Abstract
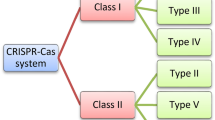
The recent thrust in research has projected the type II clustered regularly interspaced short palindromic repeats and associated protein 9 (CRISPR-Cas9) system as an avant-garde plant genome editing tool. It facilitates the induction of site-specific double-stranded DNA cleavage by the RNA-guided DNA endonuclease (RGEN), Cas9. Elimination, addition, or alteration of sections in DNA sequence besides the creation of a knockout genotype (CRISPRko) is aided by the CRISPR-Cas9 system in its wild form (wtCas9). The inactivation of the nuclease domain generates a dead Cas9 (dCas9), which is capable of targeting genomic DNA without scissoring it. The dCas9 system can be engineered by fusing it with different effectors to facilitate transcriptional activation (CRISPRa) and transcriptional interference (CRISPRi). CRISPR-Cas thus holds tremendous prospects as a genome-manipulating stratagem for a wide gamut of crops. In this article, we present a brief on the fundamentals and the general workflow of the CRISPR-Cas system followed by an overview of the prospects of bioinformatics in propelling CRISPR-Cas research with a special thrust on the available databases and algorithms/web-accessible applications that have aided in increasing the usage and efficiency of editing. The article also provides an update on the current regulatory landscape in different countries on the CRISPR-Cas edited plants to emphasize the far-reaching impact of the genomic editing technology.

© 2019, This is the work of a U.S. government)

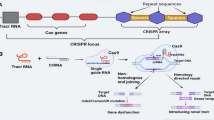
(Reproduced from Zhao et al. (2021) under provisions of Creative Commons License, http://creativecommons.org/licenses/by/4.0/, © Zhao et al. 2021). b Examples of modifications in the Cas enzyme system. (A) The binding of normal CRISPR-Cas9/sgRNA complex results in DSB, (B) Mutated Cas9 protein, without DNA cutting activity (dCas9), can still bind at the specific sgRNA-guided site on DNA to inhibit the transcription by blocking the progression of the RNA polymerase. (C) The dCas9 fused transcriptional activators can be utilised to switch on or enhance transcription at the target site. (D) The dCas9 fused histone modifiers or DNA methylation enzymes can introduce site-specific epigenetic changes at the target site. (Reproduced from Horodecka & Düchler (2021) under the provisions of Creative Commons License, http://creativecommons.org/licenses/by/4.0/, Copyright: © 2021 by the authors. Licensee MDPI, Basel, Switzerland)


(Reproduced from Chincinska et al. (2022) under the provisions of Creative Commons License, http://creativecommons.org/licenses/by/4.0/ © The Author(s) 2022)
Similar content being viewed by others
References
Agaba J (2019) Why South Africa and Sudan lead the continent in GMO crops. Alliance Science, Ithaca
Ahmad S, Tang L, Shahzad R, Mawia AM, Rao GS, Jamil S, Wei C, Sheng Z, Shao G, Wei X, Hu P (2021a) CRISPR-based crop improvements: a way forward to achieve zero hunger. J Agric Food Chem 69(30):8307–8323
Ahmad S, Shahzad R, Jamil S, Tabassum J, Chaudhary MAM, Atif RM, Iqbal MM, Monsur MB, Lv Y, Sheng Z, Ju L (2021) Regulatory aspects, risk assessment, and toxicity associated with RNAi and CRISPR methods. In: CRISPR and RNAi systems, pp 687–721. Elsevier
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nat 576:149–157. https://doi.org/10.1038/s41586-019-1711-4
Babar U, Nawaz MA, Arshad U, Azhar MT, Atif RM, Golokhvast KS, Tsatsakis AM, Shcerbakova K, Chung G, Rana IA (2020) Transgenic crops for the agricultural improvement in Pakistan: a perspective of environmental stresses and the current status of genetically modified crops. GM Crops Food 11:1–29. https://doi.org/10.1080/21645698.2019.1680078
Bae S, Park J, Kim JS (2014) Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinform 30(10):1473–1475. https://doi.org/10.1093/bioinformatics/btu048
Bao A, Burritt DJ, Chen H, Zhou X, Cao D, Tran LP (2019) The CRISPR-Cas9 system and its applications in crop genome editing. Crit Rev Biotechnol 3:321–336. https://doi.org/10.1080/07388551.2018.1554621
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819):1709–1712. https://doi.org/10.1126/science.1138140
Benítez Candia N, Fernández Ríos D, Vicién C (2020) Paraguay’s path toward the simplification of procedures in the approval of GE crops. Front Bioeng Biotechnol 8:1023. https://doi.org/10.3389/fbioe.2020.01023
Beyene G, Chauhan RD, Villmer J, Husic N, Wang N, Gebre E, Girma D, Chanyalew S, Assefa K, Tabor G, Gehan M, McGrone M, Yang M, Lenderts B, Schwartz C, Gao H, Gordon-Kamm W, Taylor NJ, MacKenzie DJ (2022) CRISPR-Cas9-mediated tetra-allelic mutation of the ‘Green Revolution’ SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef). Plant Biotechnol J 20:1716–1729. https://doi.org/10.1111/pbi.13842
Biswas A, Staals RHJ, Morales SE, Fineran PC, Brown CM (2016) CRISPRDetect: a flexible algorithm to define CRISPR arrays. BMC Genom. https://doi.org/10.1186/s12864-016-2627-0
Bratlie S, Halvorsen K, Myskja BK, Mellegård H, Bjorvatn C, Frost P, Heiene G, Hofmann B, Holst‐Jensen A, Holst‐Larsen T, Malnes RS (2019) A novel governance framework for GMO. EMBO Rep; 20:2–5. https://doi.org/10.15252/embr.201947812
Brazelton VA, Zarecor S, Wright DA, Wang Y, Liu J, Chen K, Yang B, Lawrence-Dill CJ (2015) A quick guide to CRISPR sgRNA design tools. GM Crops Food 6(4):266–276. https://doi.org/10.1080/21645698.2015.1137690
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321(5891):960–964. https://doi.org/10.1126/science.1159689
Chen B, Dai E (2020) A regulatory review of genetically modified organisms in China. Lexology. Available Online at: https://www.lexology.com/library/detail.aspx?g=f0e011c4-6621-438b-99fb-7576d40c440e (Accessed 2 Nov 2020).
Chincinska IA, Miklaszewska M, Sołtys-Kalina D (2023) Recent advances and challenges in potato improvement using CRISPR-Cas genome editing. Planta 257(1):1–27. https://doi.org/10.1007/s00425-022-04054-3
Colognori D, Trinidad M, Doudna JA (2023) Precise transcript targeting by CRISPR-Csm complexes. Nat Biotechnol. https://doi.org/10.1038/s41587-022-01649-9
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR-Cas systems. Sci 339(6121):819–823. https://doi.org/10.1126/science.1231143
Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46(W1):W246–W251. https://doi.org/10.1093/nar/gky425
Cui Y, Cheng X, Chen Q, Song B, Chiu A, Gao Y, Dawson T, Chao L, Zhang W, Li D, Zeng Z, Yu J, Li Z, Fei T, Peng S, Li W (2021) CRISP-view: a database of functional genetic screens spanning multiple phenotypes. Nucleic Acids Res 49(D1):D848–D854. https://doi.org/10.1093/nar/gkaa809
Debbarma J, Saikia B, Singha DL, Das D, Keot AK, Maharana J, Velmurugan N, Arunkumar KP, Reddy PS, Chikkaputtaiah C (2023) CRISPR-Cas9-mediated mutation in XSP10 and SlSAMT genes impart genetic tolerance to fusarium wilt disease of tomato (Solanum lycopersicum L.). Genes 14(2):488. https://doi.org/10.3390/genes14020488
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340):602–607. https://doi.org/10.1038/nature09886
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32(12):1262–1267. https://doi.org/10.1038/nbt.3026
Dondanville TW, Dougherty ML (2020) Porousness and Peru’s moratorium on genetically modified organisms: stakeholder epistemologies and neoliberal science. Environ Sociol 6:107–119. https://doi.org/10.1080/23251042.2019.1690726
Duensing N, Sprink T, Parrott WA, Fedorova M, Lema MA, Wolt JD, Bartsch D (2018) Novel features and considerations for ERA and regulation of crops produced by genome editing. Front Bioeng Biotechnol 6:79. https://doi.org/10.3389/fbioe.2018.00079
Eckerstorfer MF, Engelhard M, Heissenberger A, Simon S, Teichmann H (2019) Plants developed by new genetic modification techniques: comparison of existing regulatory frameworks in the EU and Non- EU countries. Front Bioeng Biotechnol 7:26. https://doi.org/10.3389/fbioe.2019.00026
El-Mounadi K, Morales-Floriano ML, Garcia-Ruiz H (2020) Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front Plant Sci 11:56. https://doi.org/10.3389/fpls.2020.00056
Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F (2016) Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci Rep. https://doi.org/10.1038/srep35766
Fernandes V (2020) India’s genome-editing draft guidelines are needlessly restrictive. India: smart Indian agriculture. FOEF (2018) Biotechnology: In brief. Available online at: https://www.bafu.admin.ch/bafu/en/home/topics/biotechnology/in-brief.html (Accessed 17 March 2020).
Frank H, Charpentier E (2016) CRISPR-Cas: biology, mechanisms and relevance. Phil Trans R Soc B 371:20150496–20150496. https://doi.org/10.1098/rstb.2015.0496
Friedrichs S, Takasu Y, Kearns P, Dagallier B, Oshima R, Schofield J, Moreddu C (2019) Policy considerations regarding genome editing. Trends Biotechnol 37(10):1029–1032. https://doi.org/10.1016/j.tibtech.2019.05.005
Fritsche S, Poovaiah C, MacRae E, Thorlby G (2018) A New Zealand perspective on the application and regulation of gene editing. Front Plant Sci 9:1323. https://doi.org/10.3389/fpls.2018.01323
Gakpo JO (2019) Egypt poised to again lead Africa in ag biotech innovation. Alliance Science, Ithaca
Gakpo JO (2020) Burkina Faso renews commitment to GM crops with Bt cowpea. Alliance Science, Ithaca
Garcia Ruiz MT, Knapp AN, Garcia-Ruiz H (2018) Profile of genetically modified plants authorized in Mexico. GM Crops Food 9(3):152–168. https://doi.org/10.1080/21645698.2018.1507601
Gasiunas G, Young JK, Karvelis T, Kazlauskas D, Urbaitis T, Jasnauskaite M, Grusyte MM, Paulraj S, Wang PH, Hou Z, Dooley SK, Cigan M, Alarcon C, Chilcoat ND, Bigelyte G, Curcuru JL, Mabuchi M, Sun Z, Fuchs RT, Schildkraut E, Weigele PR, Jack WE, Robb GB, Venclovas Č, Siksnys V (2020) A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat Commun 11(1):5512. https://doi.org/10.1038/s41467-020-19344-1
Gatica-Arias A (2020) The regulatory current status of plant breeding technologies in some Latin American and the Caribbean countries. Plant Cell Tissue Organ Cult 141:229–242. https://doi.org/10.1007/s11240-020-01799-1
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A T to G C in genomic DNA without DNA cleavage. Nat 551(7681):464–471. https://doi.org/10.1038/nature24644
Ge Z, Zheng L, Zhao Y, Jiang J, Zhang EJ, Liu T, Qu LJ et al (2019) Engineered xCas9 and SpCas9‐NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol J 17(10):1865
Gebretsadik K, Kiflu A (2018) Challenges and opportunities of genetically modified crops production; future perspectives in Ethiopia, review. Open Agric J 12:240–250. https://doi.org/10.2174/1874331501819010240
Geng Y, Deng Z, Sun Y (2016) An insight into the protospacer adjacent motif of Streptococcus pyogenes Cas9 with artificially stimulated RNA-guided-Cas9 DNA cleavage flexibility. RSC Adv 6(40):33514–33522. https://doi.org/10.1039/C6RA02774A
Gong Z, Cheng M, Botella JR (2021) Non-GM genome editing approaches in crops. Front Genome Ed 3:817279. https://doi.org/10.3389/fgeed.2021.817279
Grissa I, Vergnaud G, Pourcel C (2007a) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. https://doi.org/10.1093/nar/gkm360
Grissa I, Vergnaud G, Pourcel C (2007b) The CRISPRdb database and tools to display CRISPRs and generate dictionaries of spacers and repeats. BMC Bioinform. https://doi.org/10.1186/1471-2105-8-172
Gupta A, Hua L, Zhang Z, Yang B, Li W (2022) CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol J 21(3):536–548. https://doi.org/10.1111/pbi.13969
Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. https://doi.org/10.1186/s13059-016-1012-2
Herrera FF, Domené-Painenao O, Cruces JM (2017) The history of agroecology in Venezuela: a complex and multifocal process. Agroecol Sustain Food Syst 41:401–415. https://doi.org/10.1080/21683565.2017.1285842
Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172(6):1239–1259. https://doi.org/10.1016/j.cell.2017.11.032
Hodgkins A, Farne A, Perer S, Grego T, Parry-Smith DJ, Skarnes WC, Iyer V (2015) WGE: a CRISPR database for genome engineering. Bioinform 31(18):3078–3080. https://doi.org/10.1093/bioinformatics/btv308
Horodecka K, Düchler M (2021) CRISPR-Cas9: principle, applications, and delivery through extracellular vesicles. Int J Mol Sci 22(11):6072. https://doi.org/10.3390/ijms22116072
Hough SH, Ajetunmobi A, Brody L, Humphryes-Kirilov N, Perello E (2016) Desktop genetics. Personal Med 13(6):517–521. https://doi.org/10.2217/pme-2016-0068
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffin LA, Bao G, Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832. https://doi.org/10.1038/nbt.2647
Hua K, Tao X, Han P, Wang R, Zhu JK (2019) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant 12(7):1003-1014
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433. https://doi.org/10.1128/jb.169.12.5429-5433.1987
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR-Cas9. BMC Biotechnol 15(1):1–10. https://doi.org/10.1186/s12896-015-0131-2
Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46(1):505–529. https://doi.org/10.1146/annurev-biophys-062215-010822
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Sci 337(6096):816–821. https://doi.org/10.1126/science.1225829
Jorasch P (2020) Will the EU stay out of step with science and the rest of the world on plant breeding innovation? Plant Cell Rep 39:163–167. https://doi.org/10.1007/s00299-019-02482-2
Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel M, Barretto RPJ, Ranu A, Macrae RK, Faure G, Ioannidi EI, Krajeski RN, Bruneau R, Huang MW, Yu XG, Li JZ, Walker BD, Hung DT, Greninger AL, Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang F (2020) Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med 383(15):1492–1494. https://doi.org/10.1056/NEJMc2026172
Karavolias, N (2022) CRISPR in agriculture: 2022 in Review. https://innovativegenomics.org/news/crispr-agriculture-2022/ (Accessed on 5th March 2023).
Karlson D, Mojica JP, Poorten TJ, Lawit SJ, Jali S, Chauhan RD, Pham GM, Marri P, Guffy SL, Fear JM, Ochsenfeld CA, Lincoln Chapman TA, Casamali B, Venegas JP, Kim HJ, Call A, Sublett WL, Mathew LG, Shariff A, Watts JM, Mann M, Hummel A, Rapp R (2022) Targeted mutagenesis of the multicopy myrosinase gene family in allotetraploid brassica juncea reduces pungency in fresh leaves across environments. Plants 11(19):2494. https://doi.org/10.3390/plants11192494
Katam R, Hasanvand F, Teniyah V, Noel J, Gottschalk V (2022) Biosafety issue related to genome editing in plants using CRISPR-Cas9. Genome editing: current technology advances and applications for crop improvement. Springer International Publishing, Cham, pp 289–317
Kaur K, Tandon H, Gupta AK, Kumar M (2015) CrisprGE: a central hub of CRISPR-Cas-based genome editing. Database. https://doi.org/10.1093/database/bav055
Kavuri NR, Ramasamy M, Qi Y, Mandadi K (2022) Applications of CRISPR-Cas13-Based RNA editing in plants. Cells 11(17):2665. https://doi.org/10.3390/cells11172665
Kelly L (2019) Clarifying the regulation of genome editing in Australia: situation for food. Transgenic Res 28:161–164. https://doi.org/10.1007/s11248-019-00159-w
Komen J, Tripathi L, Mkoko B, Ofosu DO, Oloka H, Wangari D (2020) Biosafety regulatory reviews and leeway to operate: case studies from Sub-Sahara Africa. Front Plant Sci 11:130. https://doi.org/10.3389/fpls.2020.00130
Konwarh R (2020) Can CRISPR-Cas technology be a felicitous stratagem against the COVID-19 Fiasco? prospects and hitches. Front Mol Biosci 7:557377. https://doi.org/10.3389/fmolb.2020.557377
Kupferschmidt K (2018) EU verdict on CRISPR crops dismays scientists. Sci 361(6401):435–436. https://doi.org/10.1126/science.361.6401.435
Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44(W1):W272–W276. https://doi.org/10.1093/nar/gkw398
Lander ES (2016) The heroes of CRISPR. Cell 164(1–2):18–28. https://doi.org/10.1016/j.cell.2015.12.041
Lei J, Sun Y (2016) Assemble CRISPRs from metagenomic sequencing data. Bioinform 32(17):i520–i528. https://doi.org/10.1093/bioinformatics/btw456
Lenoir WF, Lim TL, Hart T (2018) PICKLES: The database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res 46(D1):D776–D780. https://doi.org/10.1093/nar/gkx993
Li C, Chu W, Gill RA, Sang S, Shi Y, Hu X, Zhang B (2022) Computational tools and resources for CRISPR-Cas genome editing. Genom Proteom Bioinform. https://doi.org/10.1016/j.gpb.2022.02.006
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R (2016) Editing DNA methylation in the mammalian genome. Cell 167(1):233–247. https://doi.org/10.1016/j.cell.2016.08.056
Liu X, Ding Q, Wang W, Pan Y, Tan C, Qiu Y, Ma C (2022) Targeted deletion of the first intron of the Wx b allele via CRISPR-Cas9 significantly increases grain amylose content in rice. Rice 15:1–12. https://doi.org/10.1186/s12284-021-00548-y
Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL (2017) CRISPR-P 20: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10(3):530–532. https://doi.org/10.1016/j.molp.2017.01.003
Ly DNP, Iqbal S, Fosu-Nyarko J, Milroy S, Jones MG (2023) Multiplex CRISPR-Cas9 gene-editing can deliver potato cultivars with reduced browning and acrylamide. Plants 12(2):379. https://doi.org/10.3390/plants12020379
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, Van Der Oost J, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13(11):722–736. https://doi.org/10.1038/nrmicro3569
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas C, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, Oost J, Barrangou R, Koonin EV (2020) Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18(2):67–83. https://doi.org/10.1038/s41579-019-0299-x
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 39(6121):823–826. https://doi.org/10.1126/science.1232033
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala KC (2016) DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904. https://doi.org/10.3389/fpls.2016.01904
Matsushita A, Goto H, Takahashi Y, Tsuda M, Ohsawa R (2020) Consideration of familiarity accumulated in the confined field trials for environmental risk assessment of genetically modified soybean (Glycine max) in Japan. Transgenic Res 29:229–242. https://doi.org/10.1007/s11248-020-00193-z
Meyer A, Dastgheib-Vinarov S (2021) The future of food? CRISPR-edited agriculture. Food and Drug Law Institute. https://www.fdli.org/2021/11/the-future-of-food-crispr-edited-agriculture/#_edn7 (Accessed on 5th Mar 2023)
Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM (2015) CRISPR-Cas mutagenesis of soybean and Medicago truncatula using a new web tool and a modified Cas9 enzyme. GM Crops Food 6(4):243–252. https://doi.org/10.1080/21645698.2015.1106063
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: A CRISPR-Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. https://doi.org/10.1093/nar/gku410
Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ (2015) CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12(10):982–988. https://doi.org/10.1038/nmeth.3543
Muzhinji N, Ntuli V (2021) Genetically modified organisms and food security in Southern Africa: conundrum and discourse. GM Crops Food 12:25–35. https://doi.org/10.1080/21645698.2020.1794489
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: Software for designing CRISPR-Cas guide RNA with reduced off-target sites. Bioinform 31(7):1120–1123. https://doi.org/10.1093/bioinformatics/btu743
News (2022a) FDA makes low-risk determination for marketing of products from genome-edited beef cattle after safety review. FDA news release. https://www.fda.gov/news-events/press-announcements/fda-makes-low-risk-determination-marketing-products-genome-edited-beef-cattle-after-safety-review
News, (2022a) GABA-enriched tomato is first CRISPR-edited food to enter market. Nat Biotechnol 40:9–11. https://doi.org/10.1038/d41587-021-00026-2
News, (2022b) Japan embraces CRISPR-edited fish. Nat Biotechnol 40:10. https://doi.org/10.1038/s41587-021-01197-8
Nikolić I, Samardžić J, Stevanović S, Miljuš-Đukić J, Milisavljević M, Timotijević G (2023) CRISPR-Cas9-targeted disruption of two highly homologous Arabidopsis thaliana DSS1 genes with roles in development and the oxidative stress response. Int J Mol Sci 24(3):2442. https://doi.org/10.3390/ijms24032442
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156(5):935–949. https://doi.org/10.1016/j.cell.2014.02.001
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361(6408):1259–1262. https://doi.org/10.1126/science.aas9129
Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA (2014) Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21(6):528–534. https://doi.org/10.1038/nsmb.2820
O’Brien A, Bailey TL (2014) GT-Scan: identifying unique genomic targets. Bioinform 30(18):2673–2675. https://doi.org/10.1093/bioinformatics/btu354
Oliveros JC, Franch M, Tabas-Madrid D, San-León D, Montoliu L, Cubas P, Pazos F (2016) Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res 44(W1):W267–W271. https://doi.org/10.1093/nar/gkw407
Pan C, Li G, Malzahn AA, Cheng Y, Leyson B, Sretenovic S, Gurel F, Coleman G, Qi Y (2022) Boosting plant genome editing with a versatile CRISPR-Combo system. Nat Plants 8(5):513–525. https://doi.org/10.1038/s41477-022-01151-9
Park J, Kim JS, Bae S (2016) Cas-database: web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-Cas9. Bioinform 32(13):2017–2023. https://doi.org/10.1093/bioinformatics/btw103
Pasquito LN (2019) The biosafety regulatory approach and governance mechanism of GM crops in the Philippines. Iapa Proc Conf 2019:620–649. https://doi.org/10.30589/proceedings.2019.290
Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A (2017) GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol 35(4):347–349. https://doi.org/10.1038/nbt.3804
Pliatsika V, Rigoutsos I (2015) “Off-Spotter”: Very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR-Cas guide RNAs. Biol Direct. https://doi.org/10.1186/s13062-015-0035-z
Pourcel C, Touchon M, Villeriot N, Vernadet JP, Couvin D, Toffano-Nioche C, Vergnaud G (2020) CRISPRCasdb: a successor of CRISPRdb containing CRISPR arrays and Cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res 48(D1):D535–D544. https://doi.org/10.1093/nar/gkz915
Rauscher B, Heigwer F, Breinig M, Winter J, Boutros M (2017) GenomeCRISPR: a database for high-throughput CRISPR-Cas9 screens. Nucleic Acids Res 45(D1):D679–D686. https://doi.org/10.1093/nar/gkw997
Ren C, Li H, Liu Y, Li S, Liang Z (2022) Highly efficient activation of endogenous gene in grape using CRISPR/dCas9-based transcriptional activators. Hortic Res 9:uhab037. https://doi.org/10.1093/hr/uhab037
Ronda C, Pedersen LE, Hansen HG, Kallehauge TB, Betenbaugh MJ, Nielsen AT, Kildegaard HF (2014) Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol Bioeng 111(8):1604–1616. https://doi.org/10.1002/bit.25233
Sánchez MA (2020) Chile as a key enabler country for global plant breeding, agricultural innovation, and biotechnology. GM Crop Food 11:130–139. https://doi.org/10.1080/21645698.2020.1761757
Schmidt SM, Belisle M, Frommer WB (2020) The evolving landscape around genome editing in agriculture. EMBO Rep; 21:19–22. https://doi.org/10.15252/embr.202050680
Schiml S, Fauser F, Puchta H (2016). CRISPR/Cas-Mediated Site-Specific Mutagenesis in Arabidopsis thaliana Using Cas9 Nucleases and Paired Nickases. Methods Mol Biol 1469:111–122. https://doi.org/10.1007/978-1-4939-4931-1_8
Shah P, Magar ND, Barbadikar KM (2022) Current technological interventions and applications of CRISPR-Cas for crop improvement. Mol Biol Rep 49(6):5751–5770. https://doi.org/10.1007/s11033-021-06926-5
Shahzad R, Jamil S, Ahmad S, Nisar A, Khan S, Amina Z, Kanwal S, Aslam HMU, Gill RA, Zhou W (2021) Biofortification of cereals and pulses using new breeding techniques: current and future perspectives. Front Nutri 665
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Kondo A (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35(5):441–443. https://doi.org/10.1038/nbt.3833
Skennerton CT, Imelfort M, Tyson GW (2013) Crass: identification and reconstruction of CRISPR from unassembled metagenomic data. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt183
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Sci 351(6268):84–88. https://doi.org/10.1126/science.aad5227
Smyth SJ (2017) Canadian regulatory perspectives on genome engineered crops. GM Crop Food 8:35–43. https://doi.org/10.1080/21645698.2016.1257468
Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, Tian L, Gonzalez VE, Xu J, Jung IY, Melenhorst JJ, Plesa G, Shea J, Matlawski T, Cervini A, Gaymon AL, Desjardins S, Lamontagne A, Salas-Mckee J, Fesnak A, Siegel DL, Levine BL, Jadlowsky JK, Young RM, Chew A, Hwang WT, Hexner EO, Carreno BM, Nobles CL, Bushman FD, Parker KR, Qi Y, Satpathy AT, Chang HY, Zhao Y, Lacey SF, June CH (2020) CRISPR-engineered T cells in patients with refractory cancer. Science 367(6481):eaba7365. https://doi.org/10.1126/science.aba7365
Stemmer M, Thumberger T, Del Sol KM, Wittbrodt J, Mateo JL (2015) CCTop: an intuitive, flexible and reliable CRISPR-Cas9 target prediction tool. PLoS ONE. https://doi.org/10.1371/journal.pone.0124633
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507(7490):62–67. https://doi.org/10.1038/nature13011
Subburaj S, Chung SJ, Lee C, Ryu SM, Kim DH, Kim JS, Bae S, Lee GJ (2016) Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep 35(7):1535–1544. https://doi.org/10.1007/s00299-016-1937-7
Tek MI, Calis O, Fidan H, Shah MD, Celik S, Wani SH (2022) CRISPR-Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front Plant Sci 13:5234. https://doi.org/10.3389/fpls.2022.1081506
Turnbull C, Lillemo M, Hvoslef-Eide TA (2021) Global regulation of genetically modified crops amid the gene edited crop boom–a review. Front Plant Sci 12:630396. https://doi.org/10.3389/fpls.2021.630396
Van Vu T, Sung YW, Kim J, Doan DTH, Tran MT, Kim J-Y (2019) Challenges and perspectives in homology-directed gene targeting in monocot plants. Rice 12:95. https://doi.org/10.1186/s12284-019-0355-1
Waltz E (2016a) Gene-edited CRISPR mushroom escapes US regulation. Nature 532:293. https://doi.org/10.1038/nature.2016.19754
Waltz E (2016b) CRISPR-edited crops: free to enter market, skip regulation. Nat Biotechnol 34(6):582–583. https://doi.org/10.1038/nbt0616-582
Wang JY, Doudna JA (2023) CRISPR technology: a decade of genome editing is only the beginning. Science 379(6629):8643. https://doi.org/10.1126/science.add8643
Whelan AI, Lema MA (2015) Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food 6(4):253–265. https://doi.org/10.1080/21645698.2015.1114698
Whelan AI, Gutti P, Lema MA (2020) Gene editing regulation and innovation economics. Front Bioeng Biotechnol 8:303. https://doi.org/10.3389/fbioe.2020.00303
Wilson LOW, Reti D, O’Brien AR, Dunne RA, Bauer DC (2018) High activity target-site identification using phenotypic independent CRISPR-Cas9 core functionality. CRISPR J 1(2):182–190. https://doi.org/10.1089/crispr.2017.0021
Wilson LOW, Hetzel S, Pockrandt C, Reinert K, Bauer DC (2019) VARSCOT: Variant-aware detection and scoring enable sensitive and personalized off-target detection for CRISPR-Cas9. BMC Biotechnol. https://doi.org/10.1186/s12896-019-0535-5
Wolt JD (2017) From personalized medicine to personalized nutrition: closing the gap. Appl Trans Genom 12:3–8. https://doi.org/10.1016/j.atg.2017.03.001
Wolt JD, Wolf C (2018) Policy and governance perspectives for regulation of genome edited crops in the United States. Front Plant Sci 9:1–12. https://doi.org/10.3389/fpls.2018.01606
Woo J, Kim J, Kwon S, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164. https://doi.org/10.1038/nbt.3389
Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R, Zhang F, Sharp PA (2014) Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32(7):670–676. https://doi.org/10.1038/nbt.2889
Yang L, Machin F, Wang S, Saplaoura E, Kragler F, (2023) Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat Biotechnol 41:958–967. https://doi.org/10.1038/s41587-022-01585-8
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829. https://doi.org/10.1101/gr.074492.107
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771. https://doi.org/10.1016/j.cell.2015.09.038
Zhang D, Tang S, Xie P, Yang D, Wu Y, Cheng S, Du K, Xin P, Chu J, Yu F, Xie Q (2022a) Creation of fragrant sorghum by CRISPR-Cas9. J Integr Plant Biol 64(5):961–964. https://doi.org/10.1111/jipb.13232
Zhang S, Wu S, Hu C, Yang Q, Dong T, Sheng O, Deng G, He W, Dou T, Li C, Sun C, Yi G, Bi F (2022b) Increased mutation efficiency of CRISPR-Cas9 genome editing in banana by optimized construct. PeerJ 10:e12664. https://doi.org/10.7717/peerj.12664
Zhang Y, Guo W, Chen L, Shen X, Yang H, Fang Y, Ouyang W, Mai S, Chen H, Chen S, Hao Q, Yuan S, Zhang C, Huang Y, Shan Z, Yang Z, Qiu D, Zhou X, Cao D, Li X, Jiao Y (2022c) CRISPR-Cas9-mediated targeted mutagenesis of GmUGT enhanced soybean resistance against leaf-chewing insects through flavonoids biosynthesis. Front Plant Sci 13:74. https://doi.org/10.3389/fpls.2022.802716
Zhang Q, Ye Y (2017) Not all predicted CRISPR-Cas systems are equal: Isolated Cas genes and classes of CRISPR-like elements. BMC Bioinform. https://doi.org/10.1186/s12859-017-1512-4
Zhang J, Kasciukovic T, White MF (2012) The CRISPR associated protein Cas4 Is a 5' to 3' DNA exonuclease with an iron-sulfur cluster. PLoS One. 7(10):e47232. https://doi.org/10.1371/journal.pone.0047232. Epub 2012 Oct 8. PMID: 23056615; PMCID: PMC3466216.
Zhao Z, Li C, Tong F, Deng J, Huang G, Sang Y (2021) Review of applications of CRISPR-Cas9 gene-editing technology in cancer research. Biol Proceed Online 23(1):1–13. https://doi.org/10.1186/s12575-021-00151-x
Zhu LJ, Holmes BR, Aronin N, Brodsky MH (2014) CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS ONE. https://doi.org/10.1371/journal.pone.0108424
Acknowledgements
The first author gratefully acknowledges the help rendered by Ms Rashmi Wanjari, Senior Research Fellow, and Mr Piyush Ghoshe, Project Assistant of DBT—Citrus—NER project at ICAR—National Institute of Biotic Stress Management, Raipur, India in corrections and formatting of the manuscript as per the journal’s instructions. The first author was a recipient of the Indian National Science Academy’s Visiting Scientist Fellowship at ICGEB, New Delhi.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors provided critical feedback and have equal contributions in the writing of the review manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflicts of interest
All the authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kharbikar, L., Konwarh, R., Chakraborty, M. et al. 3Bs of CRISPR-Cas mediated genome editing in plants: exploring the basics, bioinformatics and biosafety landscape. Physiol Mol Biol Plants 29, 1825–1850 (2023). https://doi.org/10.1007/s12298-023-01397-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-023-01397-3