Abstract
Mapping wetland ecosystems at the species level provides critical information for understanding the nutrient cycle, carbon sequestration, retention and purification of water, waste treatment and pollution control. However, wetland ecosystems are threatened by climate variability and change and anthropogenic activities; thus, their assessment and monitoring have become critical to inform proper management interventions. Contemporary studies show that satellite-based Earth observation (EO) has significant potential for achieving this task. While many multispectral EO data are freely and readily available, its broad spectral bands limit its utility in differentiating subtle differences among similar plant species. In contrast, hyperspectral data has a high spectral resolution, which is superior in discerning minute differences in similar plant species. However, this data is associated with high dimensionality and multicollinearity, which negatively affect the performance of traditional, parametric classification algorithms. To this end, machine algorithms are often preferred to classify hyperspectral data due to their robustness to various data distributions and noise. The current study compared the performance of three advanced machine learning classifiers, i.e., Support Vector Machine (SVM), Random Forest (RF), and Partial Least Squares Discriminant Analysis (PLS-DA), in discriminating four dominant wetland plant species, i.e., Crocosmia sp., Grasses, Agapanthus sp. and Cyperus sp. using simulated hyperspectral data from an upcoming sensor, i.e., nSight-2. The results revealed that SVM is superior, with an overall accuracy of 93.18% (and class-wise accuracies > 85%). In comparison, there were minor differences in the performances of RF and PLS-DA, i.e., 84.09% and 83.63%, respectively. Overall, the results demonstrated that all the evaluated classifiers could achieve acceptable mapping accuracies. However, SVM is more robust, providing exceptional accuracies, and should be considered for operational mapping once the sensor is in space.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wetland ecosystems are among the most productive systems in the world. They offer huge ecological, social, and economic benefits. It has been shown that they support over one billion people globally through the provision of various ecosystem goods and services (Amler et al. 2015). The estimated financial value of the ecosystem goods and services provided by wetlands such as recreation, education, scientific research, photography, fishing, hunting, and bird-viewing (Ola and Benjamin 2019), is US$4.9 trillion annually across the globe (Walter and Mondall, 2019). Monitoring, conservation, and management of wetland ecosystems at varying scales are planned and executed. However, these efforts are hampered by the scarcity of accurate, reliable, and up-to-date spatial information about wetland ecosystems in some parts of the world, particularly in Africa (Adam et al. 2010). For example, at the international level, wetland ecosystem conservation measures have culminated in the demand for large integrated monitoring and reporting frameworks such as the Ramsar Convention of 1971. This Convention aims at the wise use of wetlands with emphasis on the wetland ecosystems and protection of Red Data species within this ecosystem (Dixon et al. 2016). Moreover, it provides for cooperation regarding the conservation, preservation, and management of wetlands and their sustainable use at national and international scales. Wetland ecosystem functions are critical in supporting eleven of the seventeen United Nations Sustainable Development Goals (UN-SDG), also known as Agenda 2030. Other efforts include the Strategic Plan for Biodiversity 2011–2020 and the Post-2020 Global Biodiversity Framework of the Convention on Biological Diversity (Rebelo et al. 2018). All these initiatives aim at achieving environmental sustainability and the health of humanity while eradicating poverty. The success in meeting these targets hinges on a thorough understanding of the current and emerging pressures on wetland ecosystems and their various components, like the wetland plants, through a robust suite of monitoring strategies. Wetland plants play an important role in the provision of food, habitat, and sanctuaries of endemic and endangered animals. They improve water quality and abate floods. As such, the quest for accurate and reliable wetland plant species mapping is increasingly needed for understanding terrestrial processes such as surface energy balance, biogeochemical cycles, biomass distributions, carbon budgets, and climate change modelling (Mahdavi et al. 2018).
While the potential of remote sensing has been widely demonstrated, discriminating wetland plant species using this technique is not without challenges. Spectral analysis of wetland plant species is affected by high intra-class and low inter-class variability (Adam et al. 2010). High spatial heterogeneity and temporal dynamics, resulting from seasonal and daily changes in water levels make the extent and spectral separability of wetland plant species difficult (Ludwig et al. 2019; Adam et al. 2010). For example, the one plant species can give different spectral signatures owing to the seasonal and daily changes in water levels, while on the other hand, varying species may portray similar reflectance (Adam and Mutanga 2009). According to Dronova and Tadeo (2016) dead plant matter attenuates the spectral signal of vegetation, while inundation affects plant signal in the red and near-infrared regions of the electromagnetic spectrum.
The debut of space-borne hyperspectral sensors provides new prospects for plant species discrimination in complex environments such as wetlands. Spectroscopic data from these sensors offer detailed spectral information, increasing many opportunities to detect relevant spectral absorption regions that enable differentiation of subtle differences among related Earth surface targets. However, only a few hyperspectral sensors exist as pre-operational and technology demonstrator missions and thus have limited scope, e.g., Hyperion, Compact High-Resolution Imaging Spectrometry (CHRIS), Environmental Mapping and Analysis Program (EnMap), and Hyperspectral Precursor of the Application Mission (PRISMA). Therefore, home-grown sensors such as the forthcoming nSight-2 by the Space Advisory Company in South Africa have the potential to meet the data requirements for priority research areas. The sensor offers 160 linear filtered and pre-selected spectral bands in the VNIR spectral range (i.e., 400 – 900 nm). However, prior to its launch, it is imperative to evaluate the relevance of its spectral settings for many applications including wetlands species discrimination. Gasela et al. (2022) assessed the usefulness of nSight-2’s spectral settings for classifying various wetland plant species and found that it performed superior to EnMap and WorldView-2.
Despite promising results with hyperspectral data, its properties, such as many (thousands) contiguous and correlated bands, may lead to poor classification accuracy (Raczko and Zagajewski 2017). Most of these spectral bands do not add new information; instead, they burden the classifiers (Dabija et al. 2021; Elgeldawi et al. 2021). Furthermore, high dimensionality creates an imbalance between input bands and training samples, resulting in overfitting since collecting many training samples that balance with input bands is expensive. Additionally, multicollinearity and nonlinearity in hyperspectral data create challenges for many classifiers leading to poor classification accuracy. Several machine learning classifiers exist and have been tested for many applications in remote sensing. However, their performance varies according to their robustness to noisy and highly dimensional datasets as well as land cover types, among others. Some of the commonly used machine learning classifiers include Support Vector Machines (SVM), Random Forest (RF), Neural Networks (NN), Decision Trees (DT), and Partial Least Squares Discriminant Analysis (PLS-DA). For example, Stratoulias et al. (2018) used SVM and Maximum Likelihood to classify emergent wetland vegetation in Lake Balaton, Hungary. Gosh et al. (2014) used Hyperion and HyMap data in a forest in Germany to compare the performances of SVM and RF and found that their performance was similar at 71% and 72%, respectively. In another study, Raczko and Zagajewski (2017) compared SVM, RF, and NN for tree species classification using airborne hyperspectral APEX images in Karkonosze National Park in Poland, the ANN achieved the highest overall classification accuracy with 77%, compared with 68% of SVM, and 62% of RF. Elsewhere in a dense species Central European forest area, Richter et al. (2016) compared RF, SVM, and PLS-DA in tree species classification using an airborne AISA Dual imaging system, and their results showed that the PLS-DA consistently outperformed SVM and RF with an overall accuracy of 78%, followed by 73% for SVM, and 65% for RF.
In other studies, these machine learning algorithms achieved considerably high accuracies. For example, Yang et al. (2019) compared the performances of RF and Gradient Boosting Decision Trees (GBDT) and SVM in a cropland ecosystem in Jiangsu Province of East China and found that GBDT had a higher overall accuracy of 92.4% compared with RF and SVM accuracies of 91.8% and 90.5%, respectively. In another study, Britz et al. (2022) worked on the spectral-based classification of plant species groups and functional plant traits in three grassland ecosystems in Austria to compare the performances of Multi-Layer Perceptron (MLP), PLS-DA, and RF. They concluded that MLP outperformed both PLS-DA and RF, achieving an overall accuracy of 95.7%.
As shown in the studies above, there are inconsistencies in the performance of these machine learning algorithms when used with varying datasets and vegetation types, thus making it difficult to conclude the superiority of a single classifier. To ascertain the full potential of the nSight-2 sensor in the classification of wetland plant species, there is a need to explore the performances of various machine learning classifiers. The comparison of various machine learning classifiers will facilitate the choice of the optimal algorithm for future mapping of the distributions of wetland plant species using nSight-2 once it is operational. Such distribution maps are essential for optimizing management approaches and strategies to ensure sustainable wetland ecosystems. Therefore, this study compared the performances (measured by overall agreement, quantity, and allocation differences) of SVM, RF, and PLS-DA in discriminating wetland species in a Ramsar Wetland Site, located in Mpumalanga province, South Africa. In hindsight, the performances of these algorithms also illustrate their sensitivity to high dimensionality and multicollinearity because they were tested with hyperspectral data.
Study area
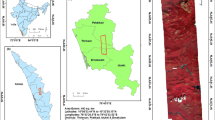
The study was conducted in Verloren Vallei Nature Reserve (VVNR, Fig. 1), which is a Ramsar site and managed by the Mpumalanga Tourism and Parks Agency (MTPA). It is located in Emakhazeni Local municipality, approximately 15 km north of Dullstroom town in the Mpumalanga Province of South Africa. The climate in the area is characterised by cold winters with the lowest temperatures of about − 13 °C between May and August, and hot and wet summers with the highest temperatures of about 29 °C and an average annual rainfall of more than 800 mm. The relief is underlain by rock outcrops and hills at an altitude of 2000 m. The VVNR is home to several red data species and birds of significance hence was declared by the Ramsar Convention as a site of international importance in 2011. Verloren Vallei Nature Reserve wetlands are significant for reducing flooding in the Lowveld of South Africa and improving water quality, by allowing a steady flow of water during the dry season. It is also a habitat for the endangered flora and fauna. It is home to the blue crane birds that have been declared endangered. Several other Red Data Species mammalian species like the striped weasel, grey rhebok, and blesbokcan also be found at VVNR. Interested readers may visit https://verlorenvalei.org.za/ for additional information and pictures.
Verloren Vallei Nature Reserve in Mpumalanga province, South Africa. Wetlands are delineated in navy blue (obtained from the Biodiversity GIS website, https://bgis.sanbi.org/). The outer shaded boundary in blue represents a sampling area (100 m buffer) for the current study. The band combination of the Sentinel-2 image in the background is B12, B8A, B4
Materials and methods
An overview of the methodology is provided in Fig. 2.
Sampling strategy and spectral measurements
Spectral measurements were recorded in the lab following a field survey conducted from the 14th to 18th of December 2020 since in-situ spectral measurements were not possible due to overcast weather conditions. We selected thirty 40 m x 40 m plots within a 100 m buffer of the wetlands across the study area using a random sampling strategy. Each plot was tagged with a GPS coordinate, using Garmin eTrex® 20 with ± 3 m GNSS accuracy. The choice of the plot distribution was based on the spatial dominance of plant species of interest, i.e., Crocosmia paniculata, Agapanthaceae, Themeda triandra, and Cyperus sp. (Fig. 3), while the plot size was selected considering the spatial resolution of the forthcoming sensor, i.e., nSight-2 (~ 20 m). Moreover, we considered the potential of applying the models to satellite images to map spatial distributions of these species in the future. Various leaves of the four dominant species in the study area, i.e., Crocosmia paniculata, Agapanthaceae, Themeda triandra, and Cyperus sp. were harvested from five randomly selected sub-plots of 1 m x 1 m within each plot. The choice of these plant species was based on their endemic feature as the cornerstone of the Verloren Vallei Nature Reserve and their wide occurrence within the wetland areas. These sub-plots represented homogeneous species per plot. The leaves were preserved for quality in plastic bags and stored in a cooler box. To capture variability per plot, several spectral measurements were captured on various parts of the leaves. The Spectral Evolution PSR-3500 spectrometer (Spectral Evolution, Inc. © 2014), used here, had a spectral range of 350 – 2500 nm, spectral resolutions of 3.5 nm, 10 nm, and 7 nm at 350 nm – 1000 nm, 1500 nm, and 2100 nm, respectively. The spectral bands in the regions: 350 – 1000 nm, at 1500 nm, and at 2100 nm have nominal spectral sampling intervals of 1.5 nm, 3.8 nm, and 2.5 nm, respectively, and were interpolated to 1 nm following Kganyago et al. (2017). A 5-watt Tungsten halogen light on the Fiber Optic Illumination Module provided an artificial light source similar to the natural light and a bifurcated cable attached to the leaf clip was used to take spectral measurements, thus masking background effects. Calibration using a white reflectance panel was performed after every five spectral measurements.
Data processing
It is critical to assess the capability of forth-coming sensors for various applications to ascertain their utility and robustness to support such applications. Therefore, in the current study, the spectral configuration for the nSight-2 sensor, a planned hyperspectral Earth observation satellite, was simulated and tested with various classifiers to determine their prospects in the context of plant species mapping. This sensor will have a spectral range from 400 to 900 nm with 160 linear filtered and pre-selected spectral bands. To simulate the spectral resolution of the nSight-2 sensor, we measured full-range spectral reflectance in 30 plots, which resulted in 2151 spectral bands sampled at an interval of 1 nm. These 2151 spectral bands were then resampled based on the spectral settings (i.e., full width at half maximum and band centers) of the nSight-2 sensor using the Hsdar R package (Lehnert et al. 2018). Using Gaussian distribution, the spectral response of each band was estimated. The pre-processed (resampled) spectral data had a spectral range of 467 – 901 nm with a mean bandwidth of 6.08 nm. The samples were split into 70% training and 30% validation datasets for classifying wetland species using various machine learning classifiers described in the next section.
Machine learning classifiers
Three machine learning classifiers were used in the current study, i.e., Random Forest (RF), Support Vector Machine (SVM) and Partial Least Squares-Discriminant Analysis (PLS-DA). The three classifiers differ in terms of their features and have strengths and weaknesses reported in the literature. Below, we describe each classifier, the required tuning parameters and their respective strengths and weaknesses as reported in the literature.
Random forest classifier
Random Forest (RF) is a tree-based classifier based on the Classification and Regression of Trees (CART) (Breiman et al., 1984). It classifies the different classes by combining results from many (i.e., hundreds of) decision trees using a voting strategy (Breinman, 2001). Each tree is trained with a random subset of variables and training samples (i.e., in-bag samples) selected using a bagging or bootstrapping resampling strategy. The in-bag samples consist of about two-thirds (i.e., 63.2%) of the total training data, and the remaining one-third of the samples (i.e., out-of-the-bag [OOB] samples, [32.8%]) is used for cross-validation (Lim et al. 2019; Zafari et al. 2019). RF requires tuning of two parameters, i.e., the number of trees (n-tree) and the number of variables randomly selected at each split (mtry). The accuracy tends to increase with the increasing n-tree (Breinman, 2001), but stabilises with no further improvements in accuracy at around 500 trees. On the other hand, the default mtry is the square root of the total number of variables in the dataset (i.e.,default is \( \sqrt{m}\)), and the lower the mtry result in diverse and less correlated trees (Probst et al. 2019). The two parameters assist in avoiding overfitting in the RF classification model. An overfit model is one that performs optimally with the training data but poorly generalises the independent test data. In the current study, a grid-search strategy – an approach for exhaustively selecting tuning parameters from all possible combinations – was used to search for optimal n-tree and mtry. The n-tree values from 100 to 1000 were tested. A pair of n-tree and mtry parameters that result in minimum OOB error is considered optimal. After testing the n-tree values from 100 to 1000 with an interval of 100, and mtry values from 1 to \( \sqrt{m}\), the optimal parameters were 500 and 2 for n-tree and mtry, respectively, which resulted in an OOB accuracy of 78.29% (KappaOOB of 0.72).
Some of the advantages of an RF classifier are that it is computationally efficient, transparent, and interpretable, does not require many parameters, and results in higher classification accuracy (Kganyago et al. 2024). Moreover, it has been proven to be robust in handling noisy data and, therefore, effective for a wide variety of classification and regression tasks (Teluguntla et al. 2018; Golrang et al., 2020). In contrast, it can result in poor performance when there is a sample imbalance between classes, thus overfitting the majority class (Breiman, 2001).
Support vector machine
Support Vector Machine (SVM) (Vapnik, 1999), on the other hand, classifies data by finding an optimal hyperplane in n-dimensional space with the highest margin between classes (Tzotsos and Argialas 2008). It defines decision boundaries using geometrical characteristics of data through a hyperplane based on support vectors rather than density estimation and uses a smaller number of training samples without overfitting (Melgani and Bruzzone 2004). SVM is highly sensitive to the type of the kernel, the size of the kernel, and parameter C (Hsu et al. 2010). We used the commonly used Radial Basis Function (RBF) kernel, which requires two tuning parameters, i.e., sigma (γ) and regularisation parameters (C). The γ parameter controls the width of the kernel facilitating the SVM to distinguish multi-modal classes in a high dimensional space, while the C parameter controls the trade-off between the maximisation of the margin between the training data vectors and decision boundaries and margin errors of the training data. Its purpose is to handle potential noise in the data, class confusion and prevent overfitting (Mountrakis et al. 2011). The smaller the value of C, the more accurate the classification (Kganyago et al. 2018). Similar to RF, the optimal tuning parameters were determined using a grid-search strategy and 5-fold cross-validation (cv). The optimal tuning parameters were 32 and 0.01136596, for γ and C, respectively, which achieved a cv accuracy of 90.54% (Kappacv of 0.87). These were determined from a range of 0.25 (i.e., 2− 2) to 32 (i.e., 25) and 0.5 (i.e., 2− 1) to 2 (i.e., 21) with an interval of 0.01 in Caret R-package (Kuhn 2008).
Its strength lies in that it is computationally fast and does not employ density estimation to discriminate classes; instead, it utilises the geometrical characteristics of data to define decision boundaries by assessing only support vectors (Melgani and Bruzzone 2004). Moreover, it does not require a priori knowledge about the statistical distribution of data, can reduce classification errors while increasing resolution, and is insensitive to highly dimensional data (Pal and Foody 2010; Mountrakis et al. 2011). However, SVM has some weaknesses, such as being computationally inefficient, highly sensitive to parameter tuning, lacking probabilistic outputs, and difficult to interpret complex decision boundaries (Ray 2024).
Partial least squares-discriminant analysis
Lastly, Partial Least Squares-Discriminant Analysis (PLS-DA) is a multivariate supervised statistical algorithm that finds a linear regression model by constructing predictive variables and response variables into a new space (Chauhan et al. 2020). The PLS-DA creates predictive and response variables with few eigenvectors from spectral data matrices (Peerbhay et al. 2013). This results in data that is correlated and characterised by predictor variables that are more than the observations. PLS-DA then finds optimum components that improve its classification performance (Peerbhay et al. 2013, 2016). The PLS-DA was used by Peerbhay et al. (2013) to test its robustness in classifying commercial tree species in KwaZulu Natal. They found that PLS-DA can significantly discriminate tree species with an overall accuracy of 88.8% using AISA Eagle bands. Although this was in a different environment, their studies ascertain that the PLS-DA can be successfully used in with hyperspectral data in vegetation mapping and monitoring. Richter et al. (2016) used airborne hyperspectral in a heterogeneous mixed forest in Central Europe to compare the performances of SVM, RF and PLS-DA in discriminating tree species; interestingly, PLS-DA outperformed both SVM and RF. Like SVM classifier, we used a grid-search strategy and 5-fold cv to search for the optimal number of components (i.e., latent variables) for PLS-DA from a total of 160 components (i.e., same as the number of spectral bands in nSight-2). The optimal model had 2 latent variables, which explained the highest variability, i.e., > 20%. PLS-DA analysis was performed using the Caret R-package (Kuhn 2008).
The strengths of PLS-DA include reducing noise in the dataset, showing the probability of a sample belonging to the class being modelled, and selecting the best variables (Li et al. 2016). Moreover, it can handle multicollinearity, missing data, and information redundancy, which is common in hyperspectral data (Peerbhay et al. 2016). Richter et al. (2016) show that the higher number of predictor variables than observations and the multicollinearity of the spectral bands risk overfitting.
Accuracy assessment
Accuracy assessment was assessed using confusion matrix and associated statistical metrics, i.e., overall accuracy (OA, Eq. 1), Producer’s accuracy (PA, Eq. 2) and User’s accuracy (UA, Eq. 3). Moreover, Allocation Difference (AD) and Quantity Difference (QD) proposed by Pontius and Millones (2011), were used instead of the Kappa co-efficient. Overall accuracy indicates the percentage of correctly classified samples (Eq. 1).
where r is the number of classes, \( {n}_{ii}\) are the diagonal elements and n represents the total number of considered samples. Although there is no universally accepted value of OA, Anderson et al. (1976) considered 85% acceptable, while Pringle et al. (2009) prefer any OA value > 70%. Therefore, the current study considers OA values > = 75%OA < = 85% as acceptable and those above 85% as exceptional.
The PA, calculated from the confusion matrix, indicates the probability that the classifier has correctly classified the samples. It is calculated by taking the total number of correct classifications for a particular class, i.e.,\( {n}_{ii}\) and dividing it by the column total, i.e., \( {n}_{icol}\) (see Eq. 2) (Verma et al. 2020). The UA is calculated by dividing the total number of correctly classified the samples for a particular class, i.e.,\( {n}_{ii}\) the row total, i.e.,\( {n}_{irow}\) (see Eq. 3).
\( {n}_{ii}\) is the number of correctly classified samples and \( {n}_{icol}\) and \( {n}_{irow}\) are the column and row totals, respectively (Verma et al. 2020). Omission Errors (OE) and Commission Errors (CE) were also calculated for each class as \( 1-PA\) and\( 1-UA\), respectively.
Quantity Difference (QD, Eq. 5) refers to the imperfect match in the class proportions between the classification and reference datasets. In contrast, Allocation Difference (AD, Eq. 6) refers to the imperfect match in the class allocations between the reference and classification datasets given their quantities (Warrens 2015).
\( {n}_{+i}\) and \( {n}_{i+}\) represent the marginal sums of the columns and rows, respectively. AD is divided into Shift and Exchange. The lower the QD and AD values the better the classification accuracy.
Results and discussion
Results
The classification results from three evaluated machine learning classifiers are presented in Table 1. As shown, the Support Vector Machines (SVM) achieved a superior Overall Accuracy (OA) with 93.18%, followed by Random Forest (RF) with 84.09%, and then Partial Least Squares-Discrimination Analysis (PLS-DA) with 83.63%. The corresponding Allocation Difference (AD) and Quantity Difference (QD) were also proportionately low for the SVM classification model, i.e., ~ 2% and < 5%, respectively, juxtaposed with the RF classification model, i.e., ~ 9% and ~ 7%, respectively. The PSL-DA classification model, on the other hand, had ~ 7% for the AD and 10% for QD. Although the PSL-DA classification model had a relatively lower OA than the RF classification model, its AD was better at 6.36% compared to the 9.09% of the RF classification model. To compare the differences caused by pair-wise and non-pair-wise class confusions, the Shift and Exchange metrics were used. The RF and PLS-DA classification models had similar Shift (i.e., pair-wise class confusions) of 4.54%, while the SVM classification model had the lowest Shift of ~ 2%. The Exchange (i.e., non-pair-wise class confusions) was 0% for SVM model and < 2% for the PLS-DA classification model, while it was 4.54% for RF classification model.
The class-wise accuracies (i.e., PA and UA) were fairly high across all models, i.e., mostly about 75%. For Crocosmia sp., SVM outperformed both RF and PLS-DA achieving PA and UA of 100%. On the other hand, there was no difference in the PA performances of RF and PLS-DA, while UA was higher in PLS-DA, i.e., 90.32%, than RF which only achieved 84.62%. Moreover, SVM outperformed both RF and PLS-DA classifiers with a PA of 100% and a UA of 86.67% for Grasses. On the other hand, RF and PLS-DA had PA above 91% and 95%, respectively. There was no significant difference in the UA of RF and PLS-DA, with only a < 1% difference. Interestingly, PLS-DA had the highest PA for Cyperus sp. (i.e., 93.10%), which is ~ 1.5% higher than the PA achieved by SVM and ~ 9% higher than RF. However, the UA of the PLS-DA was the lowest (i.e., 84.38%), while there was no difference between the UA achieved by RF and SVM. RF had the worst PA for Agapanthus sp. across the compared algorithms (i.e., 66.67%), while PLS-DA achieved the best PA (i.e., 93.33%) for the same class. Figure 4 shows the class-wise errors, i.e., Omission (OE) and commision errors (CE). As can be seen, RF and SVM models had the highest OE, i.e., > 30%, for Agapanthus sp. while the PLS-DA model had an OE of ~ 6% for the same class. Grasses had the highest CE, i.e., > 25%, across classifiers, while the RF and SVM models achieved CE of 0% for Agapanthus sp. as compared to PLS-DA model’s ~ 12%.
The class-wise performances of Random Forest (RF), Support Vector Machines (SVM), and Partial Least Squares-Discrimination Analysis (PLS-DA) in classifying wetland species. PA and UA denote the Producer’s and User’s Accuracy, respectively while OE and CE are Omission and Commission Errors respectively
Discussion
The comparison of the performance of machine learning algorithms is a critical endeavor considering that previous studies show inconsistencies in performance according to the climatic environments, vegetation and species types, and datasets. It is particularly interesting to determine the suitability of the algorithms for classifying plant species using the simulated data of forthcoming sensors in specific priority environments such as wetland sites. In many regions, including the developing regions, the wetland environments are threatened by climate variability and change, as well as human over-utilisation and conversion to other land-uses. This study found that the SVM classifier was the most suitable algorithm for classifying wetland vegetation using the resampled nSight-2 data. In contrast, the RF and PLS-DA classification models generally performed similarly, with only slight differences. The superior performance of the SVM classifier can be explained by its ability to combat overfitting. Moreover, the SVM classifier’s performance can be attributed to the high 5-fold cv accuracy, i.e., 90.54%, achieved during the training process, indicating the robustness of the trained model and its generalisation capability. On the other hand, the training (i.e., OOB) accuracy of RF was 78.29%, while the final model resulted in 84% accuracy. Therefore, the results suggest that parameterisation of the classifiers could have played a critical role in the performances of these classifiers on the test dataset. The tuning accuracies seem to influence the final performances of the classification models. This means the higher the training accuracies, the better the model performance on the independent testing data. The different training accuracies also raises questions of the influence of the resampling techniques, i.e., 5-fold cv and bagging (or bootstrapping) for SVM and RF, respectively, necessitating further investigation into their influence on the model performance.
SVM was able to reduce overlap in the spectra of the plant species under study for plant species discrimination, as depicted by low AD and QD (2.27% and 4.54%), which were lower than those of RF and PLS-DA (9.09% and 6.81% and 6.36% and 10.00%, respectively). The obtained accuracies are in agreement with the accuracies achieved by Rackzo and Zagajeweski (2017) and Dabija et al. (2021) who also found SVM performing better than RF. Rackzo and Zagajeweski (2017) compared ANN, SVM and RF in the classification of trees in Karkonosze National Park in Poland, using the Airborne Prism Experiment hyperspectral data and found that SVM performed better, achieving an accuracy of 68% when compared to the 62% achieved by RF. Dabija et al. (2021) compared the performance of SVM and RF in the classification of different vegetation classes in eastern Romania using Sentinel-2 data and found that SVM performed better with an accuracy of 88% compared with the 70% achieved by RF. The higher performance of SVM can be explained by its ability to handle small training samples, which characterised the current study. The findings here are different from a similar study (Richter et al. 2016), which found that the PLS-DA outperformed SVM and RF in forest tree species discrimination where it achieved > 78% compared to 72% of SVM and 68% of RF. Indeed, the species sought after by this study were different structurally and in a different environment, and the relatively low performance of the PLS-DA in this study can be attributed to the fact that it is a linear algorithm and, therefore may have been ineffective in identifying non-linear patterns in the dataset.
The class-wise PA and UA metrics were fairly high across all models. However, the Crocosmia class had relatively high PA and UA across the classifiers. This can be attributed its larger parcels and regular boundaries. A high PA of all the other classes except Agapanthus shows that there was no spectral confusion among those classes, while there was significant misclassification of Agapanthusus with other classes. The grasses class had > 90% PA across all the classification models, while its UA was < 75% for both RF and PLS-DA, compared with > 85% for the SVM model. This confirms the superiority of the SVM classifier in discriminating wetland plant species. While the per-class accuracies differed between classes and algorithms, they were generally high, i.e., > 80%, except Agapanthus sp. and grasses in RF and grasses in PLS-DA models. Indeed, the plant species in the current study differed in terms of their structural properties; however, the spectral measurements from leaves instead of canopies may have resulted in invariant spectral signatures in certain regions of the electromagnetic spectrum due to similar biochemical composition, e.g., water content. Therefore, the classifiers considered here could not adequately discriminate such species as indicated by the AD and QD in RF and PLS-DA (Table 1). Moreover, many spectral bands, characteristic of hyperspectral data, cause the “curse of dimensionality” where the number of variables is greater than the number of observations (i.e., p > n) and many others may be collinear. Therefore, the results obtained here indicate that RF and PLS-DA are sensitive to these phenomena, while the superior performance of the SVM classifier ascertains the findings of previous studies that showed its strength in dealing with high dimensionality and small datasets (Melgani and Bruzzone 2004; Pal and Foody 2010; Mountrakis et al. 2011). This is critical for eliminating certain processing steps such as data dimensionality reduction and feature selection, which are commonly required when dealing with hyperspectral data, thus enhancing the capability to rapidly provide information for wetland management purposes.
The results also showed that there is no significant difference between class-wise performances of RF and PLS-DA algorithms. This can be explained by the fact that PLS-DA is a multivariate method that is slightly passive in feature selection. PLS-DA does not select features of importance in the classification process. Instead, it generates and selects a few latent variables that contain the greatest variability representative of the entire dataset (Ruiz-Perez, 2020), which resulted in a proportionately better classification accuracy comparable to that of the RF classifier. While there were reasonable overall accuracies across classifiers, i.e., ~ 84 − 93%, this may be misleading for practical wetland management applications. Therefore, the PA and UA provide further insights into the capability of the algorithms with the simulated nSight-2 dataset as well as the reliability of the results. For example, the high class-wise accuracies achieved by the SVM classification model in all classes can be critical information for wetland managers, equipping them with reliable and actionable knowledge about the spatial distribution of specific species within the wetland and how they change over time. Moreover, species-specific maps can be extracted to support conservation decisions, resource allocation, and prioritisation based on the dominance or negligible occurrence and spatial distributions of certain species. This is particularly important for Verloren Vallei Nature Reserve, a Ramsar wetland site, where attractive plant species such as Crocosmia paniculata and Agapanthus are a tourism attraction due to their aesthetic flowers, while cyprus sp. occurs in the moist areas. Therefore, although both Crocosmia paniculata and Agapanthus have a status of “Least concern” in the Red List of South African Plants (Raimondo et al., 2009), the reserve managers may be interested in assessing their presence and absence and spatial changes over time against threats of alien invasive species and effect of changes in climatic patterns, heatwaves, and droughts to safeguard their tourism value. Also, changes in the spatial distribution of cyprus sp. may be indicative of changes in the wetland extent in response to climatic variability. On the other hand, the relatively poor class-wise accuracies of the Agapanthus sp. and grasses by the RF classification model suggest that the resulting species-specific maps may be misleading and extra attention may be required when utilising this classifier for these specific classes. The reported accuracies and class-wise errors provide a baseline for wetland managers to establish monitoring protocols; thus, using these machine learning classifiers can help detect changes in vegetation patterns over time, allowing for adaptive management strategies in response to evolving wetland conditions or potential threats.
Limitations
This study encountered several shortcomings that can be leveraged for further investigation. Among these was the fact that the spectral data was not acquired under field conditions and by a satellite. Instead, the data was simulated using the spectral band settings of the forthcoming sensor (i.e., nSight-2). Therefore, other sources of error may be encountered when real data is used, such as residual errors after atmospheric correction and spectral mixing; hence, the models obtained here may not be necessarily transferable to the satellite sensor measurement. Future studies should attempt to collect data under field conditions, where the spectral reflectance will be representative of various plant components and backgrounds. Also, gaussian noise may be added in line with the nSight-2’s signal-to-noise ratio to ensure that more realistic spectral observations are used instead of leaf spectra, used here. Nonetheless, the results in the current study demonstrate the potential of the upcoming nSight-2 hyperspectral sensor and SVM in classifying wetland species. It is anticipated that advanced algorithms such as deep neural networks and extreme gradient boosting may result in better results; therefore, it is recommended that future studies consider evaluating the performance of these classifiers in combination with other explanatory variables, such as vegetation indices.
Conclusion
This study aimed to compare the performances of machine learning algorithms in discriminating among four dominant wetland plant species using the simulated nSight-2 hyperspectral data. The results demonstrate that the SVM machine learning classifier had a comparatively high classification accuracy performance (93.18% overall accuracy) in discriminating among the four wetland plant species. Even though the RF classification algorithm also had a better classification accuracy (i.e., 84.09%), there was no significant difference between its performance and that of PLS-DA, which had a performance of 83.63%. Moreover, the SVM algorithm had a higher class-wise PA and UA, while there was no significant difference in the PA and UA for both RF and PLS-DA. From these results, it can be concluded that spatial information derived from the SVM machine learning classifier in conjunction with data from the forth-coming nSight-2 can support practical applications of monitoring and management of wetlands. These findings provide insights into the effectiveness of machine learning classifiers in discriminating wetland species using hyperspectral data. The information derived from these results can be used to enhance the accuracy of species-specific mapping, prioritise management efforts, and contribute to the sustainable conservation and utilisation of wetland ecosystems.
References
Adam E, Mutanga O (2009) Spectral discrimination of papyrus vegetation (Cyperus papyrus L.) in swamp wetlands using field spectrometry. ISPRS J Photogrammetry Remote Sens 64(6):612–620
Adam E, Mutanga O, Rugege D (2010) Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: a review. Wetlands Ecol Manage 18(3):281–296
Amler E, Schmidt M, Menz G (2015) Definitions and mapping of east African wetlands: a review. Remote Sens 7(5):5256–5282
Anderson JR, Hardy EE, Roach JT, Witmer RE (1976) A Land Use and Land Cover Classification System for Use with Remote Sensor Data, Geological Survey Professional Paper 964
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Britz R, Barta N, Schaumberger A, Klingler A, Bauer A, Pötsch EM, Gronauer A, Motsch V (2022) Spectral-based classification of Plant species groups and Functional Plant Parts in Managed Permanent Grassland. Remote Sens 14(5):1154
Breiman L, Friedman J, Stone CJ, Olshen RA, Classification, Trees R (1984) ; Wadsworth&Brooks/Cole Advanced Books & Software: Monterey, CA, USA, ; ISBN 978-0-412-04841-8
Chauhan S, Darvishzadeh R, Boschetti M, Nelson A (2020) Discriminant analysis for lodging severity classification in wheat using RADARSAT-2 and Sentinel-1 data. ISPRS J Photogrammetry Remote Sens 164:138–151
Dabija A, Kluczek M, Zagajewski B, Raczko E, Kycko M, Al-Sulttani AH, Tardà A, Pineda L, Corbera J (2021) Comparison of support vector machines and random forests for corine land cover mapping. Remote Sensing, 13(4), p.777
Dixon MJR, Loh J, Davidson NC, Beltrame C, Freeman R, Walpole M (2016) Tracking global change in ecosystem area: the Wetland Extent Trends Index. Biol Conserv 193:27–35
Dronova I, Taddeo S (2016) Canopy leaf area index in non-forested marshes of the California Delta. Wetlands 36(4):705–716
Gasela M, Kganyago M, De Jager G (2022) Testing the utility of the resampled nSight-2 spectral configurations in discriminating wetland plant species using Random Forest classifier. Geocarto Int, pp.1–16
Ghosh A, Fassnacht FE, Joshi PK, Koch B (2014) A framework for mapping tree species combining hyperspectral and LiDAR data: role of selected classifiers and sensor across three spatial scales. Int J Appl Earth Obs Geoinf 26:49–63
Hsu C, Chang C, Lin C (2010) A practical guide to support vector classification. National Taiwan University, Department of Computer Science, Taipei
Kganyago M, Odindi J, Adjorlolo C, Mhangara P (2017) Selecting a subset of spectral bands for mapping invasive alien plants: a case of discriminating Parthenium hysterophorus using field spectroscopy data. Int J Remote Sens 38(20):5608–5625
Kganyago M, Odindi J, Adjorlolo C, Mhangara P (2018) Evaluating the capability of Landsat 8 OLI and SPOT 6 for discriminating invasive alien species in the African Savanna landscape. Int J Appl Earth Obs Geoinf 67:10–19
Kganyago M, Adjorlolo C, Mhangara P, Tsoeleng L (2024) Optical remote sensing of crop biophysical and biochemical parameters: an overview of advances in sensor technologies and machine learning algorithms for precision agriculture. Comput Electron Agric 218:108730. https://doi.org/10.1016/j.compag.2024.108730
Kuhn M (2008) Building predictive models in R using the caret package. J Stat Softw 28:1–26
Lehnert LW, Meyer H, Obermeier WA, Silva B, Regeling B, Bendix J (2018) Hyperspectral data analysis in R: the Hsdar package.arXiv preprint arXiv: 1805.05090
Lim J, Kim KM, Jin R (2019) Tree species classification using Hyperion and Sentinel-2 Data with Machine Learning in South Korea and China. ISPRS Int J Geo-Information 8(3):150
Ludwig C, Walli A, Schleicher C, Weichselbaum J, Riffler M (2019) A highly automated algorithm for wetland detection using multi-temporal optical satellite data. Remote Sens Environ 224:333–351
Mahdavi S, Salehi B, Granger J, Amani M, Brisco B, Huang W (2018) Remote sensing for wetland classification: a comprehensive review. GIScience Remote Sens 55:623–658
Melgani F, Bruzzone L (2004) Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans Geosci Remote Sens 42:1778–1790
Mountrakis G, Im J, Ogole C (2011) Support vector machines in remote sensing: a review. ISPRS J Photogrammetry Remote Sens 66(3):247–259
Ola O, Benjamin E (2019) Preserving biodiversity and ecosystem services in west African forests, watersheds, and wetlands: a review of incentives. Forests 10(6):479
Pal M, Foody GM (2010) Feature selection for classification of hyperspectral data by SVM. IEEE Trans Geosci Remote Sens 48:2297–2307. https://doi.org/10.1109/TGRS.2009.2039484
Peerbhay KY, Mutanga O, Ismail R (2013) Commercial tree species discrimination using airborne AISA Eagle hyperspectral imagery and partial least squares discriminant analysis (PLS-DA) in KwaZulu-Natal, South Africa. ISPRS J Photogrammetry Remote Sens 79:19–28
Peerbhay K, Mutanga O, Lottering R, Ismail R (2016) Mapping Solanum mauritianum plant invasions using WorldView-2 imagery and unsupervised random forests. Remote Sens Environ 182:39–48
Pontius RG Jr, Millones M (2011) Death to Kappa: birth of quantity disagreement and allocation disagreement for accuracy assessment. Int J Remote Sens 32:4407–4429
Pringle RM, Syfert M, Webb JK, Shine R (2009) Quantifying historical changes in habitat availability for endangered species: use of pixel-and object‐based remote sensing. J Appl Ecol 46(3):544–553
Probst P, Wright MN, Boulesteix A-L (2019) Hyperparameters and tuning strategies for random forest. WIREs Data Min Knowl Discov 9:e1301. https://doi.org/10.1002/widm.1301
Raczko E, Zagajewski B (2017) Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur J Remote Sens 50(1):144–154
Ray S (2024) Learn how to use Support Vector Machines (SVM), analyticsvidhya.com (accessed on 9 Feb. 2024)
Rebelo LM, Finlayson CM, Strauch A, Rosenqvist A, Perennou C, Tøttrup C, Hilarides L, Paganini M, Wielaard N, Siegert F, Ballhorn U (2018) The use of Earth Observation for wetland inventory, assessment and monitoring: An information source for the Ramsar Convention on Wetlands. In: Ramsar Technical Report No. 10. Gland, Switzerland: Ramsar Convention Secretariat
Richter R, Reu B, Wirth C, Doktor D, Vohland M (2016) The use of airborne hyperspectral data for tree species classification in a species-rich central European forest area. Int J Appl Earth Observation Geo-information 52:464–474
Ruiz-Perez D, Guan H, Madhivanan P, Mathee K, Narasimhan G (2020) So you think you can PLS-DA? BMC Bioinformatics 21(1):1–10
Stratoulias D, Balzter H, Zlinszky A, Tóth VR (2018) A comparison of airborne hyperspectral-based classifications of emergent wetland vegetation at Lake Balaton, Hungary. Int J Remote Sens 39(17):5689–5715
Teluguntla P, Thenkabail PS, Oliphant A, Xiong J, Gumma MK, Congalton RG, Yadav K, Huete A (2018) A 30-m landsat-derived cropland extent product of Australia and China using random forest machine learning algorithm on Google Earth Engine cloud computing platform. ISPRS J Photogrammetry Remote Sens 144:325–340
Tzotsos A, Argialas D (2008) Support vector machine classification for object-based image analysis. Object-based image analysis. Springer, Berlin Heidelberg, pp 663–677
Verma N, Mishra P, Purohit N (2020) Development of a knowledge based decision tree classifier using hybrid polarimetric SAR observables. Int J Remote Sens 41(4):1302–1320
Walter M, Mondal P (2019) A rapidly assessed wetland stress index (RAWSI) using landsat 8 and Sentinel-1 radar data. Remote Sens 11(21):2549
Warrens MJ (2015) Relative quantity and allocation disagreement measures for category-level accuracy assessment. Int J Remote Sens 36(23):5959–5969
Yang L, Mansaray LR, Huang J, Wang L (2019) Optimal segmentation scale parameter, feature subset and classification algorithm for geographic object-based crop recognition using multisource satellite imagery. Remote Sens 11(5):514
Zafari A, Zurita-Milla R, Izquierdo-Verdiguier E (2019) Evaluating the performance of a random forest kernel for land cover classification. Remote Sens 11(5):575
Golrang A, Golrang AM, Yayilgan SY, Elezaj O (2020) A novel hybrid IDS based on modified NSGAIIANN and random forest. Electronics 9(4):577
Elgeldawi E, Sayed A, Galal AR, Zaki AM (2021) Hyperparameter tuning for machine learning algorithms used for arabic sentiment analysis. Informatics 8(4):79. https://doi.org/10.3390/informatics8040079
Li X, Wang S, Shi W, Shen Q (2016) Partial least squares discriminant analysis model based on variable selection applied to identify the adulterated olive oil. Food Anal Methods 9(6):1713–1718
Raczko E, Zagajewski B (2017) Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur J RemSens 50(1):144–154
Raimondo D, Staden LV, Foden W, Victor JE, Helme NA, Turner RC, Kamundi DA, Manyama PA (2009) Red list of South African plants 2009. South African National Biodiversity Institute. Pretoria, South Africa, ix + 668pp
Ghosh A, Fassnacht FE, Joshi PK, Koch B (2014) A framework for mapping tree species combining hyperspectral and LiDAR data: role of selected classifiers and sensor across three spatial scales. Int J Appl Erath Observation Geoinfo 26:49–63. https://doi.org/10.1016/j.jag.2013.05.017
Acknowledgements
The authors wish to thank the Space Advisory Company (SAC), South African Space Agency Earth Observation Directorate (SANSA-EO), Mpumalanga Tourism and Parks Agency (MTPA) for the material and financial support and precious time afforded to accomplish this research work. A further special thank you goes to Shirley Sibiya the manager at Verloren Vallei Nature Reserve and her team for their help and support during data collection.
Funding
Open access funding provided by University of Johannesburg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gasela, M., Kganyago, M. & De Jager, G. Using resampled nSight-2 hyperspectral data and various machine learning classifiers for discriminating wetland plant species in a Ramsar Wetland site, South Africa. Appl Geomat 16, 429–440 (2024). https://doi.org/10.1007/s12518-024-00560-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12518-024-00560-z