Abstract
Background
The rice Waxy (Wx) gene plays a major role in seed amylose synthesis and consequently controls grain amylose content. Wx gene expression is highly regulated at the post-transcriptional level. In particular, the GT/TT polymorphism at the 5′splicing site of its 1st intron greatly affects this intron’s splicing efficiency and defines two predominant Wx alleles, Wxa and Wxb. Wxa rice often harbours intermediate to high amylose contents, whereas Wxb rice exhibits low to intermediate amylose contents. By deleting the Wx 1st intron using CRISPR/Cas9 technology, we generate a completely novel Wx allele and further investigate how intron removal affects Wx gene expression and rice grain amylose content.
Results
CRISPR/Cas9-mediated targeted deletion of the Wx 1st intron was performed on 4 rice inbred lines: KY131 (Wxb), X32 (Wxb), X35 (Wxa) and X55 (Wxlv). Deletion of the 1st intron occurred in 8.6–11.8% of the primary transformants of these 4 inbred lines. Compared to wild-type plants, amylose content was significantly increased from 13.0% to approximately 24.0% in KY131 and X32 mutant lines, which both carried the Wxb allele. However, no significant difference in amylose content was observed between wild-type plants and X35 and X55 mutant lines, which carried the Wxa and Wxlv alleles, respectively. Wx gene expression analysis of wild-type plants and mutants yielded results that were highly consistent with amylose content results. KY131 and X32 mutants accumulated increased levels of steady mRNA transcripts compared with wild-type plants, whereas steady mRNA levels were not altered in X35 and X55 mutants compared with wild-type plants. Grain quality, including appearance quality and eating and cooking quality, which are tightly associated with amylose content, was also assessed in wild-type and mutant plants, and data were presented and analysed.
Conclusions
This study presents a novel and rapid strategy to increase amylose content in inbred rice carrying a Wxb allele. Our data strongly suggest that the 1st intron of the Wx gene regulates Wx gene expression mainly at the post-transcriptional level in rice. This finding is in contrast to a previous hypothesis suggesting that it influences Wx gene transcription. In addition, removal of the first intron generates a completely novel Wx allele. Further studies on this new Wx allele will provide invaluable insights into the regulation of Wx gene expression, which will help researchers engineer new Wx alleles to facilitate the breeding of rice cultivars with better eating and cooking quality.
Similar content being viewed by others
Background
Improving grain quality is one of the most important goals in rice (Oryza sativa L.) breeding programs. Rice quality refers to the basic characteristics of rice in commodity circulation and includes four main aspects: milling quality, appearance quality, cooking and eating quality (ECQ) and nutritional quality. Among them, appearance quality, ECQ are particularly important (Lau et al. 2015). ECQ is determined from amylose content (AC), gel consistency (GC), gelatinization temperature (GT), and viscosity (Phing Lau et al. 2016; Tian et al. 2009). Starch accounts for up to 90% of the dry weight in rice grains (Zhou et al. 2002). Amylose, a constituent of starch, is a major indicator of ECQ in rice (Fitzgerald et al. 2009; Li et al. 2016; Tian et al. 2009). Rice is divided into four types based on amylose content: glutinous (AC < 2%), soft, intermediate to low and high (AC > 25%) (Pandey et al. 2012). Generally, the higher the amylose content in rice grain is, the less sticky and harder the cooked rice is, resulting in poor taste (Jobling 2004; Juliano 1992). However, rice with too low an amylose content is too sticky and soft. Thus, rice with intermediate to low amylose content is more popular.
The Waxy (Wx) gene (LOC_Os06g04200) encodes granule-bound starch synthase I (GBSSI), which was cloned by Wang et al. in 1990 (Wang et al. 1990). The Wx gene is a major gene controlling amylose content in rice endosperm and plays a decisive role in rice ECQ (Tian et al. 2009). At present, the amylose content among cultivated rice varieties is highly diversified. Such vast differences are mainly attributed to Wx gene allele variation. At least 9 Wx natural alleles have thus far been discovered and identified in rice. These alleles include Wxa, Wxb, Wxin, Wxop/Wxhp, Wxmq, Wxmp, Wxlv, Wxla/Wxmw, and wx (Chen et al. 2008; Dobo et al. 2010; Mikami et al. 2008; Larkin and Park 2003; Teng et al. 2012; Zhang et al. 2021, 2019; Zhou et al. 2021). DNA sequence differences between Wx alleles modulate Wx gene expression or enzyme activity, consequently causing variation in AC and grain quality. Researchers have designed different strategies to improve rice quality, including both transgenic approaches and marker-assisted selection (MAS) breeding (Jin et al. 2010; Kimiko et al. 2003; Liu et al. 2005; Phing Lau et al. 2016; Terada et al., 2000; Yu et al. 2009). In particular, introducing Wxb (AC ~ 16%) and Wxin (AC ~ 20%) alleles into rice germplasm with high amylose content via MAS or traditional breeding greatly expedites rice quality improvement.
With the development of rice consumption specialization, the demand for amylose content in rice is increasingly diversified. Thus, new methods to modulate amylose content or create new Wx alleles are needed. Third-generation genome editing technology, CRISPR/Cas9 genome editing technology, first emerged approximately a decade ago (Jinek et al. 2012; Richter et al. 2012). Since then, it has rapidly evolved into a collection of diverse genome editing tools, and these genome editing tools have been widely used in the improvement of agronomic traits for various crops (Chen et al. 2019; Fiaz et al. 2019; Gao 2021). Most importantly, the T-DNA insertion site and gene editing target sites are located in different region, and homozygous edited mutants free of transgenic components can be selected through progeny separation (Zhu et al. 2020). Compared with traditional transgenic methods, CRISPR/Cas9 technology is more prone to changes in natural mutations and has more potential to be applied to production methods. Recently, CRISPR/Cas9- mediated rice Wx gene editing has been intensively reported. These studies can be grouped into 4 categories. First, researchers have created new glutinous rice by completely knocking out the Wx gene (Ma et al. 2015; Zhang et al. 2018). Second, by editing the key cis-acting elements on the Wx gene promoter, researchers have achieved fine-tuning of AC in the Wxa or Wxb background (Huang et al. 2020a; Zeng et al. 2020). Third, researchers have tried to adjust rice AC by manipulating the splicing efficiency of the Wx gene at the posttranscriptional level (Zeng et al. 2020). Fourth, researchers have used CRISPR/Cas9-mediated base editors to modulate rice AC by altering GBSSI enzyme activity (Huang et al. 2021; Xu et al. 2021). These studies have not only paved new avenues for rice AC improvement but also generated a variety of new Wx alleles and provided new insights into the regulation of rice Wx gene expression and the modulation of GBSSI enzyme activity. In addition, Li et al. (1995) demonstrated that the first intron of the rice Wx gene greatly enhances foreign gene expression in rice protoplasts. However, whether the Wx 1st intron exhibits gene expression enhancement similar to its native Wx gene remains unclear. In situ deletion of the 1st intron from its native Wx gene will allow us to closely investigate this issue.
In this study, we exploited CRISPR/Cas9 gene editing technology to remove the first intron of the Wx gene. Intron removal generated a completely novel Wx allele and significantly increased the amylose content in rice inbred with the Wxb allele, whereas the amylose content was minimally changed in rice inbred with the Wxa allele.
Results
CRISPR/Cas9-Mediated Deletion of the 1st Intron of Wx
To investigate how the Wx 1st intron regulates the amylose content of rice grains, we decided to remove the 1st intron using CRISPR/Cas9 technology. We chose four inbred rice lines with different genetic backgrounds as our testing materials. Kongyu131 (KY131) was an elite japonica inbred and carries a typical Wxb allele. The other three X32, X35 and X55 lines were indica lines inbred from our breeders that carry the Wxb, Wxa or Wxlv alleles, respectively (Additional file 2: Figure S1). The typical Wxlv allele of X55 harboured the same GT polymorphism as the Wxa allele at the well-defined GT/TT polymorphism site that differentiates Wxa and Wxb alleles (Zhang et al. 2019). Initially, we planned to delete the entire 1st intron by choosing targeted sites of the 5′ and 3′ splicing sites of the 1st intron. However, we could not find proper target sequences and protospacer adjacent motifs (PAMs) in these two regions. Instead, we designed two target sites, Target1 and Target2, which were both located within the 1st intron but close to the 5′ and 3′ splicing sites of the Wx 1st intron, respectively (Fig. 1A). Target1’s PAM sequence (CCC) was located 33 bp away from the 5′splicing site of the Wx 1st intron, and Target2’s PAM sequence (CCA) was located 51 bp away from the 3′ splicing site of the Wx 1st intron. An edit vector pZZT477 (Fig. 1B, Additional file 3: Figure S2) expressing CRISPR/Cas9, two gRNAs (guide RNA, gRNA1 for Target1 and gRNA2 for Target2), and CP4/EPSPS that conferred glyphosate resistance selection marker was constructed and delivered into rice cells through Agrobacterium-mediated transformation. A total of 314 glyphosate-resistant transgenic plants were generated for these 4 rice inbred lines (Table 1). Our PCR assays on all 314 primary transformants detected 33 mutants with deletion of large DNA fragments (~ 1.0 kb) in the targeted intron region and deletion efficiencies ranging from 8.6–11.85% (Fig. 1C, Table 1). These 33 mutants included 12 KY131 mutants, 8 X32 mutants, 5 X35 mutants and 8 X55 mutants (Fig. 1C, Table 1).
CRISPR/Ca9-mediated deletion of the Wx 1st intron. A Position and sequence of the two gRNA target sites on the genomic regions of the Wx gene. Introns are shown as lines. Exons are shown as boxes. PAM motifs (CCN) of two targeted sequences are underlined. Two primer pairs, JC-F/R used for the detection of large fragment deletions and RT-F/R used for Wx gene expression analysis via RT–qPCR, were marked at their appropriate locations. B Schematic diagram of the editing vector pZZT477’s T-DNA structure. LB, T-DNA left border; RB, T-DNA right border; 35S, CaMV35s promoter; CP4, Agrobacterium tumefaciens strain CP4 EPSP (5-enolpyruvylshikimate-3-phosphate synthase) gene; Ubi4, sugarcane ubiquitin 4 gene promoter; Cas9, CRISPR/Cas9 gene with codon optimized for expression in rice; U3, U6, rice U3 and U6 snRNA promoter. C Detection of mutations in the first intron of the Wx gene via PCR assay in the T0 generation. D Sequencing results of the first intron-deleted mutant lines. The PAM motifs are underlined, target sequences are highlighted in red, and dotted lines indicate deletions. The WT indicates the wild type, and M1, M2, and M3 are different homozygous lines in the T1 generation
We further investigated whether off-target effects occurred in our experiments. Using the online software CRISPR-P2.0 (http://crispr.hzau.edu.cn/CRISPR2/), we identified two potential off-target sites for both gRNAs used in this study (Additional file 1: Table S1). PCR amplification and DNA sequencing of predicted off-target sites were performed on both T0 mutants and some of their T1 offspring. As shown in Additional file 1: Table S1, no off-target effects were detected at the putative off-target loci in tested T0 plants their T1 offspring.
Identification of Transgene-Free, Homozygous Mutants
T1 seeds of the primary transformants with a normal seed setting rate and other agronomic traits were selected for further screening of transgene-free, homozygous mutants. As shown in Table 1, 4, 4, 4 and 2 target mutant lines were obtained for KY131, X32, X55 and X35, respectively. Sequencing these 14 lines confirmed that ~ 1040 bp was deleted from the first intron (1128 bp in size) of their Wx genes. Further analyses indicated that these 14 mutants could be categorized into 3 mutant types according to differences in the number of nucleotides deleted. These three mutant types were named M1 (− 1041 bp), M2 (− 1042 bp), and M3 (− 1043 bp), and M2 was the predominant mutant (9/14) (Fig. 1D, Table 2). Given that the 5′ and 3′ intron splicing sites and an approximately 85-bp intron sequence were still retained within the 5′UTR of all mutants, a question arose regarding whether this short intron sequence in each mutant was still recognized as an intron and would be correctly spliced out from Wx transcripts. We conducted reverse transcriptase PCR (RT-PCR) to amplify fragments containing the 5′UTR and partial 5′ coding sequence of the Wx gene from cDNA samples derived from total mRNA of wild-type and mutant immature seeds. Sequencing these amplified fragments revealed that the remaining ~ 85-bp intron sequence was retained in mature mRNA of all mutants (in both Wxa and Wxb backgrounds) (Additional file 4: Figure S3), indicating that the remaining intron sequence was no longer recognized as an intron despite it still harbouring both splicing sites. Consequently, the 5′UTRs of M1, M2 and M3 mutants would have extra 87-, 86- and 85 bp sequences, respectively, compared to that of the wild type (Additional file 4: Figure S3). Since there was no extra translation start codon ATG found in these retained-intron sequences, we speculated that the translation initiation codon of mutant transcripts would be the same as that of wild-type transcripts.
Wx Gene Expression was Significantly Increased in the KY131 and X32 Mutants
To investigate the expression of the Wx gene in intron-deleted mutants, relative Wx gene mRNA levels in 10 DAP (days after pollination) seeds of different mutant plants and WT plants were detected by RT–qPCR. The results are shown in Fig. 2A. Compared to KY131 and X32 WT plants that both carried the Wxb allele, the intron-deleted mutants of KY131 and X32 plants accumulated approximately 1–2 times more stable mRNA (Fig. 2A). Enhanced mRNA accumulation in mutants was highly correlated with increased AC in mutants. The greater the level of mature mRNA accumulation, the greater the AC (Table 2, Fig. 2), suggesting that removal of the first intron significantly affects Wx gene expression in a Wxb background. However, the relative expression levels of the Wx gene between X35 and X55 WT plants (both have the Wxa allele) and their intron-deleted mutants was minimal (Fig. 2A), demonstrating that removal of the first intron had a limited effect on Wx gene expression in a Wxa background.
Wx gene expression and amylose content of endosperm in wild-type and T1 mutants. A Relative expression level of the Wx gene in wild-type and mutant 1st-generation transgene-free homologous plants. The expression noted in WT plants was arbitrarily set to 1, and the relative expression value of mutants was obtained by comparing the mutant expression to WT expression. B Amylose content of endosperm in wild-type and 1st-generation plants of transgene-free homologous mutants. Data are presented as the mean ± SD, **indicates a significant difference at P < 0.01 (t test)
As mentioned above, the 5′UTRs of mutant M1, M2 and M3 mRNA were 87, 86 or 85 bp longer than that of WT (Additional file 4: Figure S3), and we wanted to know whether these differences influence Wx gene expression. Our Wx gene expression results and AC data demonstrated minimal change among the two X35 mutants, X35-CR-1 (M3), -CR-2 (M2), and WT X35 (Table 2, Figs. 1D, 2A), indicating that M2 and M3 mutant types had similar expression levels as WT. A similar notion could be derived from the comparison between four X55 mutants (all M2 mutant type) and WT X55 in terms of their Wx expression and AC (Table 2, Fig. 2). Thus, we concluded that the extra ~ 85/86 bp intron sequence retained in the 5′UTR of intron deletion mutants had minimal influence on Wx gene expression.
Deletion of the Wx 1st Intron Substantially Increased the Grain Amylose Content of the KY131 and X32 Mutants
The grain amylose content of transgene-free, homozygous mutants (first generation) and their corresponding WT plants was measured. The results are shown in Table 2. The amylose content of the KY131 and X32 mutants significantly increased from 13.0% to approximately 24.0%, whereas the amylose content of the X35 and X55 mutants was not significantly different (Table 2, Fig. 2B). These amylose content results were consistent with our Wx gene expression data. Significantly increased relative expression of the Wx gene was only observed in KY131 and X32 mutants that showed a significant AC increase but not in X35 and X55 mutants that showed similar ACs as their corresponding WT plants (Fig. 2A). We further analysed the grain amylose content of second-generation KY131 and X32 mutants. The results showed that the increased level of amylose content was consistent in both generations (Figs. 2B, 3A), suggesting that the AC change in mutants was genetically stable.
Amylose content and gel consistency of endosperm in wild-type and 2nd-generation mutant plants (T2). A Amylose content of endosperm in wild-type and 2nd-generation mutant plants (T2). B Gel consistency of endosperm in wild-type and 2nd-generation mutant plants (T2). Data are presented as the mean ± SD, **indicates a significant difference at P < 0.01 (t test)
Grain Quality Evaluation and Physicochemical Property Analysis of Mutant Grains
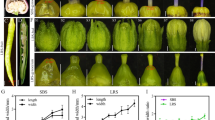
It is well established that grain amylose content is closely linked to grain quality and affects grain physicochemical properties, such as GC and GT (Tian et al. 2009). To examine whether the deletion of the first intron of the Wx gene would affect other physicochemical properties of rice grains, we measured the physicochemical properties of rice grains derived from KY131-CR-1, KY131-CR-3, X32-CR-1, and X32-CR-2 mutant lines as well as the corresponding WT KY131 and X32 lines. Compared with grains of the WT plants, grains of 4 mutant plants showed no significant difference in gelatinization temperature/ASV (alkali spreading value) (Table 3). However, their amylose content increase slightly decreased the gel consistency of rice grains (Table 3), as expected. The RVA (Rapid Visco Analyzer) pasting property values also reflected similar results (Table 3, Fig. 3B). In addition, milling quality was also similar between the grains of KY131 and X32 WT plants and their mutants (Table 3). Grain transparency evaluation demonstrated that polished rice grains of X32 intron-deleted mutants had better transparency than those of X32 WT plants, indicating that appearance quality was improved in X32 intron-deleted mutants (Table 3, Fig. 4A, B). However, the improvement of appearance quality observed in X32 mutants was not found in KY131 intron-deleted mutants (Table 3, Fig. 4C, D). We further inspected whether amylose content changes influence the structure of starch granules by scanning electron microscopy (SEM) of transverse mature endosperm sections. Our results revealed no obvious difference in the morphology of starch granules from the endosperm between mutants and wild types (Fig. 4E–P).
The appearance and morphology of starch granules of intron deleted mutants. A–D Milled rice from different mutants. A X32, B mutant X32-CR-2, C KY131, D mutant K131-CR-1. E–P SEM images showing the morphology of the starch granules. E, I, M were X32, F, J, N were mutant X32-CR-2, G, K, O were KY131and H, L, P were mutant K131-CR-1
Discussion
The AC in rice endosperm is an important factor affecting rice ECQ (Tian et al. 2009). However, vast differences in regional consumer preference, market demand and functionality make ECQ difficult to define and standardize. In general, South Asians favour long, slender rice with a high AC and hard GC, whereas Southeast Asians prefer long grains with intermediate AC and soft GC. Such differences often hinder the spread of elite varieties to other countries with rice markets that demand rice with a different AC. Developing efficient, low-cost technology to modulate grain AC will provide a reliable solution to overcome this barrier. In addition, the development of specialized rice consumption creates more diversified demands for rice AC. For example, in recent years, the market has exhibited increasing demand for rice specialized for the rice wine industry and for patients with diabetes or high blood pressure (Sun et al. 2017). Therefore, it is of great commercial value to develop biotechnological methods to accurately adjust rice grain AC to satisfy diverse consumers.
With the rapid advance of CRISPR/Cas-mediated gene editing technologies (Gao 2021), researchers have exploited these novel powerful technologies for accurate modulation of rice grain AC. Studies on knocking out the Wx gene to produce glutinous rice (Ma et al. 2015; Xu et al. 2015; Zhang et al. 2018), fine-tuning Wx gene expression via regulatory promoter editing, and modulating the enzyme activity of GBSSI via base editing have recently been reported (Huang et al. 2020a, b, 2021; Xu et al. 2021; Zeng et al. 2020). In this study, we developed a new strategy to modify rice AC. We deleted the majority of the first intron of the rice Wx gene in both Wxa and Wxb backgrounds using CRISPR/Cas9 technology. Deletion of the first intron from inbred KY131 and X32 lines with a Wxb allele significantly increased grain AC by greater than 10% (from 13.0% to approximately 24.0%) (Table 2, Fig. 2). However, such a phenomenon was not observed with X35 and X55 lines carrying a Wxa allele. The grain AC of WT X35 and X55 and their intron-deleted mutants remained approximately similar (Table 2, Fig. 2). Our method will provide an efficient and rapid method to convert intermediate AC rice cultivars (mostly with Wxb allele) to high AC cultivars, which facilitates elite rice cultivars of intermediate AC adapted to new plantation regions and consumer markets that favour high AC cultivars.
Deletion of the first intron generated a complete novel Wx allele which has other implications. First, removal of the first intron allowed us to gain insights into its regulation on Wx gene expression. Previous studies have showed that the GT/TT SNP at the 5′ splicing junction of the first intron is a major post-transcriptional regulation factor to affect rice grain AC (Cai et al. 1984; Isshiki et al. 1998; Samadder et al. 2008). By eliminating the splicing process of the first intron, we demonstrated that rice seeds accumulated approximately 1–2 times more stable mRNA, and their amylose content increased by approximately 10% (from 13.0% to approximately 24.0%) after removal of the first intron under the Wxb background (Table 2, Fig. 2). In contrast, stable mRNA accumulation was noted, and seed AC remained approximately the same after the first intron was deleted under the Wxa background (Table 2, Fig. 2). This finding provides a quantitative view of the extent to which the G to T SNP posttranscriptionally affects Wx gene expression. A previous study also demonstrated that the first intron of the rice Wx gene stimulates the expression of a foreign gene in rice protoplasts when it is placed in the 5′ UTR between the CaMV 35S promoter and GUS coding sequence, indicating that the first intron might act as a transcriptional enhancer (Li et al. 1995). This notion is plausible given that quite a few first introns with large sizes (~ 1.0 kb or more) and located within the 5′UTR of highly expressed genes, such as rice Actin1, maize Ubi1 and Adh1, indeed function as transcriptional stimulators (Rose 2008, 2019). However, our data demonstrated that rice seeds carrying the Wxa gene with or without the first intron accumulated approximately the same amount of stable mRNA and produced approximately the same amount of AC (Table 2, Fig. 2), strongly suggesting that the first intron of the rice Wx gene may not participate in transcriptional regulation. Of course, further well-designed studies are required to clarify this notion. Second, the new Wx allele without a first intron may help us in the future to further engineer new Wx alleles that could overcome the adverse effects of high temperature on rice grain quality. It has been well established that the majority of japonica rice strains carry the Wxb allele and produce rice grains with low to intermediate AC, whereas the majority of indica rice strains harbour the Wxa allele and produce rice grains with intermediate to high AC. In recent years, consumer preference has been increasingly changing to favour intermediate AC rice in the Chinese market, promoting the rapid spread of the Wxb allele into indica rice cultivars. However, previous studies have noted that under elevated temperature, Wx gene expression is down-regulated in some japonica cultivars, resulting in less GBSSI protein production and leading to lower grain AC and lower grain quality. Presumably, the selection of the first intron 5′splicing site of the Wxb gene is compromised under high temperatures (Larkin and Park 1999; Zhang et al. 2014a). The novel Wx allele created in this study may provide a promising solution to attenuate the adverse effect of high temperature on grain quality. Although removal of the first intron causes increased AC, further adjustment of the AC of the mutants with a deletion in the first intron to a suitable AC could be achievable using previously reported gene editing strategies (Huang et al. 2020a, 2021; Xu et al. 2021; Zeng et al. 2020). Engineering new Wx alleles that perform better under high temperature will be invaluable for rice production, particularly as global warming increasingly poses threats to global food security.
In conclusion, our study presented a novel and efficient strategy to modify rice grain AC, particularly to generate rice with a high AC from rice carrying a Wxb allele. By deleting the first intron, we created a completely novel Wx allele. Further analyses of the first intron deletion mutants provided new insights into the regulation of Wx gene expression. The new Wx allele could serve as an excellent material for further engineering new Wx alleles that perform better under high temperatures and improve rice eating and cooking quality.
Materials and Methods
1Plant Materials and Growth Conditions
The rice variety KY131 (KY131, Wxb allele, Oryza sativa L. ssp. japonica) is an elite inbred variety broadly cultivated in the Northeast China region. X32 (Wxb allele, Oryza sativa L. ssp. indica), X35 (Wxa allele, Oryza sativa L. ssp. indica) and X55 (Wxlv allele, Oryza sativa L. ssp. indica) are inbred lines provided by our breeders. The transgenic rice lines and transgene-free edited rice plants were all grown in a standard greenhouse (16-h light at 30 °C/8-h night at 22 °C) in the Life Science and Technology Center, China National Seed Group Co., LTD, Wuhan, China.
Construction of the CRISPR/Cas9 Vector and Plant Transformation
Before constructing an editing vector, we employed the online software CRISPR-P2.0 (http://crispr.hzau.edu.cn/CRISPR2/) to identify proper editing target sites using the Nipponbare Wx gene (LOC_Os06g04200) as a reference. Two target sites located within the 1st intron but close to the 5′ or 3′ splicing site were selected (Fig. 1A). To confirm whether these two targeted sequences were suitable for editing the Wx gene of all 4 inbred lines, we amplified and sequenced fragments, including the targeted sequences and their flanking sequences from genomic DNA of all 4 inbred lines (KY31, X32, X35, and X55), using primer sets T1-F/T1-R (for Target1) and T2-F/T2-R (for Target2) (Additional file 1: Table S2). The CRISPR/Cas9 vector pZZT477 targeting the first intron of the Wx gene was constructed as previously described (Zhang et al. 2014b). The vector used in this study was based on the vector pCambia1300 backbone. The editing vector pZZT477 contained a Cas9 expression cassette driven by the sugarcane Ubi4 promoter, a CP4–EPSPS gene cassette (as a selection marker) driven by the CaMV 35S promoter, and two sgRNA expression cassettes driven by the rice U3 or U6 snRNA promoters (Fig. 1B, Additional file 3: Figure S2). The editing vector pZZT477 was transferred into Agrobacterium tumefaciens strain EHA105 by electroporation and consequently delivered into KY131, X32, X35 and X55 cells via Agrobacterium-mediated transformation as previously described (Hiei et al. 1997).
Molecular Characterization of the Mutant Plants
Rice genomic DNA was extracted using a DNA Quick Plant System (TransGen Biotech, Beijing, China). Fifty nanograms of genomic DNA was used as a template to perform PCR amplification using Taq polymerase (Tiangen, Beijing, China). Genomic DNA of all transgenic herbicide-resistant T0 plants was first examined by PCR using the specific primers CP4-F/CP4-R to amplify the selection marker gene CP4/EPSPS. Only CP4 amplification-positive plants were selected for further editing status analysis using the primer set JC-F/JC-R (Additional file 1: Table S2). To examine the deletion details, PCR fragments amplified with the JC-F/JC-R primer set (Additional file 1: Table S2) from transgene-free homozygous plants were sequenced on an ABI3730XL capillary sequencer. To investigate the 5′UTR sequence of the mutant plant transcripts, we amplified and sequenced fragments using primers Transcript-F and Transcript-R (Additional file 1: Table S2) from first-strand cDNA samples derived from total RNA of immature seeds of intron-deleted mutants and wild-type plants. For off-target investigation, we identified two potential off-target sites (Additional file 1: Table S1) using the online software CRISPR-P2.0 (http://crispr.hzau.edu.cn/CRISPR2/). PCR amplification of fragments flanking the two potentials was performed with primer pairs Off-T1-F/R and Off-T2-F/R. Amplified fragments were consequently sequenced and analysed.
Selection of Transgene-Free Homozygous Mutants
To select transgene-free T1 plants, the rice leaves of T1 transgenic plants from selected lines were sampled for GMO testing. An Applied Biosystems 7900HT instrument was used to conduct quantitative real-time PCR for detecting transgene residues. To ensure that there were no transgenic residues in selected homozygous mutants, we used 11 primer pairs to amplify T-DNA components, including the CaMV35S promoter (CaMV35S-F/R), SsUbi4 promoter (P#SsUbi4-F/R), NOS terminator (T#NOS-F/R, NOS-F/R), OCS terminator (T#OCS-F/R), CaMV35S terminator (T#35SpolyA-F/R), Cas9 (CAS9 Os-F/R), Epsps-CP4 (CAS9 Os-F/R), and vector backbone region (pCAMBIA1301-1-vector-F/R, pCAMBIA1301-2-vector-F/R, and pCAMBIA1301-3-vector-F/R). Only homologous mutants with PCR products negative for all 11 primer pairs were selected for further analysis. All 11 primer pairs used for GMO tests are listed in Additional file 1: Table S2.
RNA Extraction and RT–PCR Analysis
Total RNA was extracted from grains after 10 days of grain filling using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative RT–PCR analysis was performed using 2 × SYBR Green PCR Master Mix on an Applied Biosystems 7900HT (Applied Biosystems). The rice Actin1 gene was used as an internal control for normalization. Sequences of primers used in qPCR, Wx-RT-F/R and OsActin7-F/R, are listed in Additional file 1: Table S2.
Scanning Electron Microscopy of Starch Granules
Rice grains were dried in an oven at 42 °C for 2 days and cooled in a desiccator. Cross-sections of the samples were manually snapped and sputter-coated with gold palladium on copper studs. Magnifications of 500 × and 2000 × were used to observe endosperm and starch granule morphology.
Evaluation of Rice Quality
The rice quality was measured following the procedure described in GB/T 15683-2008 and NY/T 83-2017, and each sample was tested thrice. RVA pasting properties were detected with a RVA according to the manufacturer’s instructions (NewPort Sci. Co., Australia).
Statistical Analysis
The data were analysed by using Student’s unpaired t test in Microsoft Excel. Differences were considered to be significant at P < 0.05 or P < 0.01.
Abbreviations
- AC:
-
Amylose content
- bp:
-
Base pairs
- CRISPR:
-
Clustered regularly interspaced palindromic repeats
- Cas :
-
CRISPR-associated protein
- ECQ:
-
Eating and cooking quality
- EPSPS:
-
5-Enolpyruvylshikimate-3-phosphate synthase
- GBSSI:
-
Granule-bound starch synthase I
- GC:
-
Gel consistency
- GT:
-
Gelatinization temperature
- gRNA:
-
Guide RNA
- GUS:
-
β-Glucoronidase
- PAM:
-
Protospacer adjacent motif
- qRT-PCR:
-
Quantitative reverse transcriptase-PCR polymerase chain reaction
- RVA:
-
Rapid visco analyzer
- SEM:
-
Scanning electron microscopy
- SNP:
-
Single nucleotide polymorphism
- UTR:
-
Untranslated region
- WT:
-
Wild type
- Wx :
-
Waxy
References
Cai XL, Wang ZY, Xing YY, Zhang JL, Hong MM (1984) Aberrant splicing of intron 1 leads to the heterogeneous 5′UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J 14:459–465
Chen KL, Wang YP, Zhang R, Zhang HW, Gao CX (2019) CRISPR/Cas genome gditing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Chen MH, Bergman CJ, Pinson SRM, Fjellstrom RG (2008) Waxy gene haplotypes: associations with pasting properties in an international rice germplasm collection. J Cereal Sci 48:781–788
Dobo M, Ayres N, Walker G, Park WD (2010) Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J Cereal Sci 52:450–456
Fiaz S, Ahmad S, Noor MA, Wang XK, Younas A, Riaz A, Riaz A, Ali F (2019) Applications of the CRISPR/Cas9 system for rice grain quality improvement perspectives and opportunities. Int J MolSci 20:888
Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14:133–139
Gao CX (2021) Genome engineering for crop improvement and future agriculture. Cell 184:1621–1635
Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Huang LC, Li QF, Zhang CQ, Chu R, Gu ZW, Tan HY, Zhao DS, Fan XL, Liu QQ (2020a) Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol J 18:2164–2166
Huang LC, Nese S, Liu QQ (2020b) Waxy editing old meets new. Trends Plant Sci 25:963–966
Huang XR, Su F, Huang S, Mei FT, Niu XM, Ma CL, Zhang H, Zhu XG, Zhu JK, Zhang JS (2021) Novel Wx alleles generated by base editing for improvement of rice grain quality. J Integr Plant Biol 63:1632–1638
Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K (1998) A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J 15:133–138
Jin L, Lu Y, Shao YF, Zhang G, Xiao P, Shen SQ, Corke H, Bao JS (2010) Molecular marker assisted selection for improvement of the eating, cooking and sensory quality of rice (Oryza sativa L.). J Cereal Sci 51:159–164
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jobling S (2004) Improving starch for food and industrial applications. Curr Opin Plant Biol 7:210–218
Juliano BO (1992) Structure, chemistry, and function of the rice grain and its fractions. Cereal Foods World 37:772–772
Kimiko I, Hiroko O, Kyoko O, Hidetaka H, Yasuhito T, Mitsui T (2003) Introduction of Wx transgene into rice wx mutants leads to both high- and low-amylose rice. Plant Cell Physiol 44:473–480
Larkin PD, Park WD (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature—sensitive and controlled by a single-nucleotide polymorphism. Plant Mol Biol 40:719–727
Lau WC, Rafii MY, Ismail MR, Puteh A, Latif MA, Ramli A (2015) Review of functional markers for improving cooking, eating, and the nutritional qualities of rice. Front Plant Sci 6:832
Li H, Prakash S, Nicholson TM, Fitzgerald MA, Gilbert RG (2016) The importance of amylose and amylopectin fine structure for textural properties of cooked rice grains. Food Chem 196:702–711
Li Y, Ma H, Zhang J, Wang Z, Hong MM (1995) Effects of the first intron of rice Waxy gene on the expression of foreign genes in rice and tobacco protoplasts. Plant Sci 108:181–190
Liu QQ, Yu HX, Chen XH, Cai XL, Tang SZ, Wang ZY, Gu MH (2005) Field performance of transgenic indica hybrid rice with improved cooking and eating quality by down-regulation of Wx gene expression. Mol Breeding 16:199–208
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano HY, Suzuki Y, Sano Y (2008) Allelic diversification at the wx locus in landraces of Asian rice. Theor Appl Genet 116:979–989
Pandey MK, Rani NS, Madhav MS, Sundaram RM, Varaprasad GS, Sivaranjani AK, Bohra A, Kumar GR, Kumar A (2012) Different isoforms of starch-synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol Adv 30:1697–1706
Larkin PD, Park WD (2003) Association of waxy gene single nucleotide polymorphisms with starch. Mol Breeding 12:335–339
Phing Lau WC, Latif MA, Rafii MY, Ismail MR, Puteh A (2016) Advances to improve the eating and cooking qualities of rice by marker-assisted breeding. Crit Rev Biotechnol 36:87–98
Richter C, Chang JT, Fineran PC (2012) Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems. Viruses 4:2291–2311
Terada R, Nakajima M, Isshiki M, Okagaki RJ, ShimamotoK WSR (2000) Antisense Waxy genes with highly active promoters effectively suppress Waxy gene expression in transgenic rice. Plant Cell Physiol 41:881–888
Rose AB (2008) Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol 326:277–290
Rose AB (2019) Introns as gene regulators: a brick on the accelerator. Front Genet 9:672
Samadder P, Sivamani E, Lu JL, Li XG, Qu RD (2008) Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Genet Genomics 279:429–439
Sun YW, Jiao G, Liu ZP, Zhang X, Li JY, Guo XP, Du WM, Du JL, Francis F, Zhao YD, Xia LQ (2017) Generation of high-amylose rice through CRISPR/Cas9- mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci 8:298
Teng B, Zeng RZ, Wang YC, Liu ZQ, Zhang ZM, Zhu HT, Ding XH, Li WT, Zhang GP (2012) Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol Breeding 30:583–595
Tian ZX, Qian Q, Liu QQ, Yan MX, Liu XF, Yan CJ, Liu GF, Gao ZY, Tang SZ, Zeng DL, Wang YH, Yu JM, Gu MH, Li JY (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA 106:21760–21765
Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM (1990) Nucleotide sequence of rice waxy gene. Nucleic Acids Res 18:5898
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:11491
Xu Y, Lin QP, Li XF, Wang FQ, Chen ZH, Wang J, Li WQ, Fan FJ, Tao YJ, Jiang YJ, Wei XD, Zhang R, Zhu QH, Bu QY, Yang J, Gao CX (2021) Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol J 19:11–13
Yu HX, Liu QQ, Xu L, Lu MF, Yang XJ, Gong ZY, Cai XL, Zhang YS, Zhang CQ, Wang ZY, Gu MH (2009) Quality characteristics and field performance of selectable marker-free transgenic rice with antisense Wx gene and improved quality derived from the elite parents of hybrid indica rice. J Cereal Sci 50:370–375
Zeng DC, Liu TL, Ma XL, Wang B, Zheng ZY, Zhang YL, Xie XR, Yang BW, Zhao Z, Zhu QL, Liu YG (2020) Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol J 18:2385–2387
Zhang CQ, Yang Y, Chen SJ, Liu XJ, Zhu JH, Zhou LH, Lu Y, Li QF, Fan XL, Tang SZ, Gu MH, Liu QQ (2021) A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J Integr Plant Biol 63:889–901
Zhang CQ, Zhu JH, Chen SJ, Fan XL, Li QF, Lu Y, Wang M, Yu HX, Yi CD, Tang SZ, Gu MH, Liu QQ (2019) Wx(lv), the ancestral allele of rice waxy gene. Mol Plant 12:1157–1166
Zhang H, Duan L, Dai JS, Zhang CQ, Li J, Gu MH, Liu QQ, Zhu Y (2014a) Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor Appl Genet 127:273–282
Zhang H, Zhang JS, Wei PL, Zhang BT, Gou F, Feng ZY, Mao YF, Yang L, Zhang H, Xu NF, Zhu JK (2014b) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang JS, Zhang H, Botella JR, Zhu JK (2018) Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol 60:369–375
Zhou H, Xia D, Zhao D, Li YH, Li PB, Wu B, Gao GJ, Zhang QL, Wang GW, Xiao JH, Li XH, Yu SB, Lian XM, He YQ (2021) The origin of Wx(la) provides new insights into the improvement of grain quality in rice. J Integr Plant Biol 63:878–888
Zhou Z, Robards K, Helliwell S, Blanchard C (2002) Composition and functional properties of rice. Int J Food Sci Tech 37:849–868
Zhu HC, Li C, Gao CX (2020) Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat Rev Mol Cell Biol 21:661–677
Acknowledgements
We were very grateful of Dr. Yizhong Cai, Ms. Jing Wei for their invaluable helps in this work.
Funding
This work was supported by The National Key Research and Development Program of China (2016YFD0102000) and “Breeding of major new varieties of main grain crops” program (2020ABA016) from Department of science and technology of Hubei Province.
Author information
Authors and Affiliations
Contributions
LX. designed the experiments and conducted partial vector construction, plant transformation, Wx gene expression analysis and other required works. DQ, WW, PY, TC, CY, XN and LH contributed in vector construct, plant transformation, molecular analysis works. WX, YR participated in grain quality analysis. LY, QY and YN were responsible for plant growth and management. LJ and MC wrote the manuscript and coordininated all the experiments. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Analysis of potential off-target effects. Table S2. Primers used in this study.
Additional file 2: Figure S1.
The Wx genotypic sequences of the four rice inbred used in the test. The two SNPs defining Wxb, Wxa and Wxlv alleles were shown in red. Int1-1: the first neucleotide of the 1st intron of the Wx gene; Ex10-115: the 115th neucleotide of the 10th exon of the Wx gene.
Additional file 3: Figure S2.
The structure diagram of vector pZZT477.
Additional file 4: Figure S3.
The 5′UTR sequences of the matured mRNAs of the Wx gene of different mutants. Sequences were derived from PCR amplification of DNA fragments covering the 5′UTR and a partial coding sequence from total RNA isolated from mutant and wild type immature seeds. Primer Transcript-F and Transcript-R (Table S2) were used to perform in PCR amplification. The extra sequence presented in each mutant type were marked in black. The translation start codon ATG were marked in red. The G/T polymorphism site that differentiates Wxa and Wxb alleles was marked in different colours.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Ding, Q., Wang, W. et al. Targeted Deletion of the First Intron of the Wxb Allele via CRISPR/Cas9 Significantly Increases Grain Amylose Content in Rice. Rice 15, 1 (2022). https://doi.org/10.1186/s12284-021-00548-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-021-00548-y