide size of 1,143 bp, the open reading frame (ORF) was determined to be 936 bp long with 312 deduced amino acids (Fig. 4). Comparison of the deduced amino acids of M. saguchitinase revealed that the ORF sequences exhibit similarity to chitinase domain family 19 and is closely related to class I chitinase.
Sequence alignment of the deduced amino acid of chitinase from M. sagu (MsChi1) with class I chitinase from Festuca arundinacea (EU837265.1), Oryza sativa (Z29961.1), Phaseolus vulgaris (AY357300.2), Pyrus pyrifolia (FJ589783.1) and Triticum aestivum (AY437443.1) is shown in Fig. 5. A highly conserved amino acid region (SHETTGG), characteristic of chitinase, was also identified in MsChi1 of M. sagu (Fig. 5) therefore strengthening the cDNA to be of chitinase origin. In silico analysis of the amino acid sequence also indicated the presence of conserved domains. A glycoside hydrolase family 19 chitinase domain, that is involved in the hydrolysis of beta-1,4-N-acetyl-d-glucosamine linkages in chitin, was predicted to be present at amino acid 75–304. A chitin binding domain that is involved in the recognition and binding to chitin was detected from amino acid 24–47. Meanwhile, catalytic residues (amino acids 136, 158 and 188) and putative sugar binding sites (amino acids 136, 158, 186, 191–192, 267 and 279) for MsChi1 were also detected and are indicated in Fig. 5 (Marchler-Bauer et al. 2011).
Multiple sequence alignment of the deduced amino acids of MsChi1 and class I chitinase from Festuca arundinacea, Oryza sativa, Phaseolusvulgaris, Pyruspyrifolia and Triticumaestivum. Identical amino acid residues are indicated by asterisks, the catalytic residues are denoted as closed triangles, amino acid for chitin binding domain are underlined and chitinase consensus sequence are indicated inbox.Dashed lines are gaps introduced to maximize the identity
References
Abd-Aziz S (2002) Sago starch and its utilisation. J Biosci Bioeng 94:526–529. doi:10.1016/S1389-1723(02)80190-6
Bishop JG, Dean AM, Mitchell-Olds T (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci USA 97:5322–5327. doi:10.1073/pnas.97.10.5322
Boller T, Gehri A, Mauch F, Vogeli U (1983) Chitinase in bean leaves: induction by ethylene, purification, properties and possible function. Planta 157:22–31
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40. doi:10.1046/j.1365-313X.1993.t01-1-00999.x
Dong JZ, Dunstan DI (1997) Endochitinase and beta-1, 3-glucanase genes are developmentally regulated during somatic embryogenesis in Picea glauca. Planta 201:189–194. doi:10.1007/BF01007703
Egertsdotter U (1996) Regulation of somatic embryo development in Norway spruce (Picea abies). Agronomie 16:603–608. doi:10.1051/agro:19961002
El-Sayed ST, Salem AM, Shehata AN, Jwanny EW (2000) Chitinase from leaves of Beta vulgaris and other higher plants. Pak J Biol Sci 3:250–256. doi:10.3923/pjbs.2000.250.256
Flach M (1984) The sago palm. A development paper presented at the FAO sponsored Expert Consultation on the sago palm and its products. Jakarta, pp 1–57
Frohman MA, Dush MK, Martin GR (1988) Rapid production of full length cDNAs from rare transcripts by amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85:8998–9002
Gasic K, Hernandez A, Korban S (2004) RNA Extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22:437a–437g
Graham LS, Sticklen MB (1994) Plant chitinases. Can J Bot 72:1057–1083
Hamel F, Bellemare G (1995) Characterization of a class I chitinase gene and of wound-inducible, root and flower-specific chitinase expression in Brassica napus. Biochim Biophys Acta 1263:212–220. doi:10.1016/0167-4781(95)00099-3
Harikrishna K, Jampates-Beale R, Milligan SB, Gasser CS (1996) An endochitinase gene expressed at high levels in the stylar transmitting tissue of tomatoes. Plant Mol Biol 30:899–911. doi:10.1007/BF00020802
Hirano S, Hayashi M, Murae K, Tsuchida H, Nishida T (1988) Chitosan and derivatives as activators of plant cells in tissues and seeds. In: Gebelein CG, Carraler CE Jr, Foster VR (eds) Applied bioactive polymeric materials. Plenum Publishing Co, New York, pp 45–49
Kim YJ, Kwak CI, Gu YY, Hwang IT, Chun JY (2004) Annealing control primer system for identification of differentially expressed genes on agarose gels. BioTech 36:424–434
Leah R, Skriver K, Knudsen S, Ruud-Hansen J, Raikhel NV, Mundy J (1994) Identification of an enhancer/silencer sequence directing the aleurone-specific expression of a barley chitinase gene. Plant J 6:579–589. doi:10.1046/j.1365-313X.1994.6040579.x
Leon J, Rojo E, Sanchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52:1–9. doi:10.1093/jexbot/52.354.1
Leung DWM (1992) Involvement of plant chitinase in sexual reproduction of higher plants. Phytochemistry 31:1899–1900. doi:10.1016/0031-9422(92)80330-H
Lotan T, Ori N, Fluhr R (1989) Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1:881–887. doi:10.1105/tpc.1.9.881
Marchler-Bauer A, Lu S, Anderson JB et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:225–229
Memelink J, Linthorst JMH, Schilperoort RA, Hoge JHC (1990) Tobacco genes encoding acidic and basic isoforms of pathogenesis-related proteins display different expression patterns. Plant Mol Biol 14:119–126. doi:10.1007/BF00018553
Neale AD, Wahleithner JA, Lund M, Bonnett HT, Kelly A, Meeks-Wagner DR, Peacock WJ, Dennis ES (1990) Chitinase, beta-1, 3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell 2:673–684
Neuhaus JM, Fritig B, Linthorst HJM, Meins FJ (1996) A revised nomenclature for chitinase genes. Plant Mol Biol Rep 14:102–104. doi:10.1007/BF02684897
Odjakova M, Hadjiivanova C (2001) The complexity of pathogen defense in plants. Bulg J Plant Physiol 27:101–109
Passarinho PA, de Vries SC (2002) Arabidopsis chitinases: a genomic survey. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville. http://www.aspb.org/publications/Arabidopsis
Passarinho PA, van Hengel AJ, Fransz PF, de Vries SC (2001) Expression pattern of the Arabidopsis thaliana AtEP3/AtchitIV endochitinase gene. Planta 212:556–567. doi:10.1007/s004250000464
Petruzzelli L, Kunz C, Waldvogel R, Meins F Jr, Leubner-Metzger G (1999) Distinct ethylene- and tissue-specific regulation of beta-1, 3 glucanases and chitinases during pea seed germination. Planta 209:195–201
Ponath Y, Vollberg H, Hahlbrock K, Kombrink E (2000) Two differentially regulated class II chitinases from parsley. Biol Chem 381:667–678. doi:10.1515/BC.2000.087
Samac DA, Shah DM (1991) Developmental and pathogen induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3:1063–1072. doi:10.1105/tpc.3.10.1063
Samac DA, Hironaka CM, Yallaly PE, Shah DM (1990) Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol 93:907–914. doi:10.1104/pp.93.3.907
Taira T, Toma N, Ichi M, Takeuchi M, Ishihara M (2005) Tissue distribution, synthesis stage, and ethylene induction of pineapple (Ananas comosus) chitinases. Biosci Biotechnol Biochem 69:852–854. doi:10.1271/bbb.69.852
Takakura Y, Ito T, Saito H, Inoue T, Komari T, Kuwata S (2000) Flower-predominant expression of a gene encoding a novel class I chitinase in rice (Oryza sativa L.). Plant Mol Biol 42:883–897. doi:10.1023/A:1006401816145
Tan SH, Roslan HA (2008) Cloning of a near complete isochorismate synthase (ICS) cDNA from Morinda citrifolia L. Pertanika J Trop Agri Sci 31:101–105
Tie APL (2004) Physico-chemical properties of starch in sago palm (Metroxylon sagu) at different growth stages. Master of Science dissertation, Universiti Sains Malaysia p 14
Trudel J, Asselin A (1989) Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem 178:362–366. doi:10.1016/0003-2697(89)90653-2
van Hengel AJ, Guzzo F, van Kammen A, de Vries SC (1998) Expression pattern of the carrot EP3 endochitinase genes in suspension cultures and in developing seeds. Plant Physiol 117:43–53
van Loon LC (1999) Occurrence and properties of plant pathogenesis related proteins. In: Datta SK, Muthukrishnan S (eds) Pathogenesis-related proteins in plants. CRC Press, Boca Raton, pp 1–19
Vasanthaiah HKN, Katam R, Sheikh MB (2008) Efficient protocol for isolation of functional RNA from different grape tissue rich in polyphenols and polysaccharides for gene expression studies. Electron J Biotechnol. doi:10.2225/vol11-issue3-fulltext-5
Wemmer T, Kaufmann H, Kirch HH, Schneider K, Lottspeich F, Thompson RD (1994) The most abundant soluble basic protein of the stylar transmitting tract in potato (Solanum tuberosum L.) is an endochitinase. Planta 194:264–273. doi:10.1007/BF00196397
Lamb CJ, Zhu Q, Yamamoto RT, Beeche A, Nelson AJ, Lawton MA (1991) Defense gene regulation. In: Rice Genetics II, International Rice Research Institute Manila, Philippines, pp 529–537
Yeboah NA, Arahira M, Nong VH, Zhang D, Kadokura K, Watanabe A, Fukazawa C (1998) A class III acidic endochitinase is specifically expressed in the developing seeds of soybean (Glycine max [L.] Merr.). Plant Mol Biol 36:407–415. doi:10.1023/A:1005921104288
Acknowledgments
This project was aided by UNIMAS Fundamental Research Grant, 01(145)/574/2006(07). The authors wish to thank Rosnah Mustafa for her comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Roslan, H.A., Anji, S.B. Characterization of inflorescence-predominant chitinase gene in Metroxylon sagu via differential display. 3 Biotech 1, 27–33 (2011). https://doi.org/10.1007/s13205-011-0004-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13205-011-0004-x