Abstract
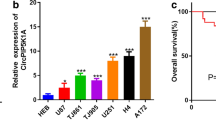
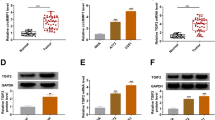
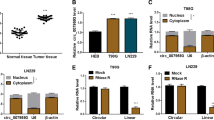
Circular RNAs (circRNAs) have been verified to play important roles in malignant tumors, including glioblastoma. The aim of this study is to explore the biological roles and underlying mechanisms of circRNA vacuolar protein sorting 18 homolog (circVPS18) in glioblastoma. A quantitative real-time polymerase chain reaction (qRT-PCR) was performed to measure the expression of circVPS18, microRNA (miR)-1299-3p, and branched-chain amino acid transaminase 1 (BCAT1). In vitro experiments were conducted using 5-ethynyl-2′-deoxyuridine (EdU), flow cytometry, transwell, and tube formation assays, respectively. Western blot was conducted to examine all protein levels. Dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were employed to confirm the interaction between miR-1229-3p and circVPS18 or BCAT1. The murine xenograft model was established to conduct in vivo assay. CircVPS18 and BCAT1 were highly expressed while miR-1229-3p was lowly expressed in glioblastoma tissues and cells. CircVPS18 knockdown inhibited glioblastoma progression by inhibiting cell proliferation, migration, invasion, and angiogenesis, and promoting cell apoptosis. Moreover, miR-1229-3p could be targeted by circVPS18; inhibition of miR-1229-3p could invert the suppressive effect of circVPS18 knockdown on glioblastoma tumorigenesis. Furthermore, BCAT1 was a target of miR-1229-3p; functionally, BCAT1 overexpression could reverse the inhibitory effects of miR-1229-3p upregulation on glioblastoma cell malignant phenotypes. Moreover, we also verified that circVPS18A could regulate BCAT1 expression by sponging miR-1229-3p. Additionally, circVPS18 silencing also restrained tumor growth and metastasis in vivo. CircVPS18 accelerated glioblastoma progression by miR-1229-3p/BCAT1 axis, providing a potential therapeutic target for glioblastoma.








Similar content being viewed by others
Availability of Data and Materials
The analyzed data sets generated during the present study are available from the corresponding author upon reasonable request.
References
Bach DH, Lee SK, Sood AK (2019) Circular RNAs in cancer. Molecular Therapy Nucleic Acids 16:118–129
Bagley SJ, Desai AS, Linette GP, June CH, O’Rourke DM (2018) CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro Oncol 20(11):1429–1438
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Butkytė S, Čiupas L, Jakubauskienė E, Vilys L, Mocevicius P, Kanopka A, Vilkaitis G (2016) Splicing-dependent expression of microRNAs of mirtron origin in human digestive and excretory system cancer cells. Clin Epigenetics 8:33
Cao Q, Shi Y, Wang X, Yang J, Mi Y, Zhai G, Zhang M (2019) Circular METRN RNA hsa_circ_0037251 Promotes Glioma Progression by Sponging miR-1229-3p and Regulating mTOR Expression. Sci Rep 9(1):19791
Chen L-L, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12(4):381–388
Cho HR, Jeon H, Park CK, Park SH, Kang KM, Choi SH (2017) BCAT1 is a new MR imaging-related biomarker for prognosis prediction in IDH1-wildtype glioblastoma patients. Sci Rep 7(1):17740
de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Großhans H (2015) Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep 16(4):500–511
Diao B, Liu Y, Zhang Y, Yu J, Xie J, Xu GZ (2017) IQGAP1-siRNA inhibits proliferation and metastasis of U251 and U373 glioma cell lines. Mol Med Rep 15(4):2074–2082
Geng Y, Jiang J, Wu C (2018) Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol 11(1):98
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Hao Z, Hu S, Liu Z, Song W, Zhao Y, Li M (2019) Circular RNAs: functions and prospects in Glioma. J Mol Neurosci 67(1):72–81
Le Rhun E, Preusser M, Roth P, Reardon DA, van den Bent M, Wen P, Reifenberger G, Weller M (2019) Molecular targeted therapy of glioblastoma. Cancer Treat Rev 80:101896
Li W, Ma Q, Liu Q, Yan P, Wang X, Jia X (2021) Circ-VPS18 knockdown enhances TMZ sensitivity and inhibits glioma progression by MiR-370/RUNX1 axis. J Mole Neurosci: MN 71(6):1234–1244
Li X, Diao H (2019) Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol 234(8):13807–13819
Li X, Yang L, Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71(3):428–442
Liu EK, Sulman EP, Wen PY, Kurz SC (2020) Novel therapies for glioblastoma. Curr Neurol Neurosci Rep 20(7):19
Liu J, Liu T, Wang X, He A (2017) Circles reshaping the RNA world: from waste to treasure. Mol Cancer 16(1):58
Lv T, Miao Y, Xu T, Sun W, Sang Y, Jia F, Zhang X (2020) Circ-EPB41L5 regulates the host gene EPB41L5 via sponging miR-19a to repress glioblastoma tumorigenesis. Aging 12(1):318–339
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M (2013) circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16(1):94
Nishibeppu K, Komatsu S, Imamura T, Kiuchi J, Kishimoto T, Arita T, Kosuga T, Konishi H, Kubota T, Shiozaki A et al (2020) Plasma microRNA profiles: identification of miR-1229-3p as a novel chemoresistant and prognostic biomarker in gastric cancer. Sci Rep 10(1):3161
Pepin G, Gantier MP (2016) microRNA Decay: refining microRNA regulatory activity. MicroRNA (shariqah, United Arab Emirates) 5(3):167–174
Quesnel A, Karagiannis GS, Filippou PS (2020) Extracellular proteolysis in glioblastoma progression and therapeutics. Biochim Biophys Acta 1874(2):188428
Shah JL, Li G, Shaffer JL, Azoulay MI, Gibbs IC, Nagpal S, Soltys SG (2018) Stereotactic radiosurgery and hypofractionated radiotherapy for glioblastoma. Neurosurgery 82(1):24–34
Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M (2020) Management of glioblastoma: state of the art and future directions. CA: a Canc J Clinic 70(4):299–312
Touat M, Idbaih A, Sanson M, Ligon KL (2017) Glioblastoma targeted therapy: updated approaches from recent biological insights. Annals of Oncology : Official J Eu Soc Med Oncol 28(7):1457–1472
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ (2010) Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 60(3):166–193
Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H (2018) CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Can Res 78(17):4812–4825
Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J, Shi Z, You Y (2021) EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro Oncol 23(4):611–624
Yang M, Li G, Fan L, Zhang G, Xu J, Zhang J (2019) Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem Biophys Res Commun 508(3):980–985
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H et al (2018) Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst 110(3):304–315
Yi C, Li H, Li D, Qin X, Wang J, Liu Y, Liu Z, Zhang J (2019) Upregulation of circular RNA circ_0034642 indicates unfavorable prognosis in glioma and facilitates cell proliferation and invasion via the miR-1205/BATF3 axis. J Cell Biochem 120(8):13737–13744
Zhang S, Liao K, Miao Z, Wang Q, Miao Y, Guo Z, Qiu Y, Chen B, Ren L, Wei Z et al (2019) CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol 21(10):1284–1296
Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL (2016) The biogenesis of nascent circular RNAs. Cell Rep 15(3):611–624
Zhao X, Cui L (2020) A robust six-miRNA prognostic signature for head and neck squamous cell carcinoma. J Cell Physiol
Zheng YH, Hu WJ, Chen BC, Grahn TH, Zhao YR, Bao HL, Zhu YF, Zhang QY (2016) BCAT1, a key prognostic predictor of hepatocellular carcinoma, promotes cell proliferation and induces chemoresistance to cisplatin. Liver International : Official J Int Assoc Stud Liver 36(12):1836–1847
Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W, Liu Z, Li Z, Zeng L, Wang L et al (2013) Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer 12:53
Author information
Authors and Affiliations
Contributions
Conceptualization and methodology: Wei Li and Yu Huang; formal analysis and data curation: Yu Huang, Qiang Chen, and Wenjin Wei; validation and investigation: Qianliang Huang, Wei Li, and Qiang Chen; writing–original draft preparation and writing–review and editing: Qianliang Huang, Wei Li, and Yu Huang; approval of the final manuscript: all authors.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Ganzhou People's Hospital of Jiangxi Province. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• CircVPS18 was highly expressed in glioblastoma tissues and cells.
• CircVPS18 downregulation suppressed the progression of glioblastoma by regulating miR-1229-3p activity.
• MiR-1229-3p exerted the anti-tumor role in glioblastoma cells by downregulating BCAT1.
• CircVPS18 acted as a miR-1229-3p sponge to regulate BCAT1 expression.
Rights and permissions
About this article
Cite this article
Huang, Q., Li, W., Huang, Y. et al. Circular RNA VPS18 Promotes Glioblastoma Progression by Regulating miR-1229-3p/BCAT1 Axis. Neurotox Res 40, 1138–1151 (2022). https://doi.org/10.1007/s12640-022-00530-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-022-00530-6