Abstract
Serious consequences due to brain tumors necessitate a timely and accurate diagnosis. However, obstacles such as suboptimal imaging quality, issues with data integrity, varying tumor types and stages, and potential errors in interpretation hinder the achievement of precise and prompt diagnoses. The rapid identification of brain tumors plays a pivotal role in ensuring patient safety. Deep learning-based systems hold promise in aiding radiologists to make diagnoses swiftly and accurately. In this study, we present an advanced deep learning approach based on the Swin Transformer. The proposed method introduces a novel Hybrid Shifted Windows Multi-Head Self-Attention module (HSW-MSA) along with a rescaled model. This enhancement aims to improve classification accuracy, reduce memory usage, and simplify training complexity. The Residual-based MLP (ResMLP) replaces the traditional MLP in the Swin Transformer, thereby improving accuracy, training speed, and parameter efficiency. We evaluate the Proposed-Swin model on a publicly available brain MRI dataset with four classes, using only test data. Model performance is enhanced through the application of transfer learning and data augmentation techniques for efficient and robust training. The Proposed-Swin model achieves a remarkable accuracy of 99.92%, surpassing previous research and deep learning models. This underscores the effectiveness of the Swin Transformer with HSW-MSA and ResMLP improvements in brain tumor diagnosis. This method introduces an innovative diagnostic approach using HSW-MSA and ResMLP in the Swin Transformer, offering potential support to radiologists in timely and accurate brain tumor diagnosis, ultimately improving patient outcomes and reducing risks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The term "brain tumor" describes the development of aberrant cells inside the brain or near it. When the tumor originates directly in the brain, it is classified as a primary tumor, whereas a secondary tumor refers to cancer cells that have spread from another part of the body and migrated to the brain. [1, 2]. There are two types of primary brain tumors: benign and malignant. Malignant tumors are cancerous and more destructive in nature [3]. Brain tumors’ characteristics, such as their size and location inside the brain, can differ greatly and cause a vast range of symptoms [4, 5].
Early brain tumor discovery is essential for successful treatment and management, as uncontrolled tumor growth can reach severe and life-threatening levels, making control and treatment more challenging [6]. Therefore, determining the diagnosis and categorization of brain tumors is crucial to ensuring the patients’ success. Researchers and scientists have made tremendous progress in creating cutting-edge tools for their identification, considering the rising occurrence of brain tumors and their major impact on persons [7]. For identifying abnormalities in brain tissues, magnetic resonance imaging (MRI) is commonly recognized as the gold standard imaging method [8, 9]. MRI is a useful tool for learning more about the shape, size, and exact location of tumors [10]. Although early and accurate detection of brain cancers is essential, manually classifying brain tumor can be challenging and time-consuming and mainly relies on the radiologists’ knowledge [11, 12].
In recent years, automated approaches utilizing machine learning algorithms have emerged as valuable tools to assist physicians in brain tumor classification, aiming to streamline the classification process and reduce dependence on radiologists [6, 7, 13, 14]. In the field of brain tumor diagnosis, researchers have made significant efforts to reduce the associated morbidity and mortality [11]. Traditionally, the manual detection of brain tumors by radiologists has proven to be burdensome because of the numerous images involved. Computer-aided diagnosis systems (CADx) have become useful tools for overcoming this difficulty by automating and streamlining the diagnostic procedure [15]. Deep learning based CADx systems have exhibited remarkable success rates in medical image analysis, cancer diagnosis, including brain tumors and other cancer types [16,17,18,19,20,21]. These systems not only aid in tumor detection and monitoring but also assist physicians in deciding on things with knowledge suitable treatment options, ultimately improving patient care [22,23,24].
In CAD applications, deep learning algorithms offer a more accurate and efficient substitute for conventional machine learning techniques, which mostly depend on manually generated features [7]. Classical machine learning approaches necessitate feature engineering by domain experts and can be time-consuming, especially with large datasets. CNNs have shown outstanding outcomes in the processing of medical images, including identifying different kinds of brain tumors [22, 25,26,27]. CNNs automatically glean pertinent characteristics from images, doing away with the necessity for feature engineering by hand [28,29,30]. CNNs have shown to be successful at extracting useful information from medical images, resulting in precise and effective categorization without the need for manually created features [31].
Furthermore, vision transformers, a distinct architecture from CNNs, have shown encouraging outcomes across various domains, including brain tumor-related diseased [32,33,34]. Vision transformers make use of the attention mechanism to record distant dependencies and relationships between image patches, enabling them to effectively model complex visual patterns. This architecture has demonstrated outstanding efficiency in applications involving natural language processing, and recently received interest in computer vision applications [35, 36]. Considering brain tumors classification, vision transformers have exhibited the ability to record both global and local image characteristics, allowing for more comprehensive and accurate analysis. Their capacity to acquire significant representations directly from unprocessed data makes them a compelling alternative for medical image analysis, offering potential advancements for diagnosing of brain tumor [12]. Further exploration and evaluation of vision transformers’ capabilities in this domain hold significant promise for enhancing brain tumor categorization systems’ precision and effectiveness.
Deep learning techniques have significantly contributed to the field of brain tumor diagnosis, with notable advancements in tumor detection, classification, and treatment planning [37]. However, there is still a need for continuous improvement in terms of accuracy, efficiency, and accessibility in brain tumor diagnosis and management. Ongoing research and innovations hold the promise of revolutionizing this field by offering more effective techniques and tools for diagnosing of brain tumors, ultimately leading to enhanced outcomes for patients. The effectiveness of deep learning methods in diagnosing various types of cancer has served as a driving force for researchers in this area [38].
Numerous research papers in the scientific literature focus on brain tumor diagnosis. Upon analyzing reviews and surveys, it becomes evident that deep learning has head to several noteworthy findings in the field of brain tumor diagnosis [6, 22, 39]. The studies state that deep learning has developed into a ground-breaking method with significant and beneficial implications for brain tumor diagnosis. Deep learning is a crucial ally in the medical industry since brain tumors’ complexity necessitates accurate and prompt diagnosis. These models may autonomously extract complex patterns and features suggestive of tumor existence and characteristics on large datasets of medical data, such as MRI. For more exact tumor delineation and more efficient treatment planning, this capability offers accurate tumor segmentation. Deep learning additionally makes it easier to classify tumor kinds and differentiate between benign and malignant tumors, both of which are essential for individualized therapeutic strategies. Deep learning’s capacity to handle enormous volumes of data with astounding speed and accuracy has the potential to increase diagnostic effectiveness, hasten treatment decisions, and ultimately improve patient outcomes. However, to ensure that these AI tools are seamlessly incorporated into clinical practice, it is necessary for AI experts and medical professionals to work closely together in order to instill confidence and interpretability, ensuring that deep learning is used as a potent decision support system rather than in place of medical expertise.
Classifying brain tumors using deep learning-based methods presents challenges, including limited labeled data availability, inter-observer variability in diagnosis, overfitting, and the need for interpretability [40]. The scarcity of labeled data necessitates the collection of diverse and well-annotated datasets to improve model performance. Addressing inter-observer variability requires establishing consensus among experts. Techniques like regularization, data augmentation, and cross-validation help mitigate overfitting. Furthermore, developing interpretable methods, such as attention maps or saliency maps, aids in understanding the reasoning behind deep learning predictions, promoting trust and acceptance in the medical community. By addressing these challenges, deep learning models can be more reliable and effective in brain tumor classification, resulting in better patient care and diagnostic accuracy.
This study presents a novel approach to address challenges in brain tumor diagnosis, emphasizing the significance of early detection for successful treatment. By introducing the Swin Transformer architecture, the study leverages its success in vision tasks and adapts it for brain tumor detection, aiming to provide rapid and accurate diagnoses with the aid of deep learning-based CAD systems.
-
We developed a model by scaling the Swin architecture based on the Swin-Base model for a set of 4-class brain MRI images. This scaled model provides improved detection accuracy with fewer parameters at the same time and is shallower than previous Swin models.
-
The proposed model improves the Swin Transformer by introducing the novel Hybrid Shifted Windows Self Attention (HSW-MSA) module, enabling better processing of overlapping brain MRI regions. This enhancement allows the model to capture fine details and long-range dependencies more effectively, leading to improved accuracy in detecting brain tumors and potentially reducing false negatives.
-
Furthermore, the paper replaces the Multi-Layer Perceptron (MLP) in the Swin Transformer with a Residual-based MLP (ResMLP). This architectural change results in higher accuracy, faster training, and improved parameter efficiency. The ResMLP’s ability to extract and represent features more efficiently contributes to the exceptional performance of the Proposed-Swin model on the brain MRI dataset.
-
The extensive evaluation demonstrates an outstanding accuracy of 99.92% achieved by the Proposed-Swin model, surpassing existing research and deep learning models. This remarkable effectiveness highlights its potential for practical application in real-world settings for accurate brain tumor diagnosis.
-
Additionally, we demonstrated the effectiveness of current, well-liked vision transformer models and CNN models using openly accessible MRI datasets to provide a thorough comparison.
The study’s design has been enhanced for improved comprehension. A thorough assessment of the literature is presented in the second section, and the straightforward technique for simple validation is highlighted in the third. Results and discussions from the experiment are covered in the fourth part. Lastly, to help the reader understand the study’s contributions, the conclusion offers a succinct summary of them.
2 Related works
Impressive progress has been made by deep learning algorithms in accurately diagnosing a variety of malignancies, which has led to substantial improvements the discipline of medical imaging. Deep learning approaches have shown encouraging results, particularly when used to analyze and diagnose MRI pictures of brain tumors. These approaches have illustrated great levels of precision in accurately identifying and categorizing brain tumors, which may lead to advancements in patient care and treatment strategy. Deep learning’s success in this area has sparked additional investigation and study with the goal of improving these algorithms’ capabilities and maximizing their potential in order to detect brain cancers. The following is a summary of several research that have been done and published in the literature on brain tumor detection.
Kumar et al. proposed a deep network model that uses ResNet50 with pooling techniques in order to overcome gradient vanishing and overfitting concerns. The effectiveness of the model is assessed using simulated studies on a public MRI dataset with three different tumor types [41]. Talukder et al. [13] presented a cutting-edge deep learning method for correctly classifying tumors utilizing transfer learning. The suggested approach entails thorough pre-processing, reconstruction of transfer learning frameworks, and tweaking. On the 3064 pictures in the Figshare MRI brain tumor dataset, various transfer learning techniques were used and assessed. The suggested framework by Rehman et al. [42] includes three experiments that classify meningioma, glioma, and pituitary brain cancers using several CNN architectures. On MRI slices from the brain tumor dataset downloaded from Figshare, transfer learning approaches are used. Increasing dataset size, lowering overfitting risk, and improving generalization are all achieved by data augmentation. The best classification and detection accuracy, up to 98.69%, was attained by the fine-tuned VGG16 architecture.
The approach suggested by Sharif et al. [43] calls for optimizing the fine-tnued Densenet201 model and applying transfer learning on imbalanced data. The average pool layer, which contains useful information about each type of tumor, is where the features of the trained model are retrieved from. But in order to improve the performance of the model for precise classification, two feature selection strategies are incorporated. To diagnose glioma brain tumors as low-grade or high-grade utilizing the MRI sequence, Mzoughi et al. [44]. presented an automatic and effective deep multi-scale 3D CNN architecture. To efficiently combine useful contextual information while reducing weights, the design uses a 3D convolutional layer with small filters. The suggested classification model by Amin et al. [26]. has seven layers, including a SoftMax layer, three convolutional layers, and three ReLU activation layers. The MRI image is segmented into a few patches, and the deep CNN is given the central pixel value of each patch. The segmentation of the image is made possible by the DNN, based on these center pixels, labels are assigned.
Amin et al. [45] suggested a technique for de-noising and enhancing input slices using a Weiner filter with multiple wavelet bands. It uses Potential Field (PF) clustering to isolate different tumor pixel subsets. Additionally, T2 MRI images, global thresholding and mathematical morphological techniques are used to identify the tumor site. For the purpose of grading and diagnosing brain tumors using MRI data, Tandel et al. [46] provided five clinically relevant multiclass datasets with various numbers of classes. In comparison to six existing machine learning classification approaches makes use of transfer learning using a CNN. On multiple datasets of MR images, a deep CNN model that was initially pre-trained as a discriminator in a GAN. The pre-training aids in the extraction of robust features and teaches the convolutional layers of the algorithm the structure of MR images. The entire deep model is then retrained as a classifier to differentiate between tumor classes once the fully connected layers are changed [47].
To categorize brain cancers in MRI data, Tabatabaei et al. [48] developed a hybrid model that integrates CNN with attention module. By taking into account both local and global variables, they developed a cross-fusion technique to merge the branches, boosting classification accuracy. The many types of brain tumors can be accurately identified by this hybrid architecture. An optimized ResNet model with a cutting-edge evolutionary approach was introduced by Mehnatkesh et al. [33]. This method automatically improves the deep ResNet model’s architecture and hyperparameters without the need for human specialists or manual architecture design, making it appropriate for classifying brain tumors. The research also introduces an improved optimization method that incorporates ideas from differential evolution strategy and multi-population operators. Deep CNN, a Dolphin-SCA based deep learning technique for enhanced accuracy and efficient classification, was introduced by Kumar et al. [49] pre-processing the raw MRI images is the first step in the procedure, which is then segmented using an improved algorithm. Then, feature extraction is carried out using statistical and power LDP features.
An automated approach for differentiating between malignant and non-cancerous brain MRIs was proposed by Amin et al. [50] The technique uses a variety of techniques to divide up potential lesions before choosing shape, texture, and intensity-based attributes for each lesion. Then, in order to compare the proposed model’s precision, a SVM classifier is applied. Swati et al. [51] proposed a deep CNN model that has already been trained is used, and a block-by-block fine-tuning approach based on transfer learning is suggested. A benchmark dataset MRI is utilized to evaluate the strategy’s efficacy. As a result of avoiding handcrafted features, requiring no preprocessing, and achieving an average accuracy of 94.82%, the method is notably more general. A CNN-based method for classifying multi-grade brain tumors was introduced by Sajjad et al. [52] First, tumor areas from MR images are segmented using deep learning approaches. Second, a significant amount of data augmentation is used to effectively train the system, addressing the issue of a shortage of data in the categorization of multi-grade brain tumors from MRI. Finally, supplemented data is used to improve a pre-trained CNN model for precise brain tumor grade classification. Deepak and Ameer [23] developed a 3-class classification issue incorporating them. The proposed classification system uses transfer learning utilizing GoogLeNet. The collected features are subsequently classified using integrated, tested classifier models.
To enhance the accuracy and efficacy of MRI data-driven diagnoses, it is evident from the summaries of research papers that there is a growing interest in exploring deep neural networks for brain tumor-related studies. Challenges such as vanishing gradient, overfitting, imbalanced data, and data heterogeneity have been effectively addressed using various strategies. Modifying well-known models like ResNet, VGG16, and Densenet201 for brain tumor classification through transfer learning consistently yields high accuracy. Increasing dataset sizes, improving generalization, and mitigating overfitting concerns have been achieved through the application of data augmentation techniques. Additionally, some studies focus on 3D CNN architectures to extract both local and global contextual information from volumetric MRI data, leading to more precise tumor grade classification. Image quality and feature extraction have been enhanced by employing preprocessing techniques like denoising and contrast augmentation. Various feature selection methods, including wavelet transforms, local binary patterns, and statistical features, have been integrated to boost the effectiveness of deep learning models.
Overall, the research outlined in these publications underscores the continual improvement in brain tumor categorization, emphasizing deep learning approaches and optimizing model architectures. These innovative methods hold significant promise for enhancing the sensitivity and accuracy of brain tumor diagnoses, ultimately benefiting patients and medical professionals. To ascertain the applicability and generalizability of these proposed approaches, further clinical research and validation may be necessary.
3 Material and methods
In this study, we introduce a cutting-edge deep learning model to diagnose brain tumors. A broad collection of brain MRI scans, a comprehensive dataset comprising three publicly available datasets, that have been painstakingly collected from various medical institutes make up the dataset used for training and evaluation. Our deep learning system makes use of cutting-edge vision transformer architecture, which has distinguished itself in tasks requiring picture understanding. The proposed method effectively detects and categorizes brain cancers with high sensitivity and specificity by fusing the strength of the vision transformer with sophisticated data augmentation and transfer learning strategies. To assure reproducibility and encourage additional study for other cancer-related diseases, complete implementation and training methods are described.
3.1 Dataset
Due to their capability to learn and generalize on vast data, deep learning models are becoming more and more popular. However, the size and quality of the training dataset strongly influence the effectiveness of these data-hungry models. The dataset is crucial in deep learning as it provides the necessary examples for the models to recognize and generalize patterns effectively. The model can extract pertinent features and make precise predictions on unobserved data with a sizable and representative dataset. Ensuring high-quality data is essential to address biases, reduce overfitting or underfitting issues, and improve performance across different subsets. For the autonomous classification of low-grade brain MRI images, several publicly available datasets exist, including Figshare [53], SARTAJ [54], and Br35H [55], which are known to be small-scale datasets. However, in this study, we utilized a publicly accessible brain MRI dataset shared on Kaggle [56], which combines and incorporates these three datasets to reveal the true capabilities of deep learning models on this task. Sample images from this dataset depicting both tumor and healthy cases are illustrated in Fig. 1.
The brain MRI dataset utilized for this study has been divided into four major tumor classes: no-tumor, glioma, meningioma, and pituitary. Malignant brain tumors include gliomas, which have an aggressive development tendency. On the other hand, meningioma tumor is a benign tumor that grows in the meninges of the brain and can go undetected for a long time without exhibiting any clear symptoms. Pituitary tumors are a specific kind of tumor that develop in the pituitary gland and can cause hormonal abnormalities. The No-tumor class, which represents healthy brain circumstances, is also a crucial point of reference for control groups. Utilizing this extensive and varied dataset, we evaluated the deep learning model’s capacity to correctly categorize each tumor type and investigated its potential as a trustworthy tool for brain tumor diagnosis.
3.2 Vision transformer
Artificial intelligence has had a big impact on deep learning, particularly in computer vision applications like face recognition, medical picture analysis, and autonomous driving. CNNs, which are specifically engineered to handle visual input, have been instrumental in this revolution. Convolutional filtering and pooling techniques allow CNNs to reduce dimensionality and recognize a variety of picture attributes. CNNs are not without limitations either, particularly when it comes to understanding relations within an image and gathering global information. Researchers have created vision transformers as a solution to this [57, 58]. Vision transformers leverage self-attention approaches to capture long-range relationships in raw data, allowing them to outperform CNNs for visual scenarios.
Unlike CNNs, vision transformers use positional embeddings and self-attention instead of convolutional layers. They are able to record both local and global information in visual sceneries because to this special technique, which makes them suitable for tasks requiring a thorough knowledge of images. Recent research has demonstrated the high performance that vision transformers may accomplish in a range of uses for visual tasks. A key development in the disciplines of computer vision and deep learning is the creation of vision transformers [59]. While CNNs remain the architecture of choice for many artificial intelligence usages, vision transformers offer an additional tactic that is highly effective in obtaining global as well as local data.
3.3 Swin transformer
The Swin Transformer, developed by Microsoft Research in 2021, is an impressive AI model designed for computer vision [60, 61]. It builds upon the Transformer model and introduces two key concepts—hierarchical feature maps and shifted window attention. These advancements help efficiently handle large-scale image data, making it a promising tool for complex computer vision tasks. The Swin Transformer utilizes hierarchical feature maps to effectively represent different levels of features in images, leading to a comprehensive understanding of context and improved comprehension of input data. The shifted window attention mechanism expands the interaction field of each block, enabling the model to capture variable-scale features more effectively.
The Swin Transformer’s four-stage architecture involves dividing the input image into patch layers, which are processed through Transformer blocks in the backbone. The resulting patches are sent to the transition block, maintaining the same number of patches. In the second stage, patch merging layers are used to create a hierarchical system by subsampling and reducing the number of tokens. Neighboring 2 × 2 patches’ features are combined to obtain a 4C-dimensional feature vector, which is transformed using linear layers while preserving a resolution of H/8 × W/8. This patch merging and feature transformation process is repeated twice in subsequent stages, resulting in output resolutions of H/16 × W/16 and H/32 × W/32, respectively. Overall, this architecture enables the Swin Transformer to effectively process image data and capture contextual information at different scales, contributing to its superior performance in various vision tasks.
The Swin Transformer Blocks (STBs) provided in Figs. 2 and 3 consist of two consecutive multi-head self-attention (MSA) modules: window-based MSA (W-MSA) and shifted window-based MSA (SW-MSA). Before each of these MSA modules, a Layer Norm (LN) layer is used. Next, there is a two-layer MLP (multilayer perceptron) with GELU non-linearity in between. Each module has a link with the LN layer. In Eqs. 1 and 2, MSA has a quadratic computational complexity with respect to the number of tokens. This configuration significantly improves the performance of the Swin Transformer and makes it more efficient compared to the standard Transformer.
Where the first part exhibits a quadratic relationship with respect to the patch number, denoted as hw, whereas the second part demonstrates a linear dependency when the value of M is constant (typically set to 7 by default). Computing global self-attention becomes prohibitively expensive for a high value of hw, whereas window-based self-attention is scalable.
In the consecutive STBs, a shifted window partitioning approach is adopted to switch between two configurations. This approach utilizes overlapping windows to introduce cross-window connections while efficiently calculating non-overlapping windows. In the first module, a regular window partitioning strategy is used, and an 8 × 8 feature map is divided into 2 × 2 windows of size 4 × 4 (M = 4). Then, the second module provides a window configuration by shifting the windows by \(\left( {\left\lfloor \frac{M}{2} \right\rfloor , \left\lfloor \frac{M}{2} \right\rfloor } \right)\) pixels from the previously partitioned windows. The Transformer blocks are computed in Eq. 3
where \({z}^{l}\) and \({\widehat{z}}^{l}\) represent the output features for block l from the (S)W-MSA module and the MLP module, respectively. W-MSA and SW-MSA refer to window-based multi-head self-attention with standard and shifted window partitioning configurations, respectively.
The Swin Transformer adopts a specialized architecture to enhance computational efficiency compared to traditional Transformer models. It achieves this by using a cyclic shifting operation between shifted token blocks (STBs). This operation divides the feature map pixels into regional blocks and cyclically shifts each block to the previous one. As a result, each block can operate with masks applied to a section of the feature map. This approach allows the Swin Transformer to process smaller blocks of data instead of the entire feature map at once, leading to more efficient feature extraction and preventing computational overhead in sliding windows.
The Swin Transformer utilizes a self-attention mechanism that incorporates relative positional bias to capture relationships between positions. The attention function involves mapping queries (Q), keys (K), and values (V) to output vectors. For each query in the Q matrix, attention weights are calculated for corresponding key-value pairs. The resulting output matrix is obtained through this computation process, which is formulates in Eq. 4
Where the query (Q), key (K), and value (V) matrices are of size \({R}^{{M}^{2}xd}\), where d represents the dimension of the query/key vectors, and \({M}^{2}\) is the number of patches in a window. In Swin Transformer, relative positions are defined along each axis within the range [− M + 1, M − 1]. The relative positional bias is parameterized as an offset matrix \(\widehat{B}\) ∈ \({R}^{(2M-1)x(2M-1)}\), and the elements of matrix B are obtained from \(\widehat{{\text{B}}}\).
The fundamental model of Swin Transformer is referred to as Swin-B. Swin-B has a comparable model size and computational complexity to ViT-B/DeiT-B. Similarly, the Swin-T and Swin-S models are designed to have computational complexities comparable to ResNet-50 (DeiT-S) and ResNet-101, respectively. The dimensions of Swin Transformer models can vary depending on various factors, including the channel size of the initial feature map (C), the layer size of the Swin Transformer block, the window size, and the expansion factor of the MLP layer.
3.4 Proposed model
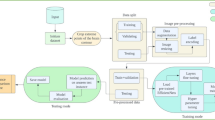
The proposed approach aims to develop a classification model based on the Swin Transformer for brain MRI images. It seeks to achieve high classification accuracy and address challenges related to distinguishing between similar lesion types and accurately identifying common ones. The proposed approach presents innovative enhancements to the Swin Transformer model for brain tumor diagnosis. Four essential elements make up the proposed approach for classifying brain tumors using the Swin Transformer architecture: scaling the model for 4-class classification of brain tumors, incorporating the Residual MLP module, incorporating hybrid shifted windows into the self-attention mechanism, and using transfer learning with data augmentation. Like other deep learning architectures, the Swin Transformer needs to have its design and parameters scaled in order to accommodate a variety of workloads and dataset sizes. Variables like model size, stage depth, and embedding dimensions can all help achieve this. For example, larger variations of the Swin Transformer, such Swin-Base, and Swin-Large, which are made for datasets like ImageNet with 1000 classes, offer improved capacity suitable for handling more difficult tasks and bigger datasets. Swin-Small, Swin-Tiny model, on the other hand, produce more useful outcomes in scenarios with fewer classes while using less resources for simpler tasks. The overall design of the Proposed-Swin Transformer model for detecting brain tumors is illustrated in Fig. 2.
In this work, the configuration of the Swin-Base model with “Embedding Dimension = 128”, “depths = (2, 2, 18, 2)”, and “Number of Heads = (4, 8, 16, 32)” was changed to “Embedding Dimension = 96”, “depths = (2, 2, 4, 2)”, and “Number of Heads = (3, 6, 12)”, leading to a more adaptable model in terms of computation, convergence speed, and cost. Notably, the increased depth in the third step of the initial configuration aligns with the Swin Transformer’s hierarchical approach and tries to capture complicated and high-level information. The proposed model (scaled model), with its integrated components, performs more effectively than other models in the context of classifying brain tumors.
By introducing residual connections into the MLP modules, the model benefits from smoother transitions between layers and improved gradient flow, facilitating the training of deeper models and achieving superior results. Moreover, the integration of hybrid shifted windows into the self-attention modules enables the model to process images at various scales and positions, leading to comprehensive feature extraction and more robust representations. By combining these learning approaches, the proposed method shows promise in creating a more comprehensive and powerful classification model for brain tumor diseases, ultimately leading to more accurate and reliable outcomes in brain tumor diagnosis and treatment.
3.4.1 Hybrid multi self-attention module
The Swin-based models consist of two different multi-head self-attention layers, W-MSA and SW-MSA. In the proposed model, Hybrid Swin Transformer blocks were introduced, employing a hybrid shifted window approach. This novel technique divides the input image into smaller patches and applies attention mechanisms to each patch, capturing relationships between features in different patches and preserving the overall context. By considering relationships among various parts of the input image, the network can maintain a broader perspective. The Swin-Tiny model, developed with hybrid transformer blocks, incorporates a hybrid self-attention module that combines traditional shifted windows with elongated rectangular shapes in horizontal and vertical directions. Unlike conventional transformer blocks, which use fully connected self-attention mechanisms, this hybrid module allows the model to flexibly capture information from windows of various sizes, addressing long-range dependencies while preserving local and detailed information. The ability to handle images at different scales and orientations enhances the model’s applicability and reduces generalization issues, potentially leading to improved performance in challenging image analysis tasks such as brain tumor detection and other medical images. Figure 3 illustrates the pure Swin Transformer block alongside the hybrid transformer blocks used in the proposed model.
The Hybrid transformer blocks in Fig. 3 consist of two self-attention modules. While the first layer of this structure remains the same as the layer in the original Swin Transformer, a more efficient layer is obtained by adding the hybrid layer, Hybrid SW-MSA, to the secondary layer, the SW-MSA layer. The HSW-MSA layer combines three different sliding window processes to enhance visual information exchange at various scales. In the first part, a SW-MSA module is applied for local pattern capture. In the second part, the input image is divided into horizontal and vertical stripe windows, enabling longer-range connections and broader context. This approach enriches the HSW-MSA’s multiple heads, facilitating comprehensive visual information exchange. It is particularly useful for improving performance in visual processing applications. The computation of hybrid Transformer blocks involves the sequential application of these two self-attentions is formulated in the Eq. 5.
where \({z}^{l}\) and \({\widehat{z}}^{l}\) represent the output features for block l from the (S) HSW-MSA module and the Res-MLP module, respectively. W-MSA and HSW-MSA refer to window-based multi-head self-attention with hybrid shifted window partitioning configurations, respectively.
3.4.2 Residual multilayer perceptron module (Res-MLP)
MLPs, short for multi-layer perceptron, are fundamental components in the standard transformer architecture [62]. Typically, a transformer includes two main building blocks: the self-attention mechanism and the MLP block. While the self-attention mechanism captures relationships between different tokens (or patches in image transformers), the MLP block processes information individually for each token. In the Swin Transformer architecture, depicted in Fig. 4, the MLPs are similar to those found in other transformer structures. However, instead of using a regular MLP, we introduced a Residual MLP module, inspired by the ResNet architecture [63] and ResMLP architecture [62], which has gained popularity recently. The proposed Res-MLP structure, a crucial component of the Proposed-Swin Transformer framework, is depicted in Fig. 4.
The Swin Transformer leverages residual connections within the MLP blocks to address the vanishing gradient issue, allowing stable and efficient training of deep architectures. The ability to skip uninformative layers through residual connections enhances the model’s capacity to learn complex representations and handle challenging tasks effectively. Moreover, the Res-MLP design not only improves expressiveness but also enhances generalization capabilities. The Swin Transformer’s ability to capture non-linear relationships between features makes it more adaptable to diverse and complex datasets. The residual connections provide resilience to changes in hyperparameter selections and architectural configurations, facilitating the model development process and supporting faster experimentation. Experimental results have shown that with these enhancements, the Swin Transformer converges faster on brain tumor data and achieves higher accuracy. As seen in Fig. 4, by adding Residual layers to the MLP structure, more effective training and stronger generalization capabilities were achieved.
4 Results and discussions
4.1 Experimental design
A Linux machine running the Ubuntu 22.04 operating system was used for this study. On an impressively powerful high-performance computer, the deep neural network models were developed and evaluated. This PC featured a 13th generation Intel Core i5 with an NVIDIA RTX 3090 GPU with 24GB of GDDR6X memory, along with 32 GB of DDR5 RAM. The most recent stable PyTorch framework with NVIDIA CUDA support was used for the experiments. In the same computing environment, each model was trained and tested, which ensured consistency by using the same parameters throughout.
4.2 Data processing and transfer learning
For deep learning algorithms, medical images need to be produced on an appropriate foundation. Data sets are frequently split into cross-validation, train-validation, or train-test sets in the literature. However, few studies actually evaluate the real performance of deep learning algorithms using the appropriate data split of train, validation, and test sets. To use the best data separation technique for assessing the performance of deep learning models, we divided the data set in our study into three separate subsets: training, validation, and testing. To assess the model’s effectiveness and lower the chance of overfitting, this division is required.
We employed a Kaggle data set that was available in separate train and test sets and was open to the public [56]. To guarantee an equitable comparison of our proposed model with others, we employed 80% of the training data for the actual training process and reserved the remaining 20% for validation purposes. The test data set was left untouched for fair comparison. Table 1 displays the data distribution for each class in the Kaggle dataset.
Table 1 summarizes the distribution of MRI dataset, categorizing samples into Glioma-tumor, Meningioma-tumor, Pituitary-tumor, and No-tumor classes. The dataset is split into three sets: Train, Validation, and Test. The dataset consists of 7023 samples in total, with the No-tumor class having the highest number of samples (2000) and the Glioma-tumor class having the fewest (1621). This dataset is essential for training and evaluating classification models.
In this study, we improved the robustness and generalization of our models using data augmentation techniques [52]. Data augmentation involved applying various transformations to the original images, creating new synthetic examples, and reducing overfitting risk. We specifically applied cropping, flipping, rotation, copy-paste, shear, and scaling during model training, effectively expanding the dataset and enhancing its ability to generalize to unseen samples. This augmentation aimed to enhance the accuracy and reliability of our models in identifying brain tumors, ultimately contributing to more efficient screening and diagnostics. The ImageNet dataset’s pre-trained weights were also used in transfer learning by [64,65,66], leveraging the model’s learned knowledge and representations from millions of diverse images. Fine-tuning the pre-trained model using our brain MRI dataset further improved its performance, saving training time, fast convergence, and computational resources.
4.3 Performance metrics
Performance metrics are of utmost importance when evaluating the effectiveness and generalizability of deep learning algorithms. They serve as essential tools in the field, enabling the assessment of models throughout training and on validation and test datasets. By utilizing these metrics, one can identify overfitting issues, gauge the effects of parameter adjustments, and gain a comprehensive understanding of the model’s overall performance. Accuracy, precision, and recall are among the widely used performance metrics in deep learning, as presented in Table 2.
False positive (FP) denotes inaccurate positive estimations, false negative (FN) denotes inaccurate negative predictions, and true negative (TN) denotes accurate negative predictions. True positive (TP) denotes accurate positive predictions. The effectiveness of binary classification models is evaluated using these metrics. Precision calculates the percentage of accurate positive forecasts to all positive predictions, whereas accuracy evaluates the ratio of correct predictions to total predictions. On the other hand, recall quantifies the ratio of correctly predicted positive cases to all actual positive examples. By determining their harmonic mean, the F1 score strikes a compromise between precision and recall, ensuring a thorough assessment of model performance. Each metric adheres to the following mathematical formula.
4.4 Training procedure
The performance of deep learning models could be enhanced by using a variety of methods and settings while they are being trained. Data augmentation and transfer learning are two efficient techniques. Furthermore, several hyper-parameters play a substantial effect in shaping a model’s performance. These parameters include input size, type of optimizer, size of batch, learning rate, and repetition of augmentation. Adjusting the learning rate has the potential to impact the model’s effectiveness, whereas weight decay serves as a preventive measure against overfitting by imposing penalties on substantial weights within the loss function. Adjusting warmup epochs and learning rate gradually increases the learning rate during initial epochs to avoid divergence during training.
In our research, we adopted a multifaceted approach to ensure the reproducibility and performance enhancement of both the proposed model and other deep learning models. The implemented techniques encompassed crucial aspects such as hyperparameter tuning, data preprocessing, transfer learning, and data augmentation. Fundamental hyperparameters, including input size, learning rates, momentum, weight decay, and optimizer selection, were consistently fine-tuned across all models, using default values to establish a standardized foundation for model training. This meticulous parameter application aimed to foster reliability and comparability in our experimental results. Additionally, data-related hyperparameters such as scale, ratio, Mixup probability, and others were carefully adjusted, introducing variability and sensitivity to diverse datasets, thereby enhancing the overall robustness of our models.
In a different vein, our approach involved tailoring specific hyperparameter values for each model to ensure optimal training conditions. For example, the input size, determining the dimensions of training images, was set at 224 × 224 (or 256 × 256 for certain models like SwinV2). The learning rate, a pivotal parameter influencing the model’s learning pace, was initialized at 0.00001. Essential parameters like initial learning rate (lr_base), learning rate cycle decay (lr_cycle_decay), and exponential moving average (EMA) decay for weights (model_ema_decay) were meticulously configured. The lr_base was specifically set to 0.1 as an effective starting point for learning rates. The lr_cycle_decay, indicating the factor by which the learning rate decreases after each training cycle, was adjusted to 0.5 for a balanced convergence and stability. The model_ema_decay, governing the update speed of weights using the EMA method, was selected as 0.9998 for a gradual and consistent adjustment.
Practical considerations such as momentum (0.9) in the Stochastic Gradient Descent (SGD) optimizer, weight decay (2.0e−05) to control overfitting, warm-up epochs (5) for a gradual learning rate increase at the start of training, and warm-up learning rate (1.0e−05) were incorporated. These hyperparameter values were meticulously chosen to strike a delicate balance between model training efficiency, stability, and robustness. The amalgamation of these finely tuned hyperparameter configurations played a pivotal role in achieving optimal model performance while maintaining consistency across experiments. Additionally, specific data-related hyperparameters, such as scale, ratio, Mixup probability, and others, were fine-tuned to ensure model sensitivity to various data characteristics.
In deep learning models, the issues of overfitting and underfitting often adversely affect the model’s generalization capability and can lead to incorrect biases. Overfitting occurs when the model excessively fits the training data and fails to generalize to test data, while underfitting arises when the model inadequately fits the training data, resulting in poor performance on test data. To address both problems collectively, we divided our dataset into three parts: training, validation, and test. We evaluated the model’s generalization performance solely on the test data. The training process was monitored for 50 epochs, and if significant improvement did not occur during this period, the training was stopped. This approach contributes to preventing overfitting and avoiding unnecessary training of the model. Additionally, due to the limited scale of MRI data, we attempted to enhance the model’s performance using transfer learning and data augmentation techniques. These strategies proved helpful in achieving better generalization with a constrained dataset.
Furthermore, to tackle these issues, regularization techniques such as dropout and weight regularization were applied to all models during the training of both the baseline models and the Proposed-Swin model. Dropout reduces overfitting by randomly disabling neurons during training, while weight regularization helps prevent excessively large weights. Default hyperparameters were set for all models to achieve a balance between model complexity and dataset size. On the other hand, underfitting is typically a problem observed in large-scale datasets, but it is not the case with our MRI dataset. To address this issue, the complexity of the Proposed-Swin model’s architecture, HSW-MSA, and ResMLP improvements were leveraged. These components enhance the model’s ability to capture complex patterns in MRI data, thereby improving diagnostic accuracy.
4.5 Results
The experimental findings of the proposed approach are presented in this part together with those of several popular CNN models and the most recent and extensively used vision transformer models that can be found in the literature. The experimental evaluations for each model were conducted exclusively on unseen data, specifically the test data that had been previously set aside. Testing the models on unseen data is the optimal choice as it demonstrates deep learning’s generalization capabilities and their applicability in real-world scenarios. Table 3 presents the experimental results on the Brain MRI dataset for the Proposed-Swin model compared to cutting-edge CNNs and vision transformer-based models.
Considering Table 3, a comparative analysis of experimental results on the brain MRI dataset reveals that the models exhibit exceptional performance in accurately classifying brain MRI images. All models demonstrate diagnostic accuracy above 98%, and when the ResNet50 model is excluded, it becomes evident that all other models achieve diagnostic accuracy well above 99%. Taking Table 3 into account, the Proposed-Swin model stands out by showcasing the highest performance, reaching 99.92% accuracy and F1-score, demonstrating a significant superiority over other models.
The Proposed-Swin model enhances its performance in brain tumor classification tasks through the integration of HSW-MSA and ResMLP structures into its architecture. HSW-MSA provides a structure that improves attention mechanisms and better understands distance relationships between features. This allows the model to adapt better to the complexity of objects and learn more general features. Additionally, the ResMLP structure, when used instead of traditional convolutional MLP structures, effectively focuses on both large and small features in MRI images, helping the model learn more comprehensive features. These two structures play a critical role in enabling the Proposed-Swin model to achieve high accuracy, precision, recall, and F1-score values. As a result, the model excels in brain tumor classification tasks, offering a more effective solution compared to other architectures.
Among other models, following Proposed-Swin in terms of the highest performance are BeiT-Base and MobileViTv2-150 models. BeiT-Base achieves an accuracy of 0.9954 and an F1-score of 0.9950, while MobileViTv2-150 similarly exhibits high performance with an accuracy of 0.9954 and an F1-score of 0.9952. On the other hand, models with the lowest performance include ResNet50 and VGG16, with accuracy and F1-score values as follows: ResNet50 (Accuracy: 0.9893, F1-score: 0.9886) and VGG16 (Accuracy: 0.9924, F1-score: 0.9917). These evaluations underscore the outstanding performance of Proposed-Swin, and its more effective solution compared to other models.
The comparative analysis also highlights the significance of considering precision, recall, and F1-score alongside accuracy to evaluate model performance comprehensively. Models like “MobileNetv3-Small” and “MobileViT-Small” exhibited remarkable precision and recall values, indicating their proficiency in correctly identifying positive samples while minimizing false positives and negatives. Such high F1-scores, coupled with competitive accuracy, are indicative of robust models with balanced performance. Additionally, it is crucial to weigh the computational efficiency of each model, especially when deploying applications in real-world settings. Models like Proposed-Swin with their exceptional performance and computational efficiency, hold promise for practical implementation in medical imaging and diagnostic systems. Among the models evaluated, the Proposed-Swin model stands out with exceptional results, achieving an outstanding metrics of 0.9992. This demonstrates the Proposed-Swin model is highly effective in accurately classifying brain MRI images, making it a promising candidate for real-world clinical applications.
Comparing the Proposed-Swin model with the other models, we can observe that it outperforms almost all of them in all metrics. For instance, the widely used VGG16 and ResNet50 models achieved an accuracy of 0.9924 and 0.9893, respectively, which are slightly lower than the Proposed-Swin model. Similarly, other state-of-the-art models, such as EfficientNetv2-Medium, MobileNetv3-Small, and DeiT3-Base, demonstrated competitive performances but were still outperformed by the Proposed-Swin model in terms of all metrics.
Moreover, the Proposed-Swin model ‘s excellent results even surpass more complex models like ViT-Base-Patch32, PiT-Base, and GcViT-Base, which suggests that the model’s design and architecture are well-suited for the brain MRI classification task. It is important to point out that the Proposed-Swin model’s exceptional performance comes with the added advantage of being computationally efficient and lightweight. This characteristic makes it highly applicable in resource-constrained environments, such as mobile applications or edge devices, without sacrificing predictive accuracy. The confusion matrix for a few Swin-based transformer model as well as a few other cutting-edge deep learning models with Proposed-Swin is shown in Fig. 5.
Considering Fig. 5 all models demonstrated high accuracy to diagnose brain tumors. The Proposed-Swin, Swin-Tiny, DeiT3-Base, and GcViT-Base models showcased consistent and impressive results, with minimal misclassifications. The ResNet50 and ConViT-Base models also performed well, albeit with slightly higher misclassification rates. MobileViT-Small and BeiT-Base models exhibited strong performances but showed a few more errors compared to the top-performing models. The performance of the Proposed-Swin model in classifying brain tumor images into four categories was outstanding, with almost all predictions being accurate. Only one misclassification was observed, where a sample from the Pituitary class was mistakenly predicted as Meningioma.
Upon observation, the No-tumor class emerges as the most successfully diagnosed class across all models, with meningioma having higher FP values and varying FN values among the models. While ResNet50 exhibits the lowest class-specific performance, the Proposed Model consistently demonstrates the highest performance across all classes. Figure 6 provides a detailed comparison of all models based on the accuracy metric in a single line graph. As seen in Fig. 6, the most successful model is the Proposed Model (Proposed-Swin), followed by MobileViTv2-150, BeiT-Base, MobileViT-Small, DeiT-Base, with ResNet50 being the least performing model. Notably, the current model, DeiT3, shows lower performance compared to its predecessor, the DeiT model. This underscores the variability in performance that each model can exhibit on medical datasets.
4.6 Efficiency of the proposed-Swin model and Swin transformer variants
In this section, we embark on a comprehensive comparison between the Proposed Model and the Swin Transformer architecture, both of which hold significant importance in the field of deep learning. Our analysis encompasses an extensive range of model variants, ranging from the compact Tiny and Small models to the more substantial Base and Large models. Furthermore, we delve into the exploration of the SwinV2 Transformer, an evolved version of the Swin Transformer that introduces varying window sizes, presenting new opportunities for fine-tuning and optimization. To ensure a rigorous evaluation, we utilize the test data from the Brain MRI dataset. Table 4 showcases a detailed comparison of these models.
Table 4 analysis reveals that all Swin-based transformer models achieve an accuracy rate of over 99.30% in correctly diagnosing brain MRI images. A noteworthy distinction is that the Proposed-Swin model beats existing tiny models in terms of accuracy and other metrics while showing comparable convergence speed and parameter count. The Proposed-Swin model achieves a much greater accuracy of 0.9992 when compared to the Swin-Tiny model, which achieves an accuracy of 0.9931, demonstrating a significant advantage.
The Proposed-Swin model performs better even when compared to the Swin-Small model. The accuracy of the Swin-Small model is 0.9939, whereas that of the Proposed-Swin model is 0.9992. Similar to this, the Proposed-Swin model still triumphs when compared to the Swin-Base model. The accuracy of the Swin-Base model is 0.9954, while that of the Proposed-Swin model is 0.9992. Additionally, the Proposed-Swin model still has an edge over the Swin-Large model. The accuracy of the Swin-Large model is 0.9947, whereas that of the Proposed-Swin model is 0.9992. The Proposed-Swin model additionally routinely exhibits improved accuracy and superior performance across a variety of parameters when compared to existing Swinv2-based models. For devices with lesser computational and memory requirements, it is a superior option because it can achieve greater performance with fewer settings. Figure 7 provides a detailed comparison of Swinv1 and Swinv2 models in a single line graph based on the accuracy metric. As seen in Fig. 7, the most successful model is the Proposed Model (Proposed-Swin), followed by the Swinv2-Window8-Tiny, Swinv2-Window8-Small, and Swin-Base models, while the Swin model with the lowest performance is the Swin-Tiny model with an accuracy of 699.31. In general, Swin-based models exhibit high accuracy in diagnosing brain tumors, ranging from 99.31 to 99.92%.
When assessing complexity, the Proposed-Swin model focuses on the HSW-MSA block and the scaled Swin-Base model. The Swin-Base model has 88 million parameters, while the scaled version, with 24 million parameters, is even lighter than the Swin-Tiny model (29M). The HSW-MSA layer, a key parameter influencer, increases model parameters by 10% when replacing the SW-MSA block. However, overall scaling and ResMLP module make the model lighter than Swin-Tiny and less complex in layer count. The HSW-MSA layer stands out by seamlessly combining three types of shifted windows. It strategically allocates 50% attention to traditional shifted windows and distributes the remaining 25% to horizontal and vertical stripe windows. This intentional partitioning allows the model to capture local and global relationships, along with direction dependencies in both horizontal and vertical axes. This enhances feature representation, demonstrating improved performance in medical image processing, particularly in exploring brain tumor features and achieving better diagnostic accuracy.
4.7 Comparison with cutting-edge methods
The rapid advancements in computer vision techniques and medical imaging present new and significant opportunities for the effective classification of brain MRI images. In this context, to assess the performance of our proposed model, we conducted a comprehensive comparison with current cutting-edge methods. Specifically, we focused on some methods that demonstrate superior success in the diagnosis of brain tumors, as highlighted in Table 5, showcasing the superior performance of our proposed model over other state-of-the-art methods.
Table 5 provides a comprehensive comparison of state-of-the-art models in the domain of brain MRI image classification, particularly focusing on the vital task of diagnosing brain tumors. Amidst the array of methodologies presented by different studies, Proposed-Swin (ViT) stands out as a pinnacle of performance in detecting brain abnormalities. The convergence of advanced computer vision techniques and medical imaging is strikingly evident in the exceptional accuracy of Proposed-Swin (ViT) on Kaggle’s dataset, reaching an impressive 99.92%. This achievement not only underscores the potential of the Swin-Based (ViT) architecture in elevating the precision of brain tumor identification but also positions it as a frontrunner in the field.
As we navigate through the intricate landscape of brain MRI classification, the diverse array of models in Table 5 reveals nuanced insights. CNN-based approaches, exemplified by Talukder et al., 2023 [13] and Tabatabaei et al., 2023 [48] on the Figshare dataset with accuracies of 99.68% and 99.30%, respectively, demonstrate noteworthy efficacy. On the Kaggle dataset, alongside Proposed-Swin (ViT), other CNN-based models such as Rahman and Islam [82], Muezzinoglu et al. [83], and Ali et al. [84] also exhibit high accuracy rates. However, it is essential to highlight that Proposed-Swin (ViT) not only surpasses these CNN-based models but excels as a benchmark for superior performance in brain tumor classification. Additionally, the comparative analysis underscores the diversity in model performances and signifies the evolving landscape of methodologies in advancing the accuracy of brain MRI-based diagnostics.
4.8 Limitations and future directions
This study introduces an advanced deep learning approach based on the Swin Transformer, but it comes with certain limitations. Among these limitations, the primary and most significant is the evaluation of the Proposed-Swin model’s performance on a brain MRI dataset composed of a combination of a few datasets due to the scarcity of publicly available datasets. Additionally, the limitation stems from the relatively small scale of the dataset for deep learning models. Assessing the model’s generalizability across different datasets, imaging characteristics, patient populations, and tumor types is challenging. Essentially, further research is needed to explore the effectiveness of the model in different datasets and clinical settings.
A second limitation is the lack of comprehensive clinical studies that validate the real-world clinical applicability of the model’s success. The model’s performance needs verification through studies involving various healthcare institutions, encompassing clinical variability, patient-specific factors, and the presence of rare tumor types. Furthermore, there is a limitation related to the tendency of deep learning models to lack interpretability. Understanding the decision-making process of the model is crucial for gaining trust from healthcare professionals.
Among the future directions of this study, the first and foremost is the multi-center validation on different datasets obtained from various healthcare institutions to enhance the Swin Model’s performance and generalizability. This multi-center validation is crucial for evaluating the model’s performance across different imaging protocols and patient demographics. Additionally, planned studies aim to demonstrate the model’s performance on different medical images. Optimizing the Swin Model for real-time applications is also a significant future direction. Improving the model’s architecture and efficient inference strategies are essential for providing timely and on-site diagnostic support to radiologists.
5 Conclusion
This study introduces a groundbreaking deep learning method using the Swin Transformer for precise brain tumor diagnosis. Addressing challenges like suboptimal imaging and diverse tumor types, we incorporated the HSW-MSA and ResMLP. Our Proposed-Swin model achieved an outstanding 99.92% accuracy on a brain MRI dataset, surpassing prior models. The Swin Transformer, enhanced by HSW-MSA and ResMLP, proves effective in improving accuracy and efficiency. Transfer learning and data augmentation bolstered model robustness. Substituting ResMLP for the traditional MLP not only enhanced accuracy but also improved training speed and parameter efficiency.
The significance of our findings lies in the potential support our method can provide to radiologists in making accurate and timely diagnoses, ultimately leading to improved patient outcomes and reduced risks associated with brain tumors. The innovative diagnostic approach introduced in this study, incorporating HSW-MSA and ResMLP in the Swin Transformer, represents a valuable contribution to the field of medical imaging and deep learning applications. As we move forward, further validation on diverse datasets and real-world clinical settings will be essential to establish the generalizability and reliability of the Proposed-Swin model. Nevertheless, our study lays a foundation for future research and developments in leveraging deep learning techniques for enhancing the diagnostic capabilities in neuroimaging, with the ultimate goal of improving patient care and outcomes in the realm of brain tumor diagnosis.
Data availability
MRI dataset can be accessed in Kaggle “https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset”.
References
Bondy ML, Scheurer ME, Malmer B et al (2008) Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer 113:1953–1968
Herholz K, Langen KJ, Schiepers C, Mountz JM (2012) Brain tumors. Semin Nucl Med 42:356–370. https://doi.org/10.1053/j.semnuclmed.2012.06.001
Ostrom QT, Barnholtz-Sloan JS (2011) Current state of our knowledge on brain tumor epidemiology. Curr Neurol Neurosci Rep 11:329–335. https://doi.org/10.1007/s11910-011-0189-8
Miller KD, Ostrom QT, Kruchko C et al (2021) Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin 71:381–406. https://doi.org/10.3322/caac.21693
Charles NA, Holland EC, Gilbertson R et al (2011) The brain tumor microenvironment. Glia 59:1169–1180. https://doi.org/10.1002/glia.21136
Liu Z, Tong L, Chen L et al (2023) Deep learning based brain tumor segmentation: a survey. Complex Intell Syst 9:1001–1026. https://doi.org/10.1007/s40747-022-00815-5
Jyothi P, Singh AR (2023) Deep learning models and traditional automated techniques for brain tumor segmentation in MRI: a review. Artif Intell Rev 56:2923–2969. https://doi.org/10.1007/s10462-022-10245-x
Solanki S, Singh UP, Chouhan SS, Jain S (2023) Brain tumor detection and classification using intelligence techniques: an overview. IEEE Access 11:12870–12886
Villanueva-Meyer JE, Mabray MC, Cha S (2017) Current clinical brain tumor imaging. Clin Neurosurg 81:397–415. https://doi.org/10.1093/neuros/nyx103
Ellingson BM, Wen PY, Van Den Bent MJ, Cloughesy TF (2014) Pros and cons of current brain tumor imaging. Neuro Oncol 16:vii2–vii11. https://doi.org/10.1093/neuonc/nou224
Xie Y, Zaccagna F, Rundo L et al (2022) Convolutional neural network techniques for brain tumor classification (from 2015 to 2022): review, challenges, and future perspectives. Diagnostics 12:1850
Ali S, Li J, Pei Y et al (2022) A comprehensive survey on brain tumor diagnosis using deep learning and emerging hybrid techniques with multi-modal MR image. Arch Comput Methods Eng 29:4871–4896
Talukder MA, Islam MM, Uddin MA et al (2023) An efficient deep learning model to categorize brain tumor using reconstruction and fine-tuning. Expert Syst Appl. https://doi.org/10.1016/j.eswa.2023.120534
Rajeev SK, Pallikonda Rajasekaran M, Vishnuvarthanan G, Arunprasath T (2022) A biologically-inspired hybrid deep learning approach for brain tumor classification from magnetic resonance imaging using improved gabor wavelet transform and Elmann-BiLSTM network. Biomed Signal Process Control. https://doi.org/10.1016/j.bspc.2022.103949
Pacal I, Kılıcarslan S (2023) Deep learning-based approaches for robust classification of cervical cancer. Neural Comput Appl. https://doi.org/10.1007/s00521-023-08757-w
Coşkun D, Karaboğa D, Baştürk A et al (2023) A comparative study of YOLO models and a transformer-based YOLOv5 model for mass detection in mammograms. Turk J Electr Eng Comput Sci 31:1294–1313. https://doi.org/10.55730/1300-0632.4048
Wang W, Pei Y, Wang SH et al (2023) PSTCNN: explainable COVID-19 diagnosis using PSO-guided self-tuning CNN. Biocell 47:373–384. https://doi.org/10.32604/biocell.2023.025905
Pacal I, Karaboga D (2021) A robust real-time deep learning based automatic polyp detection system. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2021.104519
Zhang Y-D, Govindaraj VV, Tang C et al (2019) High performance multiple sclerosis classification by data augmentation and AlexNet transfer learning model. J Med Imaging Health Inform 9:2012–2021. https://doi.org/10.1166/JMIHI.2019.2692
Wang W, Zhang X, Wang SH, Zhang YD (2022) COVID-19 diagnosis by WE-SAJ. Syst Sci Control Eng 10:325–335. https://doi.org/10.1080/21642583.2022.2045645
Pacal I (2022) Deep learning approaches for classification of breast cancer in ultrasound (US) images. J Inst Sci Technol. https://doi.org/10.21597/jist.1183679
Amin J, Sharif M, Haldorai A et al (2022) Brain tumor detection and classification using machine learning: a comprehensive survey. Complex Intell Syst 8:3161–3183. https://doi.org/10.1007/s40747-021-00563-y
Deepak S, Ameer PM (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2019.103345
Wang SH, Govindaraj VV, Górriz JM et al (2021) Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inform Fusion 67:208–229. https://doi.org/10.1016/j.inffus.2020.10.004
Chahal PK, Pandey S, Goel S (2020) A survey on brain tumor detection techniques for MR images. Multimed Tools Appl 79:21771–21814. https://doi.org/10.1007/s11042-020-08898-3
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Futur Gener Comput Syst 87:290–297. https://doi.org/10.1016/j.future.2018.04.065
Esmaeili M, Vettukattil R, Banitalebi H et al (2021) Explainable artificial intelligence for human-machine interaction in brain tumor localization. J Pers Med. https://doi.org/10.3390/jpm11111213
Zhang Y, Deng L, Zhu H et al (2023) Deep learning in food category recognition. Inform Fusion. https://doi.org/10.1016/j.inffus.2023.101859
Karaman A, Karaboga D, Pacal I et al (2022) Hyper-parameter optimization of deep learning architectures using artificial bee colony (ABC) algorithm for high performance real-time automatic colorectal cancer (CRC) polyp detection. Appl Intell. https://doi.org/10.1007/s10489-022-04299-1
Pacal I, Karaman A, Karaboga D et al (2022) An efficient real-time colonic polyp detection with YOLO algorithms trained by using negative samples and large datasets. Comput Biol Med. https://doi.org/10.1016/J.COMPBIOMED.2021.105031
Pacal I, Alaftekin M (2023) Türk İşaret Dilinin Sınıflandırılması için Derin Öğrenme Yaklaşımları. Iğdır Üniversitesi Fen Bilimleri Enstitüsü Dergisi 13:760–777. https://doi.org/10.21597/jist.1223457
Zulfiqar F, Ijaz Bajwa U, Mehmood Y (2023) Multi-class classification of brain tumor types from MR images using EfficientNets. Biomed Signal Process Control. https://doi.org/10.1016/j.bspc.2023.104777
Mehnatkesh H, Jalali SMJ, Khosravi A, Nahavandi S (2023) An intelligent driven deep residual learning framework for brain tumor classification using MRI images. Expert Syst Appl. https://doi.org/10.1016/j.eswa.2022.119087
Shamshad F, Khan S, Zamir SW et al (2023) Transformers in medical imaging: a survey. Med Image Anal 88:102802
Akinyelu AA, Zaccagna F, Grist JT et al (2022) Brain tumor diagnosis using machine learning, convolutional neural networks, capsule neural networks and vision transformers, applied to MRI: a survey. J Imaging 8:205
Celard P, Iglesias EL, Sorribes-Fdez JM et al (2023) A survey on deep learning applied to medical images: from simple artificial neural networks to generative models. Neural Comput Appl 35:2291–2323
Tummala S, Kadry S, Bukhari SAC, Rauf HT (2022) Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Curr Oncol 29:7498–7511. https://doi.org/10.3390/curroncol29100590
Karaman A, Pacal I, Basturk A et al (2023) Robust real-time polyp detection system design based on YOLO algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (ABC). Expert Syst Appl. https://doi.org/10.1016/j.eswa.2023.119741
Nazir M, Shakil S, Khurshid K (2021) Role of deep learning in brain tumor detection and classification (2015 to 2020): a review. Comput Med Imaging Graph. https://doi.org/10.1016/j.compmedimag.2021.101940
Jiang Y, Zhang Y, Lin X et al (2022) SwinBTS: a method for 3D multimodal brain tumor segmentation using Swin transformer. Brain Sci. https://doi.org/10.3390/brainsci12060797
Kumar RL, Kakarla J, Isunuri BV, Singh M (2021) Multi-class brain tumor classification using residual network and global average pooling. Multimed Tools Appl 80:13429–13438. https://doi.org/10.1007/s11042-020-10335-4
Rehman A, Naz S, Razzak MI et al (2020) A deep learning-based framework for automatic brain tumors classification using transfer learning. Circuits Syst Signal Process 39:757–775. https://doi.org/10.1007/s00034-019-01246-3
Sharif MI, Khan MA, Alhussein M et al (2022) A decision support system for multimodal brain tumor classification using deep learning. Complex Intell Syst 8:3007–3020. https://doi.org/10.1007/s40747-021-00321-0
Mzoughi H, Njeh I, Wali A et al (2020) Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J Digit Imaging 33:903–915. https://doi.org/10.1007/s10278-020-00347-9
Amin J, Sharif M, Raza M et al (2019) Brain tumor detection using statistical and machine learning method. Comput Methods Programs Biomed 177:69–79. https://doi.org/10.1016/j.cmpb.2019.05.015
Tandel GS, Balestrieri A, Jujaray T et al (2020) Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2020.103804
Ghassemi N, Shoeibi A, Rouhani M (2020) Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control. https://doi.org/10.1016/j.bspc.2019.101678
Tabatabaei S, Rezaee K, Zhu M (2023) Attention transformer mechanism and fusion-based deep learning architecture for MRI brain tumor classification system. Biomed Signal Process Control. https://doi.org/10.1016/j.bspc.2023.105119
Kumar S, Mankame DP (2020) Optimization driven deep convolution neural network for brain tumor classification. Biocybern Biomed Eng 40:1190–1204. https://doi.org/10.1016/j.bbe.2020.05.009
Amin J, Sharif M, Yasmin M, Fernandes SL (2020) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit Lett 139:118–127. https://doi.org/10.1016/j.patrec.2017.10.036
Swati ZNK, Zhao Q, Kabir M et al (2019) Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph 75:34–46. https://doi.org/10.1016/j.compmedimag.2019.05.001
Sajjad M, Khan S, Muhammad K et al (2019) Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci 30:174–182. https://doi.org/10.1016/j.jocs.2018.12.003
Brain tumor dataset. https://figshare.com/articles/dataset/brain_tumor_dataset/1512427. Accessed 30 Jul 2023
Brain Tumor Classification (MRI) | Kaggle. https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri. Accessed 30 Jul 2023
Br35H :: Brain Tumor Detection 2020 | Kaggle. https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection?select=no. Accessed 30 Jul 2023
Brain Tumor MRI Dataset | Kaggle. https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset?select=Training. Accessed 30 Jul 2023
Dosovitskiy A, Beyer L, Kolesnikov A et al (2020) An image is Worth 16 × 16 words: transformers for image recognition at scale. In: ICLR 2021—9th International Conference on Learning Representations
Pacal I (2024) Enhancing crop productivity and sustainability through disease identification in maize leaves: exploiting a large dataset with an advanced vision transformer model. Expert Syst Appl. https://doi.org/10.1016/j.eswa.2023.122099
Khan S, Naseer M, Hayat M et al (2021) Transformers in vision: a survey. ACM Comput Surv. https://doi.org/10.1145/3505244
Liu Z, Lin Y, Cao Y, et al (2021) Swin transformer: hierarchical vision transformer using shifted windows
Liu Z, Hu H, Lin Y, et al (2021) Swin transformer V2: scaling up capacity and resolution
Touvron H, Bojanowski P, Caron M, et al (2021) ResMLP: feedforward networks for image classification with data-efficient training
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2016-Decem, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Russakovsky O, Deng J, Su H et al (2015) ImageNet large scale visual recognition challenge. Int J Comput Vis 115:211–252. https://doi.org/10.1007/s11263-015-0816-y
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90. https://doi.org/10.1145/3065386
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJ, Bottou L, Weinberger KQ (eds) Advances in neural information processing systems. Curran Associates Inc
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, pp 1–14
Tan M, Le Q V (2021) EfficientNetV2: smaller models and faster training
Howard A, Sandler M, Chen B, et al (2019) Searching for mobileNetV3. In: Proceedings of the IEEE International Conference on Computer Vision. Institute of Electrical and Electronics Engineers Inc., pp 1314–1324
Mehta S, Rastegari M (2021) MobileViT: light-weight, general-purpose, and mobile-friendly vision transformer. 3
Mehta S, Rastegari M (2022) Separable self-attention for mobile vision transformers
Tu Z, Talebi H, Zhang H, et al (2022) MaxViT: multi-axis vision transformer. Lecture Notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) 13684 LNCS, pp 459–479. https://doi.org/10.1007/978-3-031-20053-3_27
Touvron H, Cord M, Douze M, et al (2020) Training data-efficient image transformers & distillation through attention, pp 1–22
Touvron H, Cord M, Ai M DeiT III : Revenge of the ViT. 1–27
Bao H, Dong L, Piao S, Wei F (2021) BEiT: BERT pre-training of image transformers
d’Ascoli S, Touvron H, Leavitt M, et al (2021) ConViT: improving vision transformers with soft convolutional inductive biases. https://doi.org/10.1088/1742-5468/ac9830
Chu X, Tian Z, Wang Y et al (2021) Twins: revisiting the design of spatial attention in vision transformers. Adv Neural Inf Process Syst 12:9355–9366
Heo B, Yun S, Han D, et al (2021) Rethinking spatial dimensions of vision transformers
Hatamizadeh A, Yin H, Heinrich G, et al (2022) Global context vision transformers
Deepak S, Ameer PM (2023) Brain tumor categorization from imbalanced MRI dataset using weighted loss and deep feature fusion. Neurocomputing 520:94–102. https://doi.org/10.1016/j.neucom.2022.11.039
Özkaraca O, Bağrıaçık Oİ, Gürüler H et al (2023) Multiple brain tumor classification with dense CNN architecture using brain MRI images. Life. https://doi.org/10.3390/life13020349
Rahman T, Islam MS (2023) MRI brain tumor detection and classification using parallel deep convolutional neural networks. Meas Sens. https://doi.org/10.1016/j.measen.2023.100694
Muezzinoglu T, Baygin N, Tuncer I et al (2023) PatchResNet: Multiple patch division-based deep feature fusion framework for brain tumor classification using MRI images. J Digit Imaging 36:973–987. https://doi.org/10.1007/s10278-023-00789-x
Ali MU, Hussain SJ, Zafar A et al (2023) WBM-DLNets: wrapper-based metaheuristic deep learning networks feature optimization for enhancing brain tumor detection. Bioengineering. https://doi.org/10.3390/bioengineering10040475
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). The authors state no financial support was received for this manuscript.
Author information
Authors and Affiliations
Contributions
Ishak Pacal is solely responsible for all aspects of this work, including conceptualization, methodology design, data collection, analysis, and the creation of visual aids. He authored the entirety of the manuscript, meticulously reviewed and edited it for clarity, and granted final approval for its publication.
Corresponding author
Ethics declarations
Conflict of interest
No competing interests declared.
Ethical approval
No ethics approval was required for this work as it did not involve human subjects, animals, or sensitive data that would necessitate ethical review.
Consent to participate
No formal consent to participate was required for this work as it did not involve interactions with human subjects or the collection of sensitive personal information.
Consent to publish
This study did not use individual person’s data.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pacal, I. A novel Swin transformer approach utilizing residual multi-layer perceptron for diagnosing brain tumors in MRI images. Int. J. Mach. Learn. & Cyber. (2024). https://doi.org/10.1007/s13042-024-02110-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13042-024-02110-w