Abstract
Circular RNA (circRNA) is a new type of endogenous non-coding RNAs (ncRNAs). circRNA regulates gene expression in many biological processes, and it also participates in the initiation and development of various diseases, including tumors, which are the focus of present research. With the development of high-throughput sequencing technique, an increasing number of circRNAs closely related to tumors have been discovered. According to numerous studies, there is a significant difference in the expressions of circRNAs among a variety of tumor tissues and para-carcinoma normal tissues. Some specifically expressed circRNAs may potentially serve as new biomarkers for tumor diagnosis and prognosis. This systemic review briefly introduces the characteristics, biogenesis, and functions of circRNAs, as well as discusses their relationship with cancer in detail. In addition, this article also describes several research strategies for circRNAs.
Similar content being viewed by others
Background
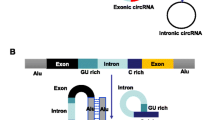
More than 70% human genomes are transcribed, and protein-coding genes only account for 1–2%. Most transcripts are non-coding RNAs (ncRNAs) [1]. Circular RNA (circRNA) is a type of new ncRNA different from linear RNA as it is a continuous covalently closed loop without the 5′-cap structure and the 3′-poly A tail. Most circRNAs are universal, stable, and conserved, and they are often specifically expressed in different tissues and developmental stages. In 1979, Hsu and Coca-Prados at Rockefeller University observed that circRNA exists in the cytoplasm of eukaryotic cells [2]. Within decades after the 1970s, circRNA has been considered as an outcome of a splicing error. With the development of RNA sequencing (RNA-seq) technology and bioinformatics in the twenty-first century, a large number of circRNAs have been discovered. There are four types of circRNAs, namely, exonic circRNA (ecircRNA), circRNAs from introns, exon-intron circRNA (EIciRNA), and intergenic circRNA [3]. More than 80% of the circRNAs are ecircRNAs, which are formed by the reverse covalent attachment of the 3′ splice donors and the 5′ splice acceptors of the precursor mRNA (pre-mRNA). circRNAs from introns are a general term for a class of circRNAs, including circular intronic RNAs (ciRNAs), excised group I introns, excised group II introns, excised tRNA introns, and intron lariats. EIciRNA is a type of circRNAs that are circularized simultaneously by exons and introns, probably similar to ecircRNAs. Intergenic circRNA is another non-exonic circRNA found by circRNA Identifier (CIRI). This integrated circRNA is formed by two intronic circRNA fragments (ICFs) flanked by GT-AG splicing signals acting as the splice donor (SD) and splice acceptor (SA) of the circular junction.
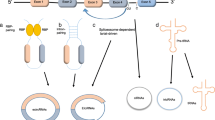
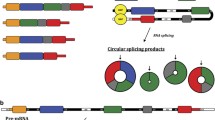
Two basic models of circRNA biogenesis have been proposed as follows: (1) intron-pairing-driven circularization, also known as direct back-splicing (Fig. 1a), is the main form of ecircRNA production, in which the flanking intronic complementary sequences of the pre-mRNA form a lariat by direct base-pairing, forming an ecircRNA when introns are removed, and (2) lariat-driven circularization, also known as exon-skipping (Fig. 1b), in which the pre-mRNA is partially folded during transcription, allowing the 3′-SD of the downstream exon to connect to the 5′-SA of the upstream exon, resulting in exon-skipping and the formation of a RNA lariat containing both exons and introns. With the removal of introns, an ecircRNA is formed. Generally, introns between circular exons will be excised. However, in some cases, these introns are retained as EIciRNAs. Some introns containing key nucleotide sequences are not decomposed by the debranching enzyme after splicing but instead independently cyclize into ciRNAs, which is called intron cyclization. In addition, there is another pattern of circRNA biogenesis that depends on RNA-binding proteins (RBPs) (Fig. 1c). The formation of circRNAs from introns and intergenic circRNAs is detailed in Fig. 2a-c and d, respectively.
Biogenesis of ecircRNAs and EIciRNAs. a Intron-pairing-driven circularization: The introns flanking inverted repeats or ALU elements form a circular RNA by base pairing, and ecircRNAs or EIciRNAs are formed by removing or retaining introns. b Lariat-driven circularization: EcircRNAs or EIciRNAs are generated by exon skipping. Exons 5′ attacks the 3′ splice site, forming an RNA lariat containing skipped exons 2 and 3 and an mRNA consisting of exon 1 and exon 4. Then, an EIciRNA and an RNA double lariat were further formed. c circRNA biogenesis depends on RBPs: RBPs bind to the sequence motifs of upstream and downstream introns, which serve as a bridge to bring flanking introns closer to each other, promoting the formation of circRNAs
Biogenesis of circRNAs from introns and intergenic circRNAs. a ciRNA: The pre-mRNA is spliced to produce an RNA lariat with a 2′,5′-phosphodiester, and the ciRNA is formed upon removal of the 3′-tail. b Excised group I introns: Exogenous guanosine (G) attacks the 5′-terminus of the intron, and exon 1 is cleaved due to transesterification. Then, the 5′-terminus of exon 2 is attacked by the 3′-hydroxyl of free exon 1, creating a linear intron. The 2′-hydroxyl close to the 3′-terminus of this linear intron attacks the phosphodiester bond near the 5′-terminus, resulting in an RNA lariat with 2′,5′-phosphodiester and a released 5′-terminus sequence. After removing the 3′-tail of the RNA lariat, a circRNA from group I introns is formed. c Excised group II introns: Pre-mRNA releases exon 2, and the 2′-hydroxyl of the intron’s 3′-terminus attacks the phosphodiester bond near the 5′-terminus, producing an RNA lariat with 2′,5′-phosphodiester and the free exon 1. d The circRNA contains two ICFs flanked by GT-AG splicing signals which act as SD and SA of the circular junction to form an integrated circRNA
circRNA has several features as follows: (1) abundance and diversity: more than 20,000 different circRNAs have been identified in eukaryotes; (2) stability: the half-life of circRNA is long because circRNA is a covalently closed circular structure without a 5′-cap and 3′-poly A tail, which is not easily degraded by exonuclease, resulting in far superior stability of circRNA than that of linear mRNA; (3) conservation: circRNA is highly conserved among different species, such as humans, mice, nematodes, zebrafish, drosophila, protists, and plants;(4) positioning: ecircRNA accounts for the majority of all circRNA types, which mainly exist in the cytoplasm, and intron-containing circRNAs including ciRNA and EIciRNA, which are mainly located in the nuclei of eukaryotes; and (5) specificity: circRNA is often specifically expressed in different tissues and different developmental stages.
circRNA has abundant biological functions and is involved in various physiological and pathological processes of tumor cells, including proliferation, apoptosis, invasion, and migration. One of the most frequently studied functions of circRNA is the microRNA (miRNA) sponge [4,5,6], namely, eliminating the miRNA’s regulation of a target gene via binding to the miRNA as a competing endogenous RNA (ceRNA) through the base complementary pairing principle. Moreover, circRNAs can also regulate gene expression at the transcriptional and post-transcriptional levels through other mechanisms, and they play a role as RBP sponges and protein scaffolds [7], some of which even have the ability to translate proteins [8, 9]. In addition, circRNAs are involved in RNAP II elongation [10, 11], alternative splicing [12], translation regulation [13], protein localization [14], histone modification [15, 16], and RNA maturation [17]. Recent studies have shown that circRNAs exert their biological functions through various mechanisms (Table 1, Fig. 3).
More and more circRNAs have been reported to be dysregulated in many human malignancies, such as lung cancer, breast cancer, gastric cancer, colorectal cancer, and liver cancer; they may serve as new diagnostic biomarkers and targets for cancer therapy. In this paper, we performed a systematic review of literature to provide information about the expression patterns and roles of circRNAs in solid tumors.
Main text
One hundred eighteen articles that investigated the expressions of circRNAs in various solid tumors were selected for the systemic review through searching PubMed, Embase, and Cochrane libraries (as of March 26, 2018), including nine cases of lung cancer, 14 cases of breast cancer, three cases of esophageal squamous cell carcinoma (ESCC), 20 cases of gastric cancer, 17 cases of colorectal carcinoma (CRC), 16 cases of hepatocellular carcinoma (HCC), 10 cases of gliomas, seven cases of bladder cancer, four cases of pancreatic cancer, five cases of osteosarcomas, and 14 cases of other tumors [two cases of ovarian cancer, one case of kidney cancer, one case of thyroid cancer, one case of basal cell carcinoma (BCC), one case of cutaneous squamous cell carcinoma (CSCC), two cases of oral squamous cell carcinoma (OSCC), one case of laryngeal squamous cell cancer (LSCC), one case of hypopharyngeal cancer, two cases of cholangiocarcinoma, one case of cervical cancer, and one case of prostate cancer].
Lung cancer
Zhao et al. screened four pairs of high-throughput circRNA microarrays of lung cancer and para-carcinoma tissues and found that 357 circRNAs are dysregulated [18]. circ-ITCH, a sponge of many oncogenic miRNAs, plays an important inhibitory role in the progression of lung cancer. Wan et al. detected the circ-ITCH levels in cancer tissues and para-carcinoma tissues in 78 lung cancer cases, and they reported that circ-ITCH expression is significantly reduced in approximately 73% of lung cancer tissues. Overexpression of circ-ITCH inhibits the proliferation of lung cancer cells and is associated with the expressions of host genes [19]. Another non-small cell lung cancer (NSCLC) study showed that the expression of circRNA_100876 (circ-CER) is significantly upregulated in cancer tissues. Such high expression of circ-CER is significantly associated with local lymph node invasion and advanced tumor. Patients with high circ-CER expression have significantly worse overall survival (OS) than those with low circ-CER expression [20]. Functional experiments have shown that circ-CER may be involved in the growth, progression, and metastasis of NSCLC cells [21]. Therefore, circ-CER may serve as a good diagnostic marker of NSCLC, and it is also a potential therapeutic target. In addition, hsa_circ_0013958 from the ACP6 gene is overexpressed in lung adenocarcinoma and correlated with tumor TNM staging [22]. Hsa_circ_0012673 is also upregulated in lung adenocarcinoma tissues and mainly located in the cytoplasm, regulating the proliferation of lung adenocarcinoma cells by adsorbing miR-22 [23]. Hsa_circ_0007385 is overexpressed in both NSCLC tissues and cells. Downregulation of hsa_circ_0007385 significantly inhibits the proliferation and invasion of NSCLC [24]. Hsa_circ_0014130 is highly expressed in NSCLC tissues and is closely related to lymph node metastasis and TNM staging, which can be used for prognostic evaluation [25]. In addition, the overexpression of hsa_circ_0000064 in lung cancer is positively correlated with T and N stage. Knockout of hsa_circ_0000064 significantly inhibits cell proliferation and promotes apoptosis [26]. The expression and function of circRNAs in lung cancer are shown in Table 2.
Breast cancer
Approximately 20% of breast cancers detected by mammography are ductal carcinomas in situ (DCIS) [27]. Some of these highly curative tumors will develop into invasive ductal carcinoma (IDC), which is life-threatening. However, the underlying determinants still remain unclear. A recent study has identified two circRNAs (hsa_circ_0122662 and hsa_circ_0001358) in five patients with DCIS/IDC and the MCF-7 invasive breast cancer cell line. Five miRNAs (miR-200b-3p, miR-200c-3p, miR-376a-3p, miR-376b-3p, and miR-429) have been confirmed to bind to hsa-circ-0001358 [28]. Further study of differentially expressed circRNAs in DCIS/IDC can aid the understanding of the molecular mechanisms underlying the progression from DCIS to IDC.
Nair et al. found 411 tumor-specific circRNAs from 885 breast cancer samples from TCGA in triple-negative (TN) breast cancer, including 256 estrogen receptor-positive (ER+), and 288 HER-2-positive (HER-2+) breast cancer cases [29]. Lu et al. found that 715 out of 1155 differentially expressed circRNAs are upregulated in breast cancer tissues compared to para-carcinoma tissues but that the other 440 circRNAs are downregulated. Validation studies have shown that hsa_circ_103110, hsa_circ_104821, and hsa_circ_104689 are upregulated in breast cancer tissues but that hsa_circ_100219, hsa_circ_006054, and hsa_circ_406697 are downregulated. The combination of hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 provides valuable insights into the diagnosis of breast cancer [30]. Some scholars believe higher diagnostic value of circRNAs in breast cancer than CEA and CA-153 [31].
circ-Foxo3 is a potential tumor suppressor that is significantly downregulated in breast cancer tissues [32] and may be involved in tumor progression [33]. Overexpression of circ-Foxo3 in the MDA-MB-231 breast cancer cell line significantly reduces cell proliferation in vitro. Subcutaneous injection of MDA-MB-231 cells transfected with circ-Foxo3 into nude mice inhibits tumor growth and promotes apoptosis [32]. A total of 25 binding sites of circ-Foxo3 for eight miRNAs (miR-22, miR-136, miR-138, miR-149, miR-433, miR-762, miR-3614-5p, and miR-3622b-5p) are detected [34], and transfection of these miRNAs into MDA-MB-231 cells can reduce apoptosis.
circ-VRK1 is also one of the downregulated circRNAs in breast cancer, especially in breast cancer stem cells (BCSCs). Upregulation of circ-VRK1 will inhibit the stemness of BCSCs [35]. In addition, circ_000911 is poorly expressed in breast cancer. In vitro experiments have confirmed that upregulation of circ_000911 increases Notch1 expression via binding to miR-449a, thereby suppressing the proliferation, invasion, and metastasis of breast cancer cells [36].
In contrast, hsa_circ_0001982, hsa_circ_0005239, and hsa_circ_0008717 are upregulated in breast cancer, and knockdown of their expressions inhibits cell proliferation and promotes apoptosis [37,38,39]. circ_0006528 is highly expressed in chemotherapy-resistant breast cancer cell lines, and the sensitivity of these cells to chemotherapy is significantly increased after knocking down circ_0006528 [40]. The expression of circ-DENND4C is increased in breast cancer cell lines under hypoxic conditions, and downregulation of circ-DENND4C inhibits the proliferation of breast cancer cells [41]. The expression and function of circRNAs in breast cancer are shown in Table 3.
ESCC
Several dysregulated circRNAs are found in ESCC, including hsa_circ_000167, hsa_circ_001059, hsa_circ_0067934, and circ-ITCH [42,43,44]. Similar to lung cancer, downregulation of circ-ITCH is also observed in 684 ESCC tissues and para-carcinoma tissues [42]. circRNAs may be associated with the radio-resistance of ESCC. In a circRNA microarray analysis of radiation-sensitive and radio-resistant cells, researchers have found that 57 significantly upregulated circRNAs and 17 downregulated circRNAs in radio-resistant ESCC cells, excluding circ-ITCH. KEGG analysis has shown that more than 400 differentially expressed target genes of circRNAs are enriched in the Wnt signaling pathway. Su et al. identified more than 3700 human circRNAs, among which hsa_circ_000167 and hsa_circ_001059 in the KYSE-150R human radiation-resistant esophageal cancer cell line are significantly different from the KYSE-150 parental cell line. These two circRNAs were confirmed by circRNA-miRNA co-expression analysis to be the most important factors in the potential circRNA/miRNA networks [44]. Xia et al. found that hsa_circ_0067934 encoded by PRKCI is upregulated in 51 cases of ESCC tissues compared to adjacent noncancerous tissues, and they reported that hsa_circ_0067934 is associated with poor tumor differentiation and advanced TNM stage. Silencing hsa_circ_0067934 by siRNA induces cell cycle arrest and inhibits proliferation and migration of ESCC cells [43]. Given that TNM staging is applied to predict patient outcomes, hsa_circ_0067934 may serve as a potential prognostic marker for ESCC. The expression and function of circRNAs in ESCC are shown in Table 4.
Gastric cancer
Several studies have examined the differential expression of circRNAs between gastric cancer and para-carcinoma tissues by circRNA microarrays [45,46,47,48,49,50]. Chen et al. found 180 circRNAs differentially expressed in gastric cancer and normal tissues using RNA-seq analysis. Among which, circ-PVT1 is upregulated, and the overexpression of circ-PVT1 suggests a better OS and disease-free survival (DFS). A luciferase reporter assay has confirmed that circ-PVT1 indirectly regulates the expression of the E2F2 transcription factor as a sponge of miR-125 family, promotes the colony formation, and is involved in cell cycle regulation [45].
Hsa_circ_0047905, hsa_circ_0087198, and hsa_circ_0138960 are also highly expressed in gastric cancer tissues. Inhibition of these circRNAs significantly suppresses the proliferation of gastric cancer cells [50]. Sui et al. found that hsa_circRNA_000792 and hsa_circRNA_400071 are upregulated in gastric cancer but that hsa_circRNA_001066, hsa_circRNA_001959, and hsa_circRNA_400066 are downregulated [49].
Hsa_circ_0000096, also known as circ-HIAT1, is downregulated in gastric cancer cells and tissues. Knockdown of hsa_circ_0000096 reduces the expression of cyclin D1, CDK6, matrix metalloproteinase (MPP)-2, and MMP-9, and it significantly inhibits cell proliferation and migration and blocks cell cycle (preventing gastric cancer cells from leaving G0/G1 phase to enter S phase), as well as inhibits tumor growth in a xenograft nude mouse model. The circRNA database shows that hsa_circ_0000096 can interact with 17 different types of miRNAs. Downregulation of hsa_circ_0000096 results in a decrease in miR-224 (a modulator of CD40) and an increase in miR-200a (targeting E-cadherin) [51].
Both circ-LPHN2 and circ-LARP4 are lowly expressed in gastric cancer tissues. The former acts as a sponge of miR-630 and inhibits the proliferation of gastric cancer cells [52]. The latter exerts biological functions by adsorbing miR-424, and it acts as an independent prognostic factor for gastric cancer [53]. Hsa_circ_002059 has also been confirmed to be downregulated in gastric cancer tissues. The low expression of hsa_circ_002059 is significantly associated with gender, age, distant metastasis, and TNM staging [54]. In particular, the postoperative level of plasma hsa_circ_002059 in gastric cancer patients is lower than that before surgery. Hsa_circ_0000190 is downregulated in gastric cancer tissues and plasma, and its low expression level is related to tumor size, lymph node and distant metastasis, and TNM staging. Hsa_circ_0000190 is considered to have better sensitivity and specificity compared to CEA and CA19-9 [55]. The low expression of hsa_circ_0001895 in gastric cancer tissues is significantly associated with histological types and grade [56]. Hsa_circ_0014717 is also lowly expressed in gastric cancer, and such downregulation is associated with distant metastasis and clinical staging. Additionally, hsa_circ_0014717 can be stably detected in gastric juice [48]. Hsa_circ_0000181, hsa_circ_0001649, hsa_circ_0000520, hsa_circ_0003159, and hsa_circ_0074362 are also lowly expressed in tissues or plasma of gastric cancer patients, and their expression is negatively correlated with distant metastasis and TNM staging. Therefore, these circRNAs may be used as diagnostic indexes to indicate if there is distant metastasis [57,58,59,60,61]. The plasma levels of hsa_circ_0001017 and hsa_circ_0061276 are also downregulated, making them suitable for the diagnosis and prognosis of gastric cancer [62]. Moreover, hsa_circ_0000745 is expressed at a higher level in gastric cancer tissues than normal tissues, and its expression in plasma of gastric cancer patients is also higher than that of healthy controls. Hsa_circ_0000745 expression in gastric cancer tissues and plasma is associated with tumor differentiation and lymph node metastasis, respectively, and plasma hsa_circ_0000745 combined with CEA has a greater diagnostic value for gastric cancer [63]. The expression of circRNA may be a predictor of early recurrence in patients with radical resection of stage III gastric cancer [64], but the major shortcoming of this previous study was that the follow-up time was too short. The expression and function of circRNAs in gastric cancer are shown in Table 5.
CRC
Bachmayr-Heyda et al. found that 11 circRNAs are upregulated and 28 circRNAs are downregulated in CRC tissues compared to para-carcinoma tissues by RNA-seq analysis [65]. In addition, the ratio of some circRNAs (circ3204/USP3, circ0817/CUL5, circ7374/TNS4, and circ6229/METTL3) to linear RNA is lower in CRC tissues than in normal tissues. Zhang et al. [66] also found that there are more circRNAs downregulated in CRC tissues and that the most significant downregulated circRNA is derived from the PTK2 tumor suppressor gene. This phenomenon can be attributed to the high stability of circRNA as it accumulates in non-proliferating cells and is dispersed in daughter cells of proliferating cells [65].
Similar to lung cancer, circ-ITCH is also significantly downregulated in CRC tissues. circ-ITCH is a sponge of some miRNAs which downregulate many target genes involved in G1/S transition, including miR-7, miR-20a, and miR-214. Overexpression of circ-ITCH reduces the proliferation of SW480 and HCT116 cells. Therefore, circ-ITCH may have anti-proliferative effects in CRC [67].
The downregulation of Hsa_circ_001988 in CRC is significantly related to differentiation and neural invasion of cancer cells. Because nerve invasion is a definite negative prognostic factor in CRC patients, hsa_circ_001988 may be a promising prognostic biomarker of patients with CRC [68]. In addition, hsa_circ_0001649, hsa_circ_0003906, and circRNA derived from ITCH78 are also downregulated in CRC, and the first two circRNAs are related to the pathological differentiation of CRC and may be used as diagnostic indicators of CRC [69, 70].
In contrast, the expression of ciRS-7 is upregulated in CRC, and it is the most significantly upregulated circRNA, deriving from METTL3, a m6A methyltransferase gene [65]. A large study by Weng et al., including 153 trial cohorts and 165 validation cohorts, has also confirmed the upregulation of ciRS-7 in CRC tissues. The high expression of ciRS-7 is positively correlated with tumor size, lymph node metastasis, TNM staging, and OS of patients [71]. Knockdown of ciRS-7 inhibits the activity of miR-7 target genes, such as EGFR and IGF-1R, thereby suppressing the proliferation and invasion of CRC.
As a positive regulator of CRC cell proliferation and invasion, hsa_circ_001569 exhibits higher expression in CRC tissues than noncancerous tissues [65, 72, 73]. Hsa_circ_001569, the sponge of miR-145, increases the number of cells in S and G2/M phase and accelerates the proliferation and invasion of CRC cells by preventing miR-145 from downregulating target genes, such as E2F5, FMNL2, and BAG4 [72]. Knockdown of hsa_circ_001569 in LOVO and SW620 cells reverses the invasive ability [72].
Hsa_circ_0000069 is also overexpressed in CRC tissues. siRNA-mediated knockdown of hsa_circ_0000069 inhibits the proliferation, migration, and invasion of HT-29 cells and induces G0/G1 phase arrest [74]. The expression of circRNAs derived from STIL60 and BANP79 genes in CRC tissues is also higher than that in normal tissues. Knockdown of circ-BANP reduces the proliferation and colony formation of the HCT116 and HT29 cell lines. Moreover, the expression of p-Akt is also decreased, suggesting that the PI3K-Akt pathway may be involved in circ-BANP-induced cell proliferation [75]. Zeng et al. found that circ-HIPK3 is overexpressed in CRC cells and tissues, and they reported that the prognosis of CRC patients with high circ-HIPK3 expression is poor. Knockdown of circ-HIPK3 significantly inhibits the proliferation, invasion, and metastasis of CRC. Additionally, changes in cell function induced by circ-HIPK3 are reversed by miR-7 [76]. The high expression of hsa_circ_0007534 in CRC tissues is associated with lymph node metastasis and tumor staging. Interfering with the expression of hsa_circ_0007534 significantly inhibits the proliferation and promotes the apoptosis of CRC cells [77]. Hsa_circ_000984 is also significantly overexpressed in CRC tissues and cells, and knockdown of hsa_circ_000984 reduces the proliferation, invasion, and metastasis of CRC cells. Moreover, competitively binding to miR-106b as a ceRNA, hsa_circ_000984 effectively upregulates CDK6 expression, thereby affecting the function of CRC cells [78]. Hsa_circ_0020397 is highly expressed in CRC cells, which promotes CRC cell proliferation and invasion. In addition, hsa_circ_000984 upregulates the expression of oncogenic TERT and PD-L1 by binding to miR-106b [79].
Hsiao et al. found that circCDK13, circCCNB, and circCCDC66 are upregulated in CRC tissues. The expression of circ-CCDC66 has been detected in various tumor cell lines. circ-CCDC66 acts as a miRNA sponge to protect MYC mRNA from the miRNA-33b- and miR-93-mediated degradation, and it is involved in cell proliferation, migration, and invasion [80].
Comparison of three syngeneic CRC cell lines with different KRAS mutation status, including DLD-1, DKO-1, and DKs-8, has shown that extracellular circRNAs are more abundant than intracellular circRNAs, and most circRNAs are downregulated in the KRAS-mutated CRC cell lines [81]. circRNA is associated with KRAS mutations, and it is a promising biomarker of CRC, especially for KRAS-mutated CRC. The expression and function of circRNAs in CRC are shown in Table 6.
HCC
Studies have shown that circRNAs are involved in the development of HCC, although the mechanism remains unclear. Shang et al. found 61 circRNAs expressed differentially between HCC and adjacent tissues, including 26 upregulated and 35 downregulated circRNAs [82]. Fu et al. identified 527 circRNAs in HCC and reported that most of them are downregulated in HCC. The two most significantly downregulated circRNAs are hsa_circ_0004018 encoded by SMYD4 and hsa_circ_0003570 encoded by FAM53B, which are associated with the clinicopathological features of HCC [83, 84]. Hsa_circ_0001649 [85], which is derived from the SHPRH gene, and circZKSCAN1 [86], which is derived from the ZKSCAN1 gene, are also significantly downregulated in HCC tissues. Tumor number, cirrhosis, vascular invasion, microvascular infiltration (MVI), and tumor grade are the major factors associated with the circ-ZKSCAN1 expression level. Overexpression of circZKSCAN1 inhibits HCC progression both in vitro and in vivo. The circRNA encoded by the MTO1 gene is also downregulated in HCC tissues. As a sponge of miR-9, circ-MTO1 can inhibit HCC progression by upregulating the expression of p21. Decreased expression of circ-MTO1 is associated with poor outcome in HCC patients, and intratumoral administration of circ-MTO1 siRNA promotes HCC growth in vivo, indicating that circ-MTO1 is a potential prognostic marker and therapeutic target for HCC [87]. The low expression of circ-ITCH in HCC is also associated with a shorter OS [88]. Hsa_circ_0001649 was also lowly expressed in HCC tissues compared to normal tissues, and the expression level of hsa_circ_0001649 is related to tumor size and tumor thrombus. Knockdown of hsa_circ_0001649 increases the levels of MMP9, MMP10, and MMP13, suggesting that it may be a protective factor for HCC metastasis [85] and can be used as a potential diagnostic and prognostic marker. Moreover, hsa_circ_0001445 (cSMARCA5), hsa_circ_0005986, and hsa_circ_0067531 [89] are also downregulated in HCC. CSMARCA5 is regulated by DHX9, and it promotes the expression of TIMP3 and inhibits the proliferation and metastasis of HCC cells by adsorbing miR-17 and miR-181b [90]. HCC patients with cSMARCA5 low expression are usually accompanied by shorter OS and RFS. The expression level of hsa_circ_0005986 in HCC cell lines, including HepG2, Huh7, SMMC7721, HCCLM3, MHCC97H, and MHCC97L, is significantly lower than that in the L02 normal liver cell line. Both hsa_circ_0005986 and Notch1 mRNA can bind to miR-129-5p, and downregulation of hsa_circ_0005986 releases miR-129-5p to decrease the level of Notch1 mRNA, accelerating the cell proliferation by promoting G0/G1 to S phase transition [91].
By using a circRNA chip, Huang et al. identified 189 circRNAs significantly upregulated and 37 circRNAs downregulated in HCC compared to adjacent tissues. circRNA_100338 acts as an endogenous sponge of miR-141-3p in HCC to regulate the invasion function of hepatoma cells, and its high expression is closely related to a poorer OS and PFS of HCC patients [92]. ciRS-7 (hsa_circ_0001946) is significantly upregulated in HCC tissues [93] and is negatively correlated with miR-7 expression [94]. Overexpression of ciRS-7 is a risk factor for MVI in the liver. When ciRS-7 is knocked down, miR-7 is released and proliferation and invasion of HCC cells are also significantly inhibited [93, 94]. However, Xue et al. found that ciRS-7 is downregulated in HCC cells and tissues [95]. Zheng et al. reported that circRNA encoded by exon 2 of HIPK3 is upregulated in HCC tissues. circ-HIPK3 is a highly stable circRNA which can adsorb and inactivate a variety of miRNAs, including miR-124, the well-known tumor suppressor [4]. Therefore, targeting circ-HIPK3 may inhibit the growth of HCC cells in patients. Hsa_circ_0067934, which is highly expressed in HCC tissues and cells, can directly adsorb miR-1324, affect the expression level of FZD5, and regulate the Wnt/β-catenin signaling pathway. Knockdown of has_circ_0067934 significantly inhibits the proliferation, invasion, and metastasis of Hep3B and HuH7 cells and induces apoptosis [96]. Hsa_circ_0005075 is considered to be closely related to cell adhesion, which is an important part of tumor cell proliferation and metastasis. A recent study showed that the expression level of hsa_circ_0005075 is significantly different between HCC and normal liver tissues and is related to HCC tumor size. Larger tumor sizes correlated with higher expression of hsa_circ_0005075. Thus, hsa_circ_0005075 has the potential to become an ideal biomarker for HCC. Hsa_circ_0005075 has binding sites for four miRNAs, namely, miR-23a-5p, miR-23b-5p, miR-93-3p, and miR-581, in which miR-23b-5p is a key factor [82]. The specific molecular mechanism of hsa_circ_0005075 as the miR-23b-5p sponge to regulate the development of HCC remains to be further studied. The combination of circRNAs and traditional biomarkers of HCC will have greater diagnostic value. The expression and function of circRNAs in HCC are shown in Table 7.
Brain glioma
Song et al. selected 476 differentially expressed circRNAs from the RNA-seq data of 46 cases with brain glioma. The expression levels of circ_COL1A2, circ_PTN, circ_VCAN, circ_PLOD2, circ_SMO, circ_CLIP2, circ_GLIS3, and circ_EPHB4 in glioblastoma (GBM) are significantly higher than those in normal tissues. These circRNAs may act as miRNA sponges, which in turn increase the expressions of certain genes involved in pathological processes. The circRNA derived from the VCAN gene is associated with the development of gliomas, and it is upregulated in oligodendrogliomas and GBM [97]. Barbagallo et al. [98] found that miR-671-5p is overexpressed in GBM cells and tissues, which is associated with downregulation of ciRS-7. Hsa_circ_0046701 is highly expressed in glioma tissues. Silencing hsa_circ_0046701 upregulates miR-142, resulting in a decrease of ITGB8 and inhibition of cell proliferation and invasion. The hsa_circ_0046701/miR-142/ITGB8 axis may contribute significantly to the development of gliomas [99]. circ-SHKBP1 is highly expressed in high-grade gliomas, and knockdown of circ-SHKBP1 significantly inhibits cell proliferation and metastasis. The regulation of cell function by circ-SHKBP1 is achieved by targeted uptake of miR-544a/miR-379 and upregulation of FOXP1/FOXP2 [100].
In addition, the expression levels of circZNF292 [101] and TTBK2 gene-derived circRNAs [102] in gliomas are downregulated and upregulated, respectively. Under hypoxic conditions, cZNF292 is a circRNA expressed in endothelial cells involved in glioma cell proliferation and tube formation. Silencing cZNF292 inactivates the Wnt/β-catenin signaling pathway in U87MG and U251 cells, thereby arresting cell cycle and inhibiting cell proliferation [101]. Hsa_circ_022705 (circ-FBXW7) is lowly expressed in glioma tissues and cells, and it is positively correlated with the prognosis of patients with glioma. In addition, hsa_circ_022705 encodes the FBXW7-185aa protein. Upregulation of FBXW7-185aa significantly inhibits the proliferation of tumor cells, while silencing this protein promotes the malignant phenotype [103]. circ-SMARCA5 is significantly downregulated in gliomas and negatively correlated with the histological grade of gliomas. Overexpression of circ-SMARCA5 significantly decreases the metastatic capacity of U87MG cells. circ-SMARCA5 has abundant binding motifs with several RBPs, and it can directly bind to SRSF1 and regulate its expression [104]. circ-SHPRH is abundantly expressed in normal human brain, but it is significantly downregulated in GBM. It has the ability to encode the SHRH-146aa protein with the help of an open reading frame (ORF) driven by an internal ribosome entry site (IRES). Overexpression of SHRH-146aa in U251 and U373 GBM cells reduces the malignant phenotype [105].
The expression of circ-TTBK2 is increased in glioma tissues, which promotes cell proliferation, migration, and invasion but inhibits apoptosis [102]. In addition, circ-BRAF is significantly reduced in glioma patients with a higher pathological grade. The high expression of circ-BRAF is an independent predictive marker for PFS and OS in glioma patients [106]. In the future, more research is needed to reveal the regulatory mechanism of circRNAs in gliomas. The expression and function of circRNAs in gliomas are shown in Table 8.
Bladder cancer
Bladder cancer is a common urinary system malignancy, especially in males [107]. The circRNAs in bladder cancer show an overall upregulation, indicating that there are more upregulated circRNAs than downregulated circRNAs in bladder cancer tissues [108,109,110]. High-throughput microarray analysis has been used to identify six circRNAs that are differentially expressed in bladder cancer and normal tissues as follows: circPTK2 (hsa_circ_0005273), circTCF25 (hsa_circ_0041103), circBC048201 (hsa_circ_0061265), and circZFR (hsa_circ_0072088) are significantly upregulated and circTRIM24 (hsa_circ_0082582) and circFAM169A (hsa_circ_0007158) are downregulated [108]. The gene expression profiles of the linear transcripts corresponding to the overexpressed circRNAs are favorable for protein modification, binding, and intracellular metabolic processes, while those downregulated circRNAs are favorable for molecular function and catalytic activity [111]. Overexpression of circTCF25 increases CDK6 expression via adsorbing miR-103a-3p and miR-107, and it promotes cell proliferation and migration [108]. Both circRNA-MYLK and vascular endothelial growth factor (VEGF) A are significantly upregulated in bladder cancer. circRNA MYLK directly binds to miR-29a and reduces its ability to target VEGFA, a molecule that activates the VEGFA/VEGFR2 signaling pathway. Functionally, overexpression of circRNA MYLK promotes cell proliferation, migration, tube formation, and rearrangement of cytoskeleton [112].
Several circRNAs are downregulated in bladder cancer. For example, circ-ITCH is downregulated in bladder cancer tissues and cell lines, and patients with low circ-ITCH expression are significantly associated with a shorter OS. Overexpression of circ-ITCH inhibits the malignant biological behavior of bladder cancer cells, such as proliferation, migration, invasion, and metastasis, by upregulating p21 and PTEN through the uptake of miR-17 and miR-224 [113]. The expression of circ-BCRC4 in bladder cancer tissues is also lower than that in adjacent normal tissues. Overexpression of circ-BCRC4 inhibits the level of miR-101, thereby upregulating EZH2 expression, which promotes the apoptosis and inhibits the activity of T24T and UMUC3 cells [114]. circ-HIPK3 is expressed at a low level in bladder cancer cells and tissues, and it is negatively associated with differentiation, infiltration, and lymph node metastasis. Overexpression of circ-HIPK3 effectively inhibits growth, migration, invasion, and angiogenesis. circ-HIPK3 contains two key binding sites for miR-558, which significantly adsorbs miR-558, thereby inhibiting HPSE expression [110]. The expression and function of circRNAs in bladder cancer are shown in Table 9.
Pancreatic cancer
In pancreatic ductal adenocarcinoma (PDAC), 351 differentially expressed circRNAs (including 209 upregulated and 142 downregulated) between cancer tissues and normal tissues are identified by microarray analysis. Hsa_circ_0000977 is abnormally upregulated in pancreatic cancer tissues, and silencing hsa_circ_0000977 inhibits cell proliferation and induces cell cycle arrest. The interaction of hsa_circ_0000977, hsa-miR-874-3p, and PLK1A has been verified by dual luciferase reporter assay and fluorescence in situ hybridization (FISH) assay, and inhibition of hsa_circ_0000977 can reduce the expression of PLK1. In animal experiments, silencing hsa_circ_0000977 inhibits tumor growth [115]. circ-LDLRAD3 is overexpressed in cells, tissues, and plasma samples of pancreatic cancer patients. High expression of circ-LDLRAD3 is significantly correlated with venous or lymphatic infiltration and metastasis, and it may be used as a diagnostic biomarker for pancreatic cancer [116]. Hsa_circ_0000284 (circRNA_100782) is significantly upregulated in PDAC tissues, which is a sponge of miR-124. Knockdown of circRNA_100782 inhibits the proliferation and colony formation of BxPC3 cells by downregulating the target genes of miR-124, namely, IL6R and STAT3 [117]. In addition, hsa_circ_0005785 has binding sites for miR-181a and miR-181b [118]. Since miR-181a and miR-181b are associated with the growth/migration and gemcitabine resistance of pancreatic cancer cells, respectively, hsa_circ_0005785 may be involved in PDAC progression and gemcitabine resistance. The expression and function of circRNAs in pancreatic cancer are shown in Table 10.
Osteosarcoma
The circRNA encoded by oncogenic KCNH1 is upregulated in osteosarcoma tissues and cells, and it promotes cell proliferation, invasion, and metastasis [119]. The circRNA encoded by UBAP2 is the most prominently upregulated circRNA in osteosarcoma tissues, and patients with high circUBAP2 expression are often associated with a poor OS. In vitro and in vivo experiments have shown that circ-UBAP2 promotes osteosarcoma cell growth and inhibits apoptosis [120]. Hsa_circ_0001564 is significantly overexpressed in osteosarcoma tissues and cells. Knockdown of hsa_circ_001564 significantly inhibits osteosarcoma cell proliferation by inducing G0/G1 cell cycle arrest and promotes apoptosis of HOS and MG-63 cells. Hsa_circ_0001564 mediates tumorigenesis as a sponge of miR-29c-3p, and miR-29c can reverse the oncogenic effects of hsa_circ_001564 [121]. Hsa_circ_0009910 is also overexpressed in osteosarcoma cells. Knockdown of circ_0009910 inhibits the proliferation of osteosarcoma cells, leading to cell cycle arrest and apoptosis. However, inhibition of miR-449a eliminates this effect. As the sponge of miR-449a, circ_0009910 upregulates the functional target gene IL6R and promotes the development of osteosarcoma [122]. In osteosarcoma cells and tissues, hsa_circRNA_103801 is upregulated, while hsa_circRNA_104980 is downregulated. The potential target miRNAs for hsa_circRNA_103801 include hsa-miR-338-3p, hsa-miR-370-3p, and hsa-miR-877-3p, which are involved in the HIF-1, Rap1, PI3K-Akt, VEGF, and angiogenesis pathways. The potential target miRNAs for hsa_circRNA_104980 are hsa-miR-660-3p and hsa-miR-1298-3p, which participate in the tight junction pathway [123]. The expression and function of circRNAs in osteosarcoma are shown in Table 11.
Other tumors
The expression and function of circRNAs in other tumors are shown in Table 12.
Ovarian cancer
Ahmed et al. identified a total of 67,580 candidate circRNAs from primary and metastatic lesions of three patients with stage IIIC ovarian cancer, and they confirmed that the differential expression of circRNAs between primary and metastatic ovarian cancer is more pronounced than corresponding mRNAs [124]. Bachmayr-Heyda et al. found that the levels of circRNAs in immortalized normal ovarian epithelial cells are generally lower than those of ovarian cancer cells [65] because immortalized normal ovarian epidermal cells proliferate faster than ovarian cancer cells, resulting in reduced accumulation of circRNAs. In addition, the levels of circRNAs in miliary tumors are lower than those of non-miliary tumors; however, it cannot be explained by the difference in proliferation rates.
Kidney cancer
In clear cell renal cell carcinoma (ccRCC) tissues, the circRNA (circ-HIAT1) derived from hippocampus-rich gene transcription protein 1 (HIAT1) is downregulated. Compared with metastatic ccRCC, circ-HIAT1 is expressed at a higher level in non-metastatic ccRCC. In addition, the OS rate of ccRCC patients with high expression of circ-RIAT1 is superior to that of patients with low circ-RIAT1. circ-HIAT1 can directly bind to miR-29a-3p, miR-29c-3p, and miR-195-5p and upregulate the expression of CDC42. Different from the classical function of miRNA sponges, circ-HIAT1 acts as a “miRNA reservoir,” which increases miRNA stability, thereby reversing androgen receptor (AR)-mediated ccRCC migration and invasion. Inhibition of the miR-29a-3p/29c-3p/195-5p signaling pathway by circ-HIAT1 inhibits the migration and invasion of ccRCC cells [125]. The AR/circHIAT1/CDC42 signaling pathway may become a new therapeutic target for metastatic ccRCC.
Thyroid cancer
Compared with normal thyroid tissues, researchers have found 88 significantly upregulated circRNAs and 10 downregulated circRNAs in papillary thyroid cancer (PTC) tissues. Based on the miRNA response elements (MREs) of these dysregulated circRNAs, a network of circRNA-miRNA interactions has been constructed by Cytoscape. The downregulated circRNA, hsa_circRNA_100395, has potential for interaction with two cancer-associated miRNAs, namely miR-141-3p and miR-200a-3p, suggesting that hsa_circRNA_100395-miR-141-3p/miR-200a-3p may be involved in the pathogenesis of PTC [126]. However, this hypothesis needs further verification.
BCC
Sand et al. identified 23 upregulated circRNAs and 48 downregulated circRNAs in BCC tissues [127]. Hsa_circ_0022383 and hsa_circ_0022392 are the most significantly downregulated circRNAs and derived from FADS2 gene. Similarly, the two most significantly upregulated circRNAs are also encoded by the same host gene, namely, LINC00340.
CSCC
Sand et al. [128] identified 143 upregulated circRNAs and 179 downregulated circRNAs in CSCC. The two most significantly downregulated circRNAs (also most significantly downregulated in BCC) are from FADS2 gene, and the two most significantly upregulated circRNAs are encoded by LARP1B gene.
OSCC
Numerous circRNAs are differentially expressed in OSCC tissues and adjacent tissues. At present, 280 circRNAs have been identified with more than a twofold difference in OSCC, including 139 upregulated and 141 downregulated circRNAs. Among these circRNAs, hsa_circ_0013339 (circRNA_100290) is derived from the SLC30A7 gene, and it is upregulated by approximately sevenfold in OSCC compared to normal tissues. circRNA_100290 is a sponge of the miR-29 family. Functional analysis has revealed that knockdown of circRNA_100290 reduces CDK6 expression, induces G1/S arrest, and significantly inhibits the proliferation of SCC9 cell lines. In a nude mouse model, interference with circRNA_100290 also reduces tumor growth [129]. Moreover, circ-DOCK1 regulates BIRC3 expression through competitively binding to miR-196a as a ceRNA, and it is involved in the apoptosis of OSCC cells [130].
LSCC
In 698 dysregulated circRNAs in LSCC (302 upregulated cases and 396 downregulated cases), hsa_circ_100855 and hsa_circ_104912 are the two most significantly up- and downregulated circRNAs, respectively. High expression of hsa_circ_100855 and low expression of hsa_circ_104912 are associated with T3–4 stage, cervical lymph node metastasis, and later clinical stage of LSCC [131]. Researchers believe that the above circRNAs are involved in the initiation and development of LSCC and that they may be helpful for the diagnosis and prognosis in clinical practice.
Hypopharyngeal squamous cell carcinoma (HSCC)
Cao et al. showed that 2392 circRNAs are differentially expressed between HSCC and normal tissues [132]. Of these circRNAs, 1304 are upregulated, including hsa_circ_0024108, hsa_circ_0058106, and hsa_circ_0058107, while 1088 are downregulated, including hsa_circ_0001189, hsa_circ_0002260, and hsa_circ_0036722. However, the functions of these circRNAs in HSCC remain unexplored.
Cholangiocarcinoma
The expression of ciRS-7 (Cdr1as) in cholangiocarcinoma tissues is higher than that in adjacent normal tissues. Overexpression of ciRS-7 is closely related to later TNM staging, lymph node infiltration, and postoperative recurrence. The OS of cholangiocarcinoma patients with high ciRS-7 expression is inferior to that of patients with low ciRS-7 expression. Based on multivariate analysis, ciRS-7 is an independent prognostic biomarker with excellent sensitivity and specificity for cholangiocarcinoma [133]. Hsa_circ_0001649 is abnormally down-expressed in cholangiocarcinoma cells and tissues, and it is related to tumor size and differentiation grade of cholangiocarcinoma. Overexpression of hsa_circ_0001649 inhibits cell proliferation, migration, and invasion but induces apoptosis of KMBC and Huh-28 cells. Silencing hsa_circ_0001649 leads to the opposite effect. Therefore, hsa_circ_0001649 may be a potential diagnostic and therapeutic target for cholangiocarcinoma [134].
Cervical cancer
circRNA-000284 is significantly upregulated in cervical cancer cells. It promotes the proliferation and invasion of cervical cancer cells and that knockdown of circRNA-000284 causes G0/G1 cell cycle arrest, resulting in inhibition of cell proliferation and invasion. miR-506 is a miRNA related to circRNA-000284, and circRNA-000284 positively regulates the expression of Snail-2 which is a target gene of miR-506. However, co-expression of a miR-506 mimic or Snail-2 silencing vector eliminates the oncogenic effect of circRNA-000284. Thus, circRNA-000284 is expected to be a new therapeutic target for cervical cancer [135].
Prostate cancer
The role of circRNAs in prostate cancer is rarely explored. circ-SMARCA5 is significantly upregulated in prostate cancer tissues, and it promotes cell cycle process and inhibits apoptosis [136], acting as an oncogene.
Discussion
Recently, the clinical significance of circRNAs in a variety of tumors has been explored. circRNAs are generally superior to the corresponding linear RNAs in terms of stability. In addition, they represent the characteristics at different stages of tumor development [83, 84, 86, 91]. In addition, circRNAs can compensate for the defect of low organ specificity of traditional biomarkers. circ-CER may serve as a diagnostic marker for NSCLC, and its overexpression is significantly associated with local lymph node invasion, advanced tumor, and poor survival [20]. The combination of hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 is helpful for the diagnosis of breast cancer [30]. Hsa_circ_0067934, a potential prognostic marker for ESCC, is overexpressed in ESCC tissues and correlates with poor differentiation and more advanced TNM staging. The upregulation of circ-PVT1 and downregulation of circ-LARP4 in gastric cancer are independent prognostic factors [45, 53], and circ-PVT1 overexpression predicts better OS and DFS. The downregulation of circMTO1, circ-ITCH, and cSMARCA5 [87, 88, 90] or upregulation of circRNA_100338 in HCC [92] is associated with poor prognosis. Overexpression of ciRS-7 in HCC tissue is a risk factor for MVI [94], and the expression of hsa_circ_0005075 in HCC is positively correlated with tumor size, suggesting that it may be a potential biomarker of HCC. Overexpression of ciRS-7 in cholangiocarcinoma is significantly correlated with later TNM staging, lymph node infiltration, and postoperative recurrence, and it may be an independent negative prognostic biomarker with good sensitivity and specificity [133]. Hsa_circ_0001649 has been reported to have potential diagnostic and prognostic value in gastric cancer, CRC, HCC, and cholangiocarcinoma [57, 69, 85, 134], and it may be a sensitive indicator for distant metastasis in gastric cancer and HCC. High expression level of circ-BRAF is an independent marker for good prognosis in glioma patients [106]. Overexpression of circ-LDLRAD3 in pancreatic cancer is significantly correlated with venous and lymphatic infiltration as well as distant metastasis, and it is also a potential diagnostic marker for pancreatic cancer [116].
Another advantage of circRNAs is that they can be easily and reproducibly detected in human blood, saliva, and gastric juices, thus increasing its potential as a biomarker [48, 137,138,139]. In general, many circRNAs are expressed much higher in blood than the corresponding linear RNAs. Therefore, plasma circRNAs may provide additional information that cannot be revealed by routine RNA detection. For example, hsa_circ_002059, hsa_circ_0001017, and hsa_circ_0061276 can be stably detected in the plasma of gastric cancer patients [54]. These circRNAs are expected to become convenient diagnostic biomarkers for gastric cancer. Hsa_circ_0000190 is expressed at a low level both in gastric cancer plasma and tissues, and it is related to tumor size, lymph node and distant metastasis and TNM staging. The sensitivity and specificity of hsa_circ_0000190 as a diagnostic marker for gastric cancer are even better than that of CEA and CA19-9 [55]. The plasma level of hsa_circ_0000745 in gastric cancer patients is related to lymph node metastasis, and it has a good diagnostic value in combination with CEA [63]. The contents of circRNAs in exosomes are enriched more than twofold compared to their intracellular levels [140]. Bahn et al. found 422 circRNAs involved in intercellular signaling and inflammatory responses by bioinformatics analysis in human cell-free saliva [138]. Shao et al. demonstrated that hsa_circ_0014717 is stably detected in gastric juice, not affected by freeze-thaw for eight cycles or storage at 4 °C for 8 h [48]. The expression patterns and characteristics of circRNAs give them the potential to serve as a good biomarker in a variety of tumors.
The use of circRNAs as a therapeutic target or therapeutic vector will be a future trend. In therapeutic strategies targeting oncogenic circRNAs, exogenous siRNAs that are fully complementary to the back-splice junction can be used. Alternatively, it is possible to interfere with back-splicing by antisense oligonucleotides that are complementary to the back-splice signals in the precursor mRNA. It is necessary to avoid interfering with the expression of homologous linear mRNA. Another strategy is to induce tumor suppressor circRNA expression through gene therapy. In addition, some prefabricated circRNAs independent of nuclear splicing and output may also be used for the treatment of tumors. circRNAs have extremely high stability and the ability to adsorb miRNAs and proteins, suggesting that they can serve as delivery vehicles for certain treatments. circRNAs containing binding sites with oncogenic miRNAs and/or proteins can control the proliferation of tumor cells or induce apoptosis. Some strategies may help to achieve more precise treatment, for example, restricting the expression of circRNAs to certain types of cells by cell-specific promoters or designing different combinations of circRNAs and miRNAs and/or protein binding sites according to sponge maps to target specific carcinogenic factors. Because some circRNAs serve as a template for protein expression [8, 9, 141], cassettes containing tumor suppressor proteins can convert circRNAs into an effective treatment method for tumors.
At present, the exploration of the correlation between circRNAs and various diseases, including tumors, has become a new research field. Various methods have been developed for detecting circRNAs expression and biological functions. RNA-seq and microarrays are used to determine target circRNAs. The expression of circRNAs is mainly verified by real-time quantitative polymerase chain reaction (RT-qPCR), micro-drop digital PCR, Northern blotting, and FISH. For functional studies, overexpression and knockdown of genes are commonly used to regulate the expression of circRNAs. Bioinformatics predictions, luciferase reporter assays, RNA immunoprecipitation, and RNA pull-down experiments combined with mass spectrometry are utilized to reveal the interactions of circRNA-miRNA and circRNA-protein. M6A, IRES, and ORF in circRNAs can be predicted by bioinformatics analysis to investigate the protein-encoding ability of circRNAs. Ribosomal imprinting, ribosome IP, m6A IP, Western blotting, and mass spectrometry are commonly used in validation studies [142]. The development of high-throughput RNA-seq technology and bioinformatics methods enables accurate identification and quantification of circRNAs. Some online databases, such as circBase, CircInteractome, CircNet, and Circ2Traits, provide the basic information and potential regulatory networks of circRNAs. Continuous improvement of statistics and calculation methods will aid in a clearer and more comprehensive understanding of the expression patterns of circRNAs.
Conclusions
The present review introduced the biogenesis, characteristics, functions, and clinical value of circRNAs. circRNAs are closely related to a variety of physiological conditions and involved in certain diseases with a high degree of tissue and cell specificity. The biogenesis of circRNAs is a strictly controlled biological process, rather than a random splicing error. Although scientists have initially proposed a synthetic model for circRNAs, more research is needed to fully explore the mechanisms of circRNA generation, including the secondary structure of circRNAs, the initiation of novel circRNAs, the relationship among homologous RNA isomers, and the crosstalk between circRNAs and other molecules.
circRNAs have an extremely wide range of biological functions. As a miRNA sponge, circRNA makes the ceRNA network more complete and complicated. However, ceRNA does not represent all the functions of circRNAs. In the future, it is necessary to explore other mechanisms of circRNAs in tumors, such as regulating gene or protein activity.
The role of circRNAs in tumorigenesis and development has become the focus of oncology. circRNAs are considered as new diagnostic and prognostic biomarkers and potential therapeutic targets in the future. In the previous studies, the detection of circRNAs was mainly in tissues, but it is possible to detect the expressions of circRNAs in more accessible and less invasive samples (such as blood, urine, and saliva) or samples closely related to diseases (such as gastric juice, synovial effusion, and cerebrospinal fluid). It is necessary to develop circRNA as a clinical diagnostic biomarker based on the optimization of consistency and standardization of sample processing and detection. The combined detection of different circRNAs and traditional markers may have higher diagnostic efficiency. In addition, the potential of circRNAs in cancer therapy cannot be ignored. circRNA-targeted treatment may become a new mode of tumor therapy in the future.
Although increasingly more circRNAs have been discovered and investigated, the functions of thousands of circRNAs remain unclear. Research on circRNAs is still in its beginning stage, and only a small part of circRNA mechanism in tumorigenesis and progression has been clarified. With the efforts of scientists and application of new methods, more circRNAs will be discovered and applied in the diagnosis and treatment of related diseases, including tumors.
Abbreviations
- AR:
-
Androgen receptor
- BCC:
-
Basal cell carcinoma
- BCSCs:
-
Breast cancer stem cells
- ccRCC:
-
Clear cell renal cell carcinoma
- ceRNA:
-
Competing endogenous RNA
- circRNA:
-
Circular RNA
- CIRI:
-
circRNA identifier
- ciRNAs:
-
Circular intronic RNAs
- CRC:
-
Colorectal carcinoma
- CSCC:
-
Cutaneous squamous cell carcinoma
- DCIS:
-
Ductal carcinomas in situ
- DFS:
-
Disease-free survival
- ecircRNA:
-
Exonic circRNA
- EIciRNA:
-
Exon-intron circRNA
- ER:
-
Estrogen receptor
- ESCC:
-
Esophageal squamous cell carcinoma
- FISH:
-
Fluorescence in situ hybridization
- GBM:
-
Glioblastoma
- HCC:
-
Hepatocellular carcinoma
- HIAT1:
-
Hippocampus-rich gene transcription protein 1
- HSCC:
-
Hypopharyngeal squamous cell carcinoma
- ICFs:
-
Intronic circRNA fragments
- IDC:
-
Invasive ductal carcinoma
- IRES:
-
Internal ribosome entry site
- LSCC:
-
Laryngeal squamous cell cancer
- miRNA:
-
MicroRNA
- MPP:
-
Matrix metalloproteinase
- MREs:
-
miRNA response elements
- MVI:
-
Microvascular infiltration
- ncRNAs:
-
Non-coding RNAs
- NSCLC:
-
Non-small cell lung cancer
- ORF:
-
Open reading frame
- OS:
-
Overall survival
- OSCC:
-
Oral squamous cell carcinoma
- PDAC:
-
Pancreatic ductal adenocarcinoma
- pre-mRNA:
-
Precursor mRNA
- PTC:
-
Papillary thyroid cancer
- RBPs:
-
RNA-binding proteins
- RNA-seq:
-
RNA sequencing
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- SA:
-
Splice acceptor
- SD:
-
Splice donor
- TN:
-
Triple negative
- VEGF:
-
Vascular endothelial growth factor
References
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8.
Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40.
Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6:1167–76.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. e29
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. e27
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–9.
Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–12.
Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62.
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233.
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44:2138–46.
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490.
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–6.
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572.
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170–82.
Wang X, Zhu X, Zhang H, Wei S, Chen Y, Chen Y, et al. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun. 2018;496:1069–75.
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144:667–74.
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878.
Luo YH, Zhu XZ, Huang KW, Zhang Q, Fan YX, Yan PW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–8.
Pinder SE. Ductal carcinoma in situ (DCIS): pathological features, differential diagnosis, prognostic factors and specimen evaluation. Mod Pathol. 2010;23(Suppl 2):S8–13.
Galasso M, Costantino G, Pasquali L, Minotti L, Baldassari F, Corra F, et al. Profiling of the predicted circular RNAs in ductal in situ and invasive breast cancer: a pilot study. Int J Genomics. 2016;2016:4503840.
Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–79.
Lu L, Sun J, Shi P, Kong W, Xu K, He B, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8:44096–107.
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2017. https://doi.org/10.1016/j.cca.2017.10.011
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–70.
Lu WY. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–90.
Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–31.
Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704–18.
Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–54.
Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–76.
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–8.
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145.
Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–88.
Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1alpha-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37:4337–43.
Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–13.
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576.
Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225.
Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19.
Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469–76.
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y, Sun B, et al. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7:9060.
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–80.
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804–14.
Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16:2301–11.
Li P, Chen H, Chen S, Mo X, Li T, Xiao B, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626–33.
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9:1585–94.
Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151.
Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6.
Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–71.
Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125.
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698.
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11–20.
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299–306.
Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32(4):e22333. https://doi.org/10.1002/jcla.22333
Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3). https://doi.org/10.1002/jcla.22281
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85–96.
Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–8.
Zhang Y, Li J, Yu J, Liu H, Shen Z, Ye G, et al. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8:22936–43.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Zhang P, Zuo Z, Shang W, Wu A, Bi R, Wu J, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39:1010428317694546.
Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225.
Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–5.
Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497:122–6.
Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–93.
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–28.
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–91.
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283.
Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–8.
Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–44.
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417.
Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:118–26.
Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674–83.
Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–64.
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–50.
Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982.
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95:e3811.
Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–16.
Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S, et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018;32(2). https://doi.org/10.1002/jcla.22239
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–9.
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–37.
Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64.
Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169–77.
Zhang K, Che S, Su Z, Zheng S, Zhang H, Yang S, et al. CD90 promotes cell migration, viability and sphereforming ability of hepatocellular carcinoma cells. Int J Mol Med. 2018;41:946–54.
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–27.
Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878–88.
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428.
Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347.
Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27.
Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. 2017;16:3891–902.
Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497:626–32.
Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87.
Barbagallo D, Condorelli A, Ragusa M, Salito L, Sammito M, Banelli B, et al. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–59.
Li G, Yang H, Han K, Zhu D, Lun P, Zhao Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem Biophys Res Commun. 2018;498:254–61.
He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, et al. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2018;10:331–48.
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–55.
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10:52.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3). https://doi.org/10.1093/jnci/djx166
Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F, et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19(2). https://doi.org/10.3390/ijms19020480
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–14.
Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang H, et al. Differential expression of circular RNAs in glioblastoma multiforme and its correlation with prognosis. Transl Oncol. 2017;10:271–9.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919.
Cai D, Liu Z, Kong G. Molecular and bioinformatics analyses identify 7 circular RNAs involved in regulation of oncogenic transformation and cell proliferation in human bladder cancer. Med Sci Monit. 2018;24:1654–61.
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–59.
Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–200.
Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17.
Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19.
Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ, Xiao XY. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2017;37:886–90.
Huang WJ, Wang Y, Liu S, Yang J, Guo SX, Wang L, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70–80.
Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:45–8354.
Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783–94.
Li H, Hao X, Wang H, Liu Z, He Y, Pu M, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40:1334–44.
Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. 2017;8:25571–81.
Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–97.
Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369–75.
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495:189–96.
Liu W, Zhang J, Zou C, Xie X, Wang Y, Wang B, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43:969–85.
Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–81.
Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12.
Peng N, Shi L, Zhang Q, Hu Y, Wang N, Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS One. 2017;12:e0170287.
Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–32.
Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83:210–8.
Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–61.
Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39:951–66.
Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932–9.
Cao S, Wei D, Li X, Zhou J, Li W, Qian Y, et al. Novel circular RNA expression profiles reflect progression of patients with hypopharyngeal squamous cell carcinoma. Oncotarget. 2017;8:45367–79.
Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui YF, et al. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2018;22:365–71.
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–61.
Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604.
Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X, et al. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–23.
Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214.
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–30.
Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11:e0148407.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N (6)-methyladenosine. Cell Res. 2017;27:626–41.
Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152.
Funding
This work was financially supported by the National Key Technology R & D Program (No. 2015BAI12B12), National Natural Science Foundation of China (No. 31700792), and Changzhou Science and Technology Project (Applied Based Research, No. CJ20159021, CJ20179047).
Author information
Authors and Affiliations
Contributions
YTG, JTJ, and CPW conceived and designed the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Geng, Y., Jiang, J. & Wu, C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol 11, 98 (2018). https://doi.org/10.1186/s13045-018-0643-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13045-018-0643-z