Abstract
This paper addresses the visualization of complex information using multidimensional scaling (MDS). MDS is a technique adopted for processing data with multiple features scattered in high-dimensional spaces. For illustrating the proposed techniques, the case of viral diseases is considered. The study evaluates the characteristics of 21 viruses in the perspective of clinical information. Several new schemes are proposed for improving the visualization of the MDS charts. The results follow standard clinical practice, proving that the method represents a valuable tool to study a large number of viruses.
Similar content being viewed by others
1 Introduction
Presently, reliable and assertive data about many real-world phenomena are available for computer processing. One example consists of clinical information about viral diseases. Viruses infections are an important cause of mortality and morbidity. More than 2000 viruses were identified and many can infect humans, or animals [1]. In general, viral diseases have very diverse characteristics and complexity, and computational methods for data mining and feature extraction are relevant strategies to adopt. As usually occurs with real-world data, information is scattered, and exhibits multiple characteristics with distinct levels of relevance. Therefore, it is important to explore reliable algorithms for highlighting the main details, and to take advantage of modern computational resources to visualize the relations embedded within the data.
Herein, we adopt the multidimensional scaling (MDS) technique to compare the relationships among several viruses responsible for human diseases. New schemes for improving the visualization of the MDS charts are proposed. In what concerns selection of the “objects” under study, most are based on their impact on people and visibility in communication media (e.g., subtype H5N1 of Influenza A virus, Ebola, Chikungunya and Zika), others due to historical reasons (e.g., Rabies, Poliomyelitis, and Smallpox), and some because of their incidence and prevalence in humans (e.g., Influenza, Rhinovirus, and Norovirus). The viruses are compared by means of their characteristics and the symptoms of the diseases that they may cause in humans.
The MDS can lead to a new perspective in the study of human pathologies. MDS is a statistical technique for analyzing similarities in information that generates geometric representations for complex objects [2]. MDS appeared in the context of behavioral sciences, for understanding judgments of individuals about features in a set of objects [3, 4]. Presently, the MDS is used in real-world data, such as biological taxonomy [5], finance [6], marketing [7], sociology [8], physics [9], geophysics [10–12], communication networks [13], biology and biomedicine [14], among others [15].
The paper is organized as follows. Section 2 introduces the MDS technique. Section 3 studies and compares data characterizing the clinical effects of 21 viruses. Finally, Sect. 4 draws the conclusions.
2 Multidimensional Scaling
We consider s objects defined in a m-dim space, \(\mathcal {M}\), and a proximity measure, \(\delta _{ij}\), between objects i and j. The first step consists of calculating \(\mathbf {C}=[\delta _{ij}]\) (\(\dim(\mathbf {C})=s \times s\)), of item-to-item dissimilarities. The MDS produces a configuration \(\mathbf {X}\) (\(\dim(\mathbf {X})=s \times q\)), where the dimension \(q < m\) is chosen by the user. Thus, \(\mathbf {X}\) attempts to replicate in a low-dimensional space, \(\mathcal {Q}\), the proximities between the s elements in \(\mathcal {M}\). In general, the MDS unveils the embedded data patterns, being different from other techniques [16, 17], not only because it requires no a priori assumptions for each dimension, but also due to its good convergence [18, 19].
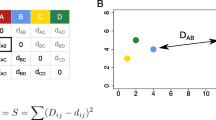
To arrive to configuration \(\mathbf {X}\), MDS evaluates different alternative values to minimize some fitness function, such as [20] the raw stress, \(\sigma ^2\):
where \(z_{ij}>0\) is a weight and \(d_{ij}\) measures the dissimilarities among the items i and j in the embedding space \(\mathcal {Q}\). Therefore, a distance measure is often adopted for implementing \(d_{ij}\) [21].
Besides (1), there are several stress measures [22], namely, the normalized raw stress, the Kruskal’s stress-1 and stress-2, and the S stress.
To assess the quality of the MDS solutions, it is used the Shepard diagram that represents the pairs \((d_{ij}, \, \delta _{ij})\). The Shepard diagram displays the outliers and residuals resulting from the MDS. A narrow scatter following the 45\(^{\circ }\) line corresponds to a good fit between \(d_{ij}\) and \(\delta _{ij}\).
Another test to the MDS quality is the stress plot that represents \(\sigma ^2\) versus q. The curve \(\sigma ^2(q)\) is monotonic decreasing and the user chooses q as a compromise between reducing \(\sigma ^2\) and having small values of q.
The MDS interpretation focuses on the emerging clusters and considers the distances between points in the produced chart. Therefore, the user can rotate, shift, or zoom the chart, while the distances remain invariant. Usually, \(q=2\), or \(q=3\), is adopted, since they allow a direct graphical representation.
3 Data Analysis and Visualization
We analyze data for \(s = 21\) viruses responsible for infectious diseases. These are \(\{\)Bird Flu, Chicken Pox, Chikungunya, Dengue Fever, Ebola, Hepatitis B, HIV, Marburg disease, Measles, MERS, Mumps, Norovirus, Polio, Rabies, Rhinovirus, Rotavirus, Rubella, SARS, Seasonal Flu, Smallpox, and Zika virus infection\(\}\), with acronyms \(\{\)BFlu, CPox, Chi, Den, Ebo, HepB, HIV, Mar, Mea, MERS, Mum, Nor, Pol, Rab, Rhi, Rot, Rub, SARS, SFlu, Sma, and ZIKV\(\}\).
For the ith virus, \(i = 1,\ldots , \, s\), we associate \(m=5\) quantitative attributes, namely, (i) the fatality rate, (ii) the average basic reproductive number, (iii) the average serial interval, (iv) the incubation period, and (v) the virus survival time outside a host. Table 1 lists the data, where the numerical values correspond to the matrix \(\mathbf {\tilde{U}} = [\tilde{u}_{ik}]\), \(i = 1,\ldots , \, s\), \(k = 1,\ldots , \, m\).
For constructing Table 1, data were obtained from several distinct sources: Influenza A virus, subtype H5N1 (or “Bird Flu”) [23–25]; Chicken Pox (varicella-zoster infection) [26–28]; Chikungunya [29–31]; Dengue Fever [32, 33]; Ebola [34–36]; Hepatitis B [37–39]; Human Immunodeficiency Virus (HIV) [40–42]; Marburg hemorrhagic fever [36, 43]; Measles [44–47]; Middle East Respiratory Syndrome (MERS) [48–50]; Mumps [51, 52]; Norovirus [53, 54]; Poliomyelitis [55–57]; Rabies [58–60]; Rhinovirus [61–63]; Rotavirus [64–67]; Rubella [46, 68]; Severe Acute Respiratory Syndrome (SARS) [49, 69]; Seasonal flu [25, 70, 71]; Smallpox [72, 73]; Zika virus disease [74, 75].
3.1 MDS Analysis using the Arc-cosine Distance
Previous to applying MDS, the data are “normalized” to avoid saturation effects of the numerical values. Therefore, the elements of each column of matrix \(\mathbf {\tilde{U}}\) are converted to the interval [0, 1], producing the data matrix \({\mathbf {U}}\). The vectors of features for item-to-item comparison correspond to the lines of \({\mathbf {U}}\) and will be denoted by \(\mathbf {u}_i\).
Various distance measures were tested for constructing the matrix \(\mathbf {C}\). Here, we present results for the arc-cosine distance, \(\delta _{ij}\), since it leads to charts that are easy to interpret. Other distances are possible and have also been used in distinct applications [6, 12], but several numerical tests confirmed that the arc-cosine leads to reliable results. Therefore, for items i and j\((i, \, j = 1,\ldots , \, s)\), we have
where \(\alpha _k>0\), \(k=1,\ldots , \, m\), represent weights specified by the user. Given expression (2), the matrix \(\mathbf {C} = [\delta _{ij}]\) can be computed for feeding the MDS.
Figure 1 represents the 2D and 3D charts (\(q=2\) and \(q=3\)) resulting from the MDS using the weights \(\alpha _k = \{5,\,2,\,1,\,1,\,1\}\), where the points represent viruses. The relationships between the items are inferred from the coordinates of the points. Objects that are similar (dissimilar) appear closer (farther) to each other in space.
With alternative distances, we capture different characteristics of the phenomena that yield distinct plots, but in general lead to identical conclusions. A “good” distance is the one that produces a MDS reflecting the real-world phenomenon in a direct and clear visualization.
Figure 2 depicts the Shepard diagram for \(q=1,\ldots , \, 5\) and the stress plot. The Shepard diagram depicts a good scatter of points around the 45\(^{\circ }\) line for \(q \ge 3\), demonstrating a good fit between the distances and the dissimilarities. The curvature of the stress plot is often adopted for deciding the value of q. In this case, we observe that \(q =2\) is insufficient, \(q=3\) seems to be a good choice, and \(q> 3\) leads to limited improvements. However, if we adopt \(q=3\) the question remains of visualizing efficiently the MDS information, since for 3D representations, we often have to zoom, shift, and rotate the MDS graph to perceive assertively the real location of the objects in space. This question will further discussed in Sect. 3.3.2.
Before continuing, two numerical aspects need to be clarified: the weights \(\alpha _k\) used and the missing data in Table 1. The weights were tuned for highlighting the importance of the features recognized as being more harmful from the medical point of view: first, the fatality rate and, second, the average basic reproductive number. However, the question remains on how to choose \(\alpha _k\). In this perspective, we performed several experiments varying the weights. Figure 3 depicts the results obtained with four distinct sets of values, namely, \(\alpha _k = \{1,\,1,\,1,\,1,\,1\},\)\(\alpha _k = \{2.5,\,1.5,\,1,\,1,\,1\}\), \(\alpha _k = \{5,\,2,\,1,\,1,\,1\}\) and \(\alpha _k = \{7.5,\,2.5,\,1,\,1,\,1\}\). For each set \(\alpha _k\), we generate one MDS chart, and afterwards, the charts are combined using Procrustes analysis [76]. Procrustes involves the operations of translation, reflection, orthogonal rotation, and scaling, to best conform the points in a given matrix under modification in relation with the points of a reference matrix.
In our case, we (i) choose the first chart for initial reference, (ii) use Procrustes to superimpose the next MDS chart into the current reference, (iii) make the current set of superimposed charts the new reference, and (iv) continue to step (ii) until all charts have been conformed. The results obtained reveal identical patterns, meaning that the method is robust to distinct values of \(\alpha _k\).
In Fig. 1, the unknown data, denoted by ‘-’ in Table 1, are considered zero. Therefore, these values do not contribute to the distance used for comparing items. Moreover, as the missing data occur only in four values of the less weighted features, their influence is not as significant as for the rest of the information. In addition, as will be shown in Sect. 3.2, the results reveal small sensitivity to possible noise in the data, which includes the uncertainty in the unknown values that were set to zero. Nonetheless, a different criterion for dealing with that problem could be adopted. Experiments with the missing data replaced not only by zero, but also by the minimum, average, and maximum values in the third and fifth columns of Table 1 led to results qualitatively similar, as depicted in Fig. 4, revealing the effectiveness of the criterion adopted.
3.2 Sensitivity Analysis
The 21 viruses were compared in the perspective of quantitative features. However, the data diverge slightly, depending on factors such as the time of the study or the operational conditions, namely, environmental conditions, geographic region, development level, or medical assistance. Therefore, we analyze here the sensitivity results with respect to the input data.
We start by adding random noise to the quantitative features, \(k=1,\ldots , \, 5\), with amplitude \(\pm 10\%\) of the values in Table 1. Moreover, any negative values are avoided by limiting numbers to zero. A sample of 50 experiments, each yielding one MDS chart, is performed and the charts are combined using the Procrustes scheme.
Figure 5 illustrates the MDS chart for \(q=2\) produced by the Procrustes algorithm. We verify that the method has low sensitivity to variations in the quantitative features, since the location of the points reveals minor variations.
3.3 Data Clustering and Visualization
The MDS interpretation focuses on the distances between points in the produced charts. For identifying clusters, we can adopt some kind of ad hoc strategy based on the direct visualization of the MDS plots, or we can implement an algorithm for obtaining an automatic clustering. In addition, the configuration, \(\mathbf {X}\), produced by the MDS tries to replicate, in the low-dimensional space, \(\mathcal {Q}\), the original proximities between pairwise elements. For \(q=2\), this leads to a direct visualization, but neglects the information described in the higher dimensional components of \(\mathbf {X}\). In this line of thought, in the next subsections, we introduce the non-hierarchical clustering algorithm K-means for automatic cluster identification and we propose a technique for an improved visualization of MDS information in the 2D space by embedding information of the extra dimensions.
3.3.1 The K-Means Clustering
Clustering is a technique that groups objects similar to each other in some sense. The K-means is a non-hierarchical clustering algorithm [77] that starts with a set of s objects, where each one is represented by a point in a q-dim space, and a certain number of clusters, K, specified in advance. The K-means groups the s objects into \(K \le s\) clusters, to minimize the sum of distances between the points and the centers of their clusters. The K-means produces a solution where objects in a cluster are close to each other and far from objects in other clusters.
An important issue in K-means is to specify K, since the notion of “good clustering” is subjective. Nevertheless, we can adopt different measures for assessing the quality of the solution, such as the Calinski-Harabasz, Davies-Bouldin, and silhouette [78].
Here, we consider the silhouette, S, to assess if an object lies “adequately” within its cluster. The silhouette varies in the interval \(S \in [\!-1,1]\), so that values close to \(\{-1,0,1\}\) correspond to \(\{incorrect, neutral, correct\}\) object assignments.
Knowing the coordinates of the \(s=21\) objects produced by the MDS in the \(q=3\) dim space, we assess the quality of the clusters in the interval \(K \in [2,6]\). Figure 6 depicts the corresponding silhouettes and the mean value for each cluster (blue marks). The optimum value is obtained \(K=4,\) corresponding to the maximum silhouette mean value \(S_M = 0.77\).
For \(K=4\), the clusters are \(\mathcal {A} = \{\)CPox, Mea, Mum, Nor, Rhi, Rot, Rub, SFlu\(\}\), \(\mathcal {B} = \{\)HepB, HIV, Rab\(\}\), \(\mathcal {C} = \{\)BFlu, Ebo, Mar, MERS, Pol, SARS, Sma\(\}\) and \(\mathcal {D} = \{\)Chi, Den, ZIKV\(\}\). These clusters are further discussed in the next subsection.
3.3.2 Improved Visualization in 2D Space
The geometrical shape of the chart produced by MDS varies significantly with the distance measure adopted for quantifying the distances between items. However, this characteristic does not precludes that we use the MDS chart taking full advantage of all its properties. Consequently, we may interpret the collection of points as “samples” of an abstract locus corresponding to the projection of the m initial dimensions into a lower dimensional (abstract) space.
We adopt a scheme that allows for a direct visualization of the MDS, while including information up to \(q=3.\) Therefore, we approximate the dimension \(x_3\) of \(\mathbf {X}\) with a contour generated by means of a linear radial basis function interpolation [79]. Moreover, we improve the identification of patterns by superimposing a tree in the MDS chart. The nodes of the tree are the \(s=21\) points representing items (viruses). In a first phase, we connect the group of points that are closer in the MDS chart producing the sets, \(\mathcal {P}\), of interconnected points (nodes). In a second phase, the sets, \(\mathcal {P}\), are compared through the distances between their constitutive nodes. The distance can be calculated taking into account any number \(p<m\) of \(\mathbf {X}\) components. A connection is established in the q-dim chart, only between the two closest nodes (i.e., \(\mathcal {P}_i\) and \(\mathcal {P}_j\)). This calculation generates a second level of interconnection, and the scheme is repeated iteratively until there is a continuous route between all points. Therefore, the interpretation of the MDS chart is based not only in the relative position of the points, but also in the structure interconnecting them.
Figure 7 depicts a projection of the MDS chart for \(q=2,\) the contour that approximates the dimension \(x_3\), and the superimposed interconnections generated by calculating the distances between objects with \(p=5\). We observe easily the four clusters identified in the previous subsection. Moreover, we verify that the proposed methodology leads to a clear visualization and produces a richer chart of the objects.
In synthesis, besides the observation based on the relative distances in 2D space, we now verify that the ZIKV has a relevant position along the \(x_3\) dimension, somehow strengthening the characteristics revealed by the Chikungunya and Dengue.
3.3.3 Discussion of the Results
The clusters \(\{\mathcal {A}, \, \mathcal {B}, \, \mathcal {C}, \, \mathcal {D}\}\) do not follow an epidemiological line of thought, but may be of medical value, since they reflect characteristics measured by health care practice. In cluster, \(\mathcal {A}\) are included viruses of Risk Group 2 that in general do not cause serious illness nor life threatening.
In cluster \(\mathcal {B}\), we find the Lentivirus that is responsible for HIV and acquired immunodeficiency syndrome (AIDS), a Risk Group 3 agent. We find also the Hepatitis B and the Rabies virus, a Lyssavirus genus and Rhabdoviridae family virus, of Risk Group 2.
In cluster \(\mathcal {C}\), we can consider two subclusters. The first subcluster includes the Ebola and Marburg viruses that belong to the Risk Group 4. In addition, in this subcluster, the agents responsible for MERS and Bird flu are classified as Risk Group 3. The second subcluster includes viruses of different Risk Groups, namely, the Polio virus and the SARS–associated coronavirus, belonging to Risk Groups 2 and 3, respectively. Smallpox is also present [80].
Cluster \(\mathcal {D}\) includes Chikungunya, considered a Risk Group 3 pathogen. Also included in \(\mathcal {D}\) are the Dengue fever virus, a Risk Group 2 arbovirus pathogenan, and ZIKV, recognized as being similar to Chikungunya and Dengue viruses.
In conclusion, we verified that the MDS provides a powerful computational visualization technique of viruses data and the obtained charts may be of medical interest in the scope of present and future viral outbreaks.
4 Conclusions
This paper discussed the computational analysis of real-world data describing viruses main quantitative characteristics. By encompassing complex scattered data, researchers have to choose between comparing all aspects and detecting the main properties. This problem represents a challenge since some information (or its absence) may lead to incomplete or eventually to incorrect conclusions. Therefore, complex information calls for computational and visualization tools capable of revealing the most relevant issues. The MDS technique was adopted, leading to substantive results that follow present-day scientific knowledge.
References
Murray PR, Rosenthal KS, Pfaller MA (2013) Medical microbiology. Elsevier Sounders, Philadelphia
Cox TF, Cox MA (2000) Multidimensional scaling. CRC Press, Boca Raton
Lawless HT, Sheng N, Knoops SS (1995) Multidimensional scaling of sorting data applied to cheese perception. Food Qual Preference 6(2):91–98
Torgerson WS (1958) Theory and methods of scaling. Wiley, New York
Vukea PR, Willows-Munro S, Horner RF, Coetzer TH (2014) Phylogenetic analysis of the polyprotein coding region of an infectious south african bursal disease virus (IBDV) strain. Infect Genet Evolut 21:279–286
Machado J, Lopes AM, Galhano AM (2015) Multidimensional scaling visualization using parametric similarity indices. Entropy 17(4):1775–1794
Cil I (2012) Consumption universes based supermarket layout through association rule mining and multidimensional scaling. Expert Syst Appl 39(10):8611–8625
Corten R (2011) Visualization of social networks in Stata using multidimensional scaling. Stata J 11(1):52–63
Hollemeyer K, Altmeyer W, Heinzle E, Pitra C (2012) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry combined with multidimensional scaling, binary hierarchical cluster tree and selected diagnostic masses improves species identification of neolithic keratin sequences from furs of the tyrolean iceman oetzi. Rapid Commun Mass Spectrom 26(16):1735–1745
Lopes AM, Machado JT (2014) Analysis of temperature time-series: embedding dynamics into the MDS method. Commun Nonlinear Sci Numer Simul 19(4):851–871
Lopes AM, Machado JT, Pinto CM, Galhano AM (2013) Fractional dynamics and MDS visualization of earthquake phenomena. Comput Math Appl 66(5):647–658
Machado JAT, Lopes AM (2013) Analysis and visualization of seismic data using mutual information. Entropy 15(9):3892–3909
Ji X, Zha H (2004) Sensor positioning in wireless ad-hoc sensor networks using multidimensional scaling. In: INFOCOM 2004. Twenty-third Annual Joint Conference of the IEEE Computer and Communications Societies, IEEE, vol. 4, pp 2652–2661
Tzagarakis C, Jerde TA, Lewis SM, Uğurbil K, Georgopoulos AP (2009) Cerebral cortical mechanisms of copying geometrical shapes: a multidimensional scaling analysis of fMRI patterns of activation. Exp Brain Res 194(3):369–380
Machado JT, Lopes AM (2014) The persistence of memory. Nonlinear Dyna 79(1):63–82
Adeshina A, Hashim R (2016) ConnectViz: accelerated approach for brain structural connectivity using Delaunay triangulation. Interdiscip Sci Comput Life Sci 8(1):53–64
Kaur H, Ahmad M, Scaria V (2016) Computational analysis and in silico predictive modeling for inhibitors of PhoP regulon in S. typhi on high-throughput screening bioassay dataset. Interdisciplinary Sciences: Computational. Life Sci 8(1):95–101
Borg I, Groenen PJ (2005) Modern multidimensional scaling: theory and applications. Springer, New York
Bronstein MM, Bronstein AM, Kimmel R, Yavneh I (2006) Multigrid multidimensional scaling. Numer Linear Algebra Appl 13(2–3):149–171
Kruskal JB (1964) Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29(1):1–27
Cha SH (2007) Comprehensive survey on distance/similarity measures between probability density functions. Int J Math Models Methods Appl Sci 4:300–307
Young FW (1987) Multidimensional scaling: history, theory, and applications. Lawrence Erlbaum Associates, Inc., Hillsdale, NJ
Osterhaus A, Jong JD, Rimmelzwaan G, Claas E (2002) H5N1 influenza in Hong Kong: virus characterizations. Vaccine 20:S82–S83
Pandit PS, Bunn DA, Pande SA, Aly SS (2013) Modeling highly pathogenic avian influenza transmission in wild birds and poultry in West Bengal, India. Scientific Reports 3
Treanor J (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Influenza (including avian influenza and swine influenza). Saunders, pp 1815–1839
Helmuth IG, Poulsen A, Suppli CH, Mølbak K (2015) Varicella in Europe—a review of the epidemiology and experience with vaccination. Vaccine 33(21):2406–2413
Ross AH, Lenchner E, Reitman G (1962) Modification of chicken pox in family contacts by administration of gamma globulin. N Eng J Med 267(8):369–376
Whitley R (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Chickenpox and herpes zoster (varicella-zoster virus), Saunders, pp 1731–1737
Charrel RN, de Lamballerie X, Raoult D (2007) Chikungunya outbreaks—the globalization of vectorborne diseases. N Eng J Med 356(8):769
Markoff L (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Alphavirus. Saunders, pp 1865–1874
Weaver SC, Forrester NL (2015) Chikungunya: evolutionary history and recent epidemic spread. Antivr Res 120:32–39
Endy TP (2014) Dengue human infection model performance parameters. J Infect Dis 209(suppl 2):S56–S60
Khan A, Hassan M, Imran M (2014) Estimating the basic reproduction number for single-strain dengue fever epidemics. Infect Dis Poverty 3(1):12
Althaus CL (2014) Estimating the reproduction number of Ebola virus (EBOV) during the 2014 outbreak in West Africa. PLOS Curr Outbreaks. doi:10.1371/currents.outbreaks.91afb5e0f279e7f29e7056095255b288
Althaus CL (2015) Ebola: the real lessons from HIV scale-up. Lancet Infect Dis 15(5):507–508
Geisbert T (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Marburg and Ebola hemorrhagic fevers (Filovirus), Saunders, pp 1995–1999
Hung HF, Wang YC, Yen AMF, Chen HH (2014) Stochastic model for hepatitis B virus infection through maternal (vertical) and environmental (horizontal) transmission with applications to basic reproductive number estimation and economic appraisal of preventive strategies. Stoch Environ Res Risk Assess 28(3):611–625
Mann J, Roberts M (2011) Modelling the epidemiology of hepatitis B in New Zealand. J Theor Biol 269(1):266–272
Thio C, Hawkins C (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Hepatitis B virus and hepatitis delta virus. Saunders, pp 1815–1839
Fettig J, Swaminathan M, Murrill C, Kaplan J (2014) Global epidemiology of HIV. Infect Dis Clin N Am 28(3):323–337
de Goede A, Vulto A, Osterhaus A, Gruters R (2014) Understanding HIV infection for the design of a therapeutic vaccine. part I: epidemiology and pathogenesis of HIV infection. In: Annales Pharmaceutiques Francaises. Elsevier
Reitz M, Gallo R (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Human immunodeficiency virus. Saunders, pp 2054–2065
Rougeron V, Feldmann H, Grard G, Becker S, Leroy E (2015) Ebola and Marburg haemorrhagic fever. J Clin Virol 64:111–119
Durrheim DN, Crowcroft NS, Strebel PM (2014) Measles-the epidemiology of elimination. Vaccine 32(51):6880–6883
Gershon AA (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Meales virus (rubeola). Saunders, pp 1967–1973
Gershon AA (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Rubella virus (German measles). Saunders, pp 1875–1880
Schlenker TL, Bain C, Baughman AL, Hadler SC (1992) Measles herd immunity: the association of attack rates with immunization rates in preschool children. JAMA 267(6):823–826
van Doremalen N, Bushmaker T, Karesh WB, Munster VJ (2014) Stability of Middle East respiratory syndrome coronavirus in milk. Emerg Infect Dis 20(7):1263–1264
McIntosh K, Perlman S (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Coronavirus, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), Saunders, pp 1731–1737
Memish Z, Assiri A, Alhakeem R, Yezli S, Almasri M, Zumla A, Al-Tawfiq J, Drosten C, Albarrak A, Petersen E (2014) Middle east respiratory syndrome corona virus, MERS-CoV. Conclusions from the 2nd scientific advisory board meeting of the WHO collaborating center for mass gathering medicine, Riyadh. Int J Infect Dis 24:51–53
Litman N, Baum S (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Mumps virus. Saunders, pp 1942–1947
Rubin S, Eckhaus M, Rennick LJ, Bamford CG, Duprex WP (2015) Molecular biology, pathogenesis and pathology of mumps virus. J Pathol 235(2):242–252
Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA (2014) Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14(8):725–730
Dolin R, Treanor JJ. Mandell (2014) Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Noroviruses and sapoviruses (caliciviruses). Saunders, pp 2122–2127
Pirtle E, Beran G (1991) Virus survival in the environment. Revue Scientifique et Technique 10:733–748
Romero J, Modlin J (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Poliovirus. Saunders, pp 2073–2079
Weinstein L, Shelokov A, Seltser R, Winchell GD (1952) A comparison of the clinical features of poliomyelitis in adults and in children. N EngJ Med 246(8):297–302
Hampson K, Dushoff J, Cleaveland S, Haydon DT, Kaare M, Packer C, Dobson A (2009) Transmission dynamics and prospects for the elimination of canine rabies. PLoS Biol 7(3): e1000,053
Matouch O, Jaros J, Pohl P (1987) Survival of rabies virus under external conditions. Veterinarni Medicina 32(11):669–674
Singh K, Rupprecht C, Bleck T (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Rabies (Rhabdoviruses). Saunders, pp 1984–1994
Dick EC, Jennings LC, Mink KA, Wartgow CD, Inborn SL (1987) Aerosol transmission of rhinovirus colds. J Infect Dis 156(3):442–448
Hendley JO, Gwaltney JM (1988) Mechanisms of transmission of rhinovirus infections. Epidemiol Rev 10(1):242–258
Turner R (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Rhinovirus, Saunders, pp 2113–2121
Dormitzer P (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Rotaviruses. Saunders, pp 1854–1864
Patel MM, Pitzer V, Alonso WJ, Vera D, Lopman B, Tate J, Viboud C, Parashar UD (2013) Global seasonality of rotavirus disease. Pediatr Infecti Dis J 32(4):e134
Pitzer VE, Atkins KE, de Blasio BF, Effelterre TV, Atchison CJ, Harris JP, Shim E, Galvani AP, Edmunds WJ, Viboud C, Patel MM, Grenfell BT, Parashar UD, Lopman BA (2012) Direct and indirect effects of rotavirus vaccination: comparing predictions from transmission dynamic models. PLoS One 7(8), e42,320
Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD (2012) 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 12(2):136–141
Reef SE, Redd SB, Abernathy E, Zimmerman L, Icenogle JP (2006) The epidemiological profile of rubella and congenital rubella syndrome in the United States, 1998–2004: the evidence for absence of endemic transmission. Clin Infect Dis 43(Supplement 3):S126–S132
Poon L, Guan Y, Nicholls J, Yuen K, Peiris J (2004) The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect Dis 4(11):663–671
Shi W, Lei F, Zhu C, Sievers F, Higgins DG (2010) A complete analysis of HA and NA genes of influenza A viruses. PLoS One 5(12), e14,454
Weinstein RA, Bridges CB, Kuehnert MJ, Hall CB (2003) Transmission of influenza: implications for control in health care settings. Clin Infect Dis 37(8):1094–1101
Milton DK (2012) What was the primary mode of smallpox transmission? implications for biodefense. Front Cell Infect Microbiol 2. doi:10.3389/fcimb.2012.00150
Theves C, Biagini P, Crubezy E (2014) The rediscovery of smallpox. Clin Microbiol Infect 20(3):210–218
Schaffner F, Medlock J, Bortel WV (2013) Public health significance of invasive mosquitoes in Europe. Clin Microbiol Infect 19(8):685–692
Service M, Ashford R (2001) Encyclopedia of Arthropod-transmitted infections of man and domesticated animals. CABI
Gower JC, Dijksterhuis GB (2004) Procrustes problems. Oxford University Press, Oxford
Jain AK (2010) Data clustering: 50 years beyond \(k\)-means. Pattern Recogn Lett 31(8):651–666
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Carr JC, Fright WR, Beatson RK (1997) Surface interpolation with radial basis functions for medical imaging. IEEE Trans Med Imaging 16(1):96–107
Borio L, Henderson D, Hynes N (2014) Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8th edn., chap. Bioterrorism: an overview. Saunders, pp 178–190
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Lopes, A.M., Machado, J.A.T. & Galhano, A.M. Computational Comparison and Visualization of Viruses in the Perspective of Clinical Information. Interdiscip Sci Comput Life Sci 11, 86–94 (2019). https://doi.org/10.1007/s12539-017-0229-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-017-0229-4