Abstract
The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) induces apoptosis via death receptor (DR) 4 or DR5 preferentially in cancer cells, and not in normal cells with relatively high decoy receptor expression. However, multiple mechanisms in cancer cells induce resistance to DRs-mediated apoptosis. Therefore, understanding of molecular mechanisms for resistance to DRs-mediated apoptosis can find the strategy to increase sensitivity. Although multiple proteins are involved in resistance to DRs-mediated apoptosis, we focus on modulation of DR5 to overcome resistance. Here, we discuss regulation of DR5 expression or activation by epigenetic modification, transcription factor at the transcriptional levels, micro RNA and RNA-binding proteins at the post-transcriptional levels, and ubiquitination and glycosylation at the post-translational levels. In addition, we also mention about relationship between localization of DR5 and death signaling activation. The purpose of this review is to help understand relationship between regulatory mechanisms of DR5 and resistance to TRAIL or DRs-targeted agonist monoclonal antibodies, and to develop innovative anti-cancer therapies through regulation of DR5 signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis, a form of cell death in which a programmed sequence of events, is most frequent cell death mode induced by anti-cancer drugs in cancer cells. The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) induces apoptosis via death receptors (DRs), DR4 or DR5, in cancer cells. In contrast, since normal cells highly express decoy receptors (DcRs), which are partially or completely lack of functional death domain (DD), ligation of TRAIL fails to induce cell death. Underlying mechanism of TRAIL-induced apoptosis is that ligation of TRAIL induces oligomerization of DRs, and Fas-associated protein with death domain (FADD) binds to DD of DRs. Death-inducing signaling complexes (DISCs) for extrinsic apoptosis are composed of DRs, cytosolic adaptor FADD, and pro-caspase-8/cFLIP. Assembly of DISC is accomplished by interaction of DDs and death effecter domains (DEDs). Procaspase-8 is recruited to DED of FADD, and then multiple procaspase-8 is assembled through DED of caspase-8. DISC formation is critical for activation of caspase-8 via cleavage of procaspase-8, and then activates effector caspases, such as caspase-3, 6, and 7, leading to induction of apoptosis in type 1 cells (Mahalingam et al. 2009). Mitochondrial pathway could be involved in TRAIL-mediated apoptosis in type 2 cells. When caspase-8 truncates Bid, it is oligomerized with Bak and Bax which induces mitochondrial membrane permeabilization. Cytochrome c released from mitochondria forms apoptosome with Apaf-1, which provide platform for recruitment and activation of procaspase-9. Activated caspase-9, like a caspase-8, triggers apoptosis via activation of effector caspases (Mahalingam et al. 2009). As mentioned above, ligation of TRAIL with DRs produces the first signal to induce apoptosis in both type 1 and type 2 cells. Therefore, dysregulation of expression and localization of DRs are resistant to TRAIL.
DR5 gene located on chromosome 8p (MacFarlane et al. 1997), and DR5 has two splice variants, long and short DR5, which is absence of 29 different amino acids in the extracellular region (Mert et al. 2017). Interestingly, Van Roosmalen et al. (2014) reviewed about DRs preference for TRAIL-induced apoptosis. These results are derived from experiments which used single recombinant TRAIL, DR4- (Tur et al. 2008), and DR5-selective TRAIL variants (van der Sloot et al. 2006). According to their research, TRAIL has different sensitivity to DRs-induced apoptosis depending on cancer cell lines. For examples, leukemic cells have a preference for DR4-induced apoptosis, and lymphoma, myeloma, and most solid tumor shows heterogeneity. In addition, Truneh et al. reported that TRAIL binding affinity to DR5 is the strongest at 37 °C, compared with other DRs and DcRs (Truneh et al. 2000). There are various mechanisms for inducing TRAIL resistance, but we discuss about enhancement of DRs-mediated apoptosis via modulation of human DR5.
Modulatory mechanisms of DR5 expression
Transcriptional regulation
Sp1 (specificity protein 1)
Sp1 activates the transcription by binding CG-rich Sp-binding sites in promoter of genes, which are related with cell growth, differentiation, apoptosis, and carcinogenesis (Vizcaino et al. 2015). DR5 is one of Sp1-regulated genes. DR5 promoter contains minimal promoter element at -198 to -166, which region also overlap with two Sp1 binding sites. These two Sp1 sites play a critical role in basal transcription activity of DR5 (Yoshida et al. 2001). In addition, anti-cancer drugs induced DR5 expression in a Sp1-dependent manner. For examples, deoxycholic acid and sodium butyrate increase DR5 expression via Sp1 transcriptional activation (Higuchi et al. 2004; Kim et al. 2004). Quercetin (3′,3′,4′,5,7-pentahydroxyflavone), a flavonoid found in fruits and vegetables, also increases Sp1-mediated DR5 expression (Kim et al. 2008). Beta-lanone, butein, piceatannol, and capsaicin sensitize TRAIL-mediated apoptosis via up-regulation of Sp1-mediated DR5 expression (Kim et al. 2010; Moon et al. 2010, 2012; Kang et al. 2011). Notch is important signaling molecules in tumorigenesis by modulation of cell differentiation, proliferation and death in cancer cells. Recently, inhibition of notch1 signaling enhances TRAIL-mediated apoptosis in glioblastoma, and these mechanisms are related with up-regulation of DR5 by JNK-mediated Sp1 activation (Fassl et al. 2015).
p53
p53 is mortal of cellular growth, division and proliferation by regulation of cell cycle arrest and apoptosis. In the initial study, p53 status seems not to be important in TRAIL-induced cell death. Since p53 status is mutated in a half of cancer cells, p53-indenepent cell death by TRAIL seems attractive. However, Wu et al. reported that DR5 is a DNA damage-inducible p53-regualted gene (Wu et al. 1997). DR5 is identified as a transcript induced by anti-cancer drugs including doxorubicin (Lowe et al. 1993), and doxorubicin activates p53-dependent signaling pathway. Therefore, Wu et al. hypothesized that DR5 is a gene controlled by p53, and found that doxorubicin increases DR5 expression in p53 wild-type cells, but not p53-mutated cells (Wu et al. 1997). The binding sites (BS) of p53 transcription factor were identified three sited in DR5 promoter region. There are BS1, BS2, and BS3, and they are located − 0.82 Kb, + 0.25 Kb (within Intron 1), and + 1.25 Kb (within Intron 2) of the ATG site, respectively. Among them, BS2 has critical roles on p53-dependnet DR5 expression (Takimoto and El-Deiry 2000). Anti-cancer drugs [etoposide, CPT-11 (Wang and El-Deiry 2003), and nutlin-3 (Hori et al. 2010)] or ionizing radiation (Sheikh et al. 1998) increase DR5 mRNA expression in a p53-dependent manner.
CHOP (CCAAT/enhancer-binding protein homologous protein)
CHOP is endoplasmic reticulum (ER) stress-induced a major transcriptional factor. CHOP induces ER-stress-mediated apoptosis depending on duration and severity of ER stress (Oyadomari and Mori 2004). ER stress inducers, including thapsigargin and tunicamycin, induce DR5 expression, and CHOP as a transcription factor plays critical roles on DR5 expression (Yamaguchi and Wang 2004; Shiraishi et al. 2005). Yamaguchi and Wang (2004) identify the CHOP binding element in the DR5 promoter between − 276 and − 264 (+ 1 represents the translation start site). Proteasome inhibitor (MG132) (Yoshida et al. 2005), farnesyltransferase inhibitor (Sun et al. 2007), silibinin (Son et al. 2007), and 15-deoxy-Δ12, 14-prostaglandin J2 (15d-PGJ2) increases CHOP-dependent DR5 transcription (Su et al. 2008). In addition, adverse effect in a cardiovascular or anti-cancer effect of cyclooxygenase-2 (COX-2) inhibitor (celecoxib and ON09310) is related with CHOP-dependent DR5 expression in a COX-2 independent manner, followed by cell death (He et al. 2008). Interestingly, although IRE1α, as an unfolded protein response (UPR) sensor transiently triggers decay of DR5 mRNAs, persistent ER stress increases CHOP-dependent DR5 expression. Up-regulated DR5 induces TRAIL-independent apoptosis via caspase-8 (Lu et al. 2014). In contrast, Glab et al. reported that ER stress by thapsigargin, tunicamycin, or subtilase cytotoxin triggers apoptosis in a DR5-independent manner, and Bim is more important on ER-stress induced cell death (Glab et al. 2017). Therefore, ER stress could increase DR5 expression via CHOP, but roles of DR5 on ER stress-induced apoptosis are controversial.
NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells)
NF-κB is composed of five Rel family members (RelA/p65, RelB, c-Rel, p50, and p52). In canonical pathways, IκB kinase (IKK) activation by stimuli induces phosphorylation and degradation of IκB. Released NF-κB dimer (predominantly p65/p50) from IκB translocate into nucleus. In non-canonical pathway, NF-κB-inducing kinase (NIK) induces activation of IΚΚα, and then phosphorylates p100, followed by processing of p100. Generated p52 from p100 binds RelB, and then translocates into the nucleus (Sun 2011). The function of NF-κB in cancer is controvercial, depending on cell types, stimuli, and regulated genes. NF-κB signaling is also involved in DR5 expression. Among NF-κB subunits, c-Rel is a critical for DR5 transcription (Ravi et al. 2001). TRAIL induces apoptosis in RelA−/− or wild-type mouse embryonic fibroblast (MEF), but not c-Rel−/− MEF, which are absence of DR5 expression. Although c-Rel activation by TNF-α increases DR5 expression, RelA is also induces anti-apoptotic proteins, such as Bcl-xL, resulted in inhibition of apoptosis by TRAIL (Ravi et al. 2001). In addtion, overexpression of RelA inhibits DR5 expression, whereas overexpression of c-Rel enhances DR5 expression in TRAIL-treated cells (Chen et al. 2003). The subunit of NF-κB has difference preferences, depending on consensus sequence of target genes (Baeuerle and Baltimore 1996). Proteasome inhibitor, apple procyanidins, Smac mimetics, and hepatitis B virus X (HBx) protein increase DR5 expresion via NF-κB activation (Chen et al. 2008; Maldonado et al. 2010; Eckhardt et al. 2013; Kong et al. 2015). Furthermore, knock down of death-associated protein kinase 2 (DAPK2) by siRNA induces NF-κB transcriptional activity, leads to the induction of DR5 expression (Schlegel et al. 2014). It reported that NF-κB binding site lies between + 385 and + 394 in the first intron of DR5 (Yoshida et al. 2001). However, NF-κB is also key signaling molecules to induce anti-apoptotic proteins, thus the role of NF-κB in DRs-mediated apoptosis must be carefully mentioned.
YY1 (Yin Yang 1)
YY1 is a 65 kDa of zinc finger transcription factor and modulates transcriptional activity of gene promoter as activator or repressor depending on interacting proteins, such as a histone deacetylases (HDAC) and a histone acetyltransferases (HAT) (Thomas and Seto 1999). In case of DR5, YY1 acts as a repressor. The binding site for the YY1 is localized between − 804 and − 794 in the promoter of DR5 (Yoshida et al. 2001; Baritaki et al. 2007a), and multiple chemotherapeutic drugs, including cisplatin, etoposide, adriamycin and vincristine, increase DR5 expression through inhibition of YY1 expression and transcriptional activity (Baritaki et al. 2007a). In addition, Raf-1 kinase inhibitor protein (RKIP) inhibits NF-κB activity, followed by inhibition of YY1 expression (Baritaki et al. 2007b). Since NF-κB directly or indirectly regulates YY1 expression (Baritaki et al. 2007a, b; Wang et al. 2007), the novel proteasome inhibitor (NPI-0052) also inhibits NF-κB activity by accumulation of p-IκB, and sequentially induces up-regulation of DR5 expression through inhibition of YY1 expression and transcriptional activity (Baritaki et al. 2008). In addition, nitric oxide (Huerta-Yepez et al. 2009) and BH3-mimetics obatoclax also sensitizes TRAIL-induced apoptosis through up-regulation of DR5 expression by inhibition of YY1 transcriptional activity (Martinez-Paniagua et al. 2011).
Others
Other transcriptional factors are also involved in DR5 expression. For examples, epithelium-specific Ets factor, family member 3 (ESE-3) increases DR5 expression by binding to purine-rich GGAA/T sequences in cooperation with CBP and p300 (Lim et al. 2006), and activating transcription factor 3 (ATF3) is a transcriptional factor for DR5 induction through ROS-ER stress pathways (Edagawa et al. 2014). In contrast, nuclear Bcl-2 nineteen kilodalton interacting protein (BNIP3) binds to the DR5 promoter, and then decreases DR5 expression (Burton et al. 2013).
Epigenetic modification
Methylation
DNA methylation is important on chromatin remodeling and gene expression, and DNA methyltransferases (DNMTs) induce cellular DNA methylation (Li et al. 2013). To identify the roles of methylation enzymes (DNMT1 and DNMT3b) in cell survival, expression of both are blocked by siRNA in human hepatocellular carcinoma cells. Double knock-down of DNMT1 and DNMT3b by siRNA increased TRAIL-treated cell death via up-regulation of DR5 mRNA and protein expression (Kurita et al. 2010). However, alteration of methylation in DR5 promoter is not detected. Therefore, TRAIL sensitization by inhibition of methylation is indirectly related with up-regulation of DR5 expression.
Histone lysine demethylase 4A (KDM4A) is a member of the Jumonji C domain-containing KDM4 subfamily of histone demethylase, which induces demethylation of histone H3 on lysine 9 and 36, and histone H1.4 on lysine 26 (Berry and Janknecht 2013). Recently, Wang et al. reported that inhibition of KDM4A induces DR5 mRNA and protein expression and enhances apoptosis in cancer cells. However, KDM4A did not directly bind to the promoter of DR5, and indirectly increases association of histone modifying enzyme complexes in promoter of CHOP expression, resulted in induction of DR5 expression (Wang et al. 2016). Since there is very little methylation in the DR5 promoter (van Noesel et al. 2002), there seems to be difficult in finding direct links between the demethylation and DR5.
Acetylation
Acetylation and deacetylation of lysine residues in nucleosomal histones is regulated by histone acetyltransferase and histone deacetylase (HDACs), respectively. Hyper-acetylation of lysine residues in nucleosomal histones increases or restores gene expression, which is associated with cell cycle arrest, cell death, and differentiation. For this reason, HDAC inhibitors are recognized as a target of cancer therapy (Newbold et al. 2016). LAQ824, a HDAC inhibitor, sensitizes TRAIL-induced apoptosis in human acute leukemia cells. The molecular mechanism of LAQ824-induced TRAIL sensitization is associated with up-regulation of DR5 mRNA and protein expression. LAQ824 acetylates histones H3 and H4 in DR5 promoter, resulted in induction of DR5 expression and DISC formation (Guo et al. 2004). In addition, depletion of HDAC2, not HDAC1, and HDAC inhibitors [trichostatin A (TSA), sodium butyrate, and suberoylanilide hydroxamic acid (SAHA)] increase TRAIL-induced apoptosis in cancer cells, but they did not show the acetylation of nucleosomal histones in DR5 gene (Nakata et al. 2004; Schuler et al. 2010). Therefore, modulation of nucleosomal histones acetylation using HDAC inhibitors could sensitize TRAIL-mediated apoptosis through up-regulation of DR5 expression.
Post-transcriptional regulation
RNA-binding protein HuR
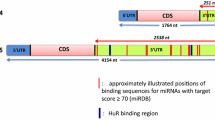
The stability of mRNAs is regulated by binding of mRNA binding proteins in the adenylate-uridylate (AU)-rich elements in the 3′-untranslated region (UTR). It has been known that the stability of DR5 mRNA is also modulated by several stimuli. For examples, the 15d-PGJ2 (Nakata et al. 2006) and thapsigargin (He et al. 2002) induces DR5 expression via stabilization of DR5 mRNA. The 3′UTR and 5′UTR of human DR5 gene has AU-rich elements. Kandasamy et al. identify the specific sequences, which are critical for DR5 mRNA stability (Kandasamy and Kraft 2008), and it is the AU-rich element from 3556 to 3587 in the 3′UTR region of human DR5 gene. Among mRNA binding proteins, only HuR binds to this AU-rich element, leads to stabilization of DR5 mRNA by proteasome inhibitor (PS-341) in LNCaP human prostate cancer cells, but not PC-3 and DU145 (Kandasamy and Kraft 2008). In addition, chloroquine also increases DR5 mRNA stability in human renal carcinoma Caki cells, but HuR is not involved (Park et al. 2016). In contrast, Pineda et al. reported that DR5 agonist induces cleavage and translocation from nucleus to cytoplasm of HuR, and then binding of HuR to 5′UTR by DR5 agonist inhibits DR5 translation in pancreatic cancer cells (Pineda et al. 2012).
MicroRNA(miR)-1246 and miR-133a
MicroRNAs are small endogenous noncoding RNA (~ 22 nucleotides), which reduces mRNA stability or inhibit translation by binding in 3′UTR of target mRNA target genes (Filipowicz et al. 2008). The miR-1246 released from irradiated cancer cells moves to recipient cells in an exosome independent manner, and induces proliferation and radio-resistance of irradiated recipient cells. They found that DR5 is direct target of miR-1246, which represses mRNA and protein expression of DR5. Extracellular miR-1246 by radiation has bystander effects, which are related with resistance in surrounding cells (Yuan et al. 2016). Although they did not investigate whether miR-1246 is involved in TRAIL resistance, the relationship between the two is predictable. However, it needs to prove through accurate experiments. In addition, miR-133a also modulates DR5 expression in glioblastoma. miR-133a suppresses DR5 expression directly by binding in 3′UTR of DR5, which is related with TRAIL resistance (Wang et al. 2017).
Others
Zhang et al. reported that levels of DR5 protein expression is critical roles on TRAIL sensitivity in multiple melanoma cells, since between mRNA and protein levels of DR5 has no correlation (Zhang et al. 2004). They identified the reason as a difference in regulation of translation. Luciferase activity with 3′UTR of DR5 in TRAIL-resistance cells was suppressed compared with that in TRAIL-sensitive cells, and a 23 base (TAAATGCTTTATTTATTT ATTTG) in AU-rich element of 3′UTR plays critical roles in translation of DR5 in TRAIL-treated cells. Although specific RNA binding proteins were not identified, at least this response is independent of HuR and HuD (Zhang et al. 2004). Therefore, studies are not enough to understand the regulatory mechanism of mRNA stabilization and translation of DR5. Modulation of DR5 mRNA stability and translation might be dependent of cell types, stimulator, RNA binding proteins and binding sites.
Post-translational regulation
Ubiquitination
Ubiquitination to the target proteins can change protein stability and functions and is mediated by three enzymes, such as E1 (ubiquitin activation), E2 (ubiquitin conjugation), and E3 (ubiquitin ligase) (Hershko and Ciechanover 1998). DR5 is also regulated by ubiquitin–proteasome pathway, and proteasome inhibitors (PS-341, MG132, and epoxomicin) increase DR5 protein expression, resulted in enhancement of TRAIL-mediated apoptosis (He et al. 2004; Liu et al. 2007). Recently, Song et al. reported that c-Casitas B-lineage lymphoma (Cbl) induces degradation of DR5 in TRAIL-treated cells (Song et al. 2010). Cbl is a multi-adaptor protein. c-Cbl, Cbl-b, and Cbl-c (Cbl-3) are identified. Cbl-b and c-Cbl have E3 ligase activity (Thien and Langdon 2005), both Cbls regulate DR5 expression. The c-Cbl binds to DR5 and induces degradation of DR5 in TRAIL-treated cells, resulted in the early phase of acquired TRAIL resistance (Song et al. 2010). In addition, bufalin is a major active ingredient of the traditional Chinese medicine ChanSu, increases DR5 expression via down-regulation of Cbl-b expression (Yan et al. 2012), and shRNA-expressing adenovirus against c-Cbl also enhances TRAIL-mediated apoptosis through induction of DR5 expression (Kim et al. 2013). Although there is not direct deubiquitinase (DUB) of DR5, the DUB also controls the expression of DR5. b-AP15 blocks ubiquitin-specific protease (USP)14 and ubiquitin carboxyl-terminal hydrolase L (UCHL) 5, which are 19S regulatory particle-associated DUBs, followed by accumulation of the ubiquitin conjugated proteins via inhibition of proteasomal function (D’arcy and Linder 2012). DR5 accumulated by b-AP15 sensitized TRAIL-mediated apoptosis (Oh et al. 2017). Therefore, ubiquitin–proteasome pathway is involved in modulation of DR5 protein expression.
Glycosylation
Glycosylation is a common post-modification that occur more than 50% of proteins, and is known to control not only protein folding but also the ability to signaling transduction by proteins. DR5 also occurs with O-glycosylation, which increases TRAIL-mediated apoptosis. O-glycosylation is mediated by N-acetyl-galactosamine (GalNAc) and N-acetyl-glucosamine (GlcNAc). Wagner et al. reported that sensitivity to TRAIL is correlated with levels of O-glycosyltransferase by N-acetyl-galactosamine transferase (GALNT)14 or GALNT3 with O-glycan processing enzymes fucosyltransferases (FUT) 3 and FUT6 depending on cell types (Wagner et al. 2007). DR5 is O-glycosylated on two stretches of serine and threonine in front of cysteine rich domains (CRD) 2 and within CRD2 and CRD3 (Micheau 2018). Interestingly, glycosylation of DR5 has no effect on TRAIL binding affinity and DR5 cell surface expression, and promotes receptor clustering and DISC formation. Furthermore, they identify that Ser201 is a primary modification site in DR5. Based on these results, Howard et al. reported that immunocytochemistry assay to detect GALNT14 and FUT3/6, but not GALNT3, can be used to distinguish patients, who have effectiveness in dulanermin- and drozitumab-based therapy (Stern et al. 2010). O-GlcNAcylation of DR5 by GlcNAc induces DR5 clustering and DISC formation, resulted in enhancement of TRAIL-induced apoptosis in non-small cell lung cancer cells (Liang et al. 2018). We summarized the molecular mechanisms and published papers that regulate expression and activation of DR5 (Fig. 1 and Table 1).
Modulatory mechanisms of DR5 expression and activation to enhance DRs-mediated apoptosis signaling. a Summary for control mechanism of DR5. b Modulatory mechanism of activation and expression of DR5. Expression of DR5 is directly or indirectly regulated by methylation and acetylation, and multiple transcription factors and RNA-binding proteins also increase or decrease DR5 expression depending on regulatory proteins and binding sites. In addition, expression and clustering of DR5 are modulated by ubiquitin–proteasome pathway and glycosylation
Modulation of DR5 localization
Lipid rafts
It has been well-known that ceramide converts raft domain in membrane into larger domains (lipid raft domains, glycosphingolipid-enrich domains or ceramide-enriched domains), which are important on initiation of receptor-specific signaling via clustering, re-organization of signaling molecules, amplification of signal, and exclusion of inhibitory signals and, thus, amplify a receptor-mediated signals (Bollinger et al. 2005; Grassme et al. 2007). Like other receptor-mediated signaling, localization of DR5 in lipid raft is important on induction of TRAIL-mediated apoptosis. Sensitivity of leukemia to TRAIL-induced apoptosis is determined by recruitment of DISC components including DR5 to the lipid raft (Min et al. 2009). In addition, reactive oxygen species-dependent acid sphingomyelinase activation by TRAIL induces ceramide release, regulated in induction of ceramide-enriched domains in plasma membrane. Clustering of DR5 in these domains plays critical roles in TRAIL-mediated apoptosis (Dumitru and Gulbins 2006). In addition, the importance of DR5 clustering in lipid raft domains in TRAIL-induced apoptosis has been reported in many studies. For examples, inhibition of COX-2 enhances TRAIL-induced apoptosis through clustering of DR5 and DISC components in ceramide-enriched caveolae (Martin et al. 2005), and DR5 clustering in lipid rafts is a mechanism of TRAIL sensitization by doxorubicin (Dumitru et al. 2007), ursodeoxycholic acid (Lim et al. 2011), oxaliplatin (Xu et al. 2009), and synthetic alkyl-lysophospholipids (ALPs) (Gajate and Mollinedo 2007). Interestingly, c-Cbl and Cbl-b also inhibit localization of DR5 in lipid rafts (Xu et al. 2009). In contrast, ceramide synthase 6 and exogenous ceramide increase TRAIL sensitivity (Voelkel-Johnson et al. 2005; White-Gilbertson et al. 2009).
Cytosolic localization by endocytosis
Before TRAIL is incorporated into the DRs, the endocytosis of DRs is sufficient to provide resistance to TRAIL (Zhang and Zhang 2008; Chen et al. 2012b), and it has been known that endocytosis of receptor is not required for TRAIL-induced apoptosis in Burkitt lymphoma B cell line (Kohlhaas et al. 2007). However, Austin et al. (2006) reported about positive feedback mechanism of TRAIL-induced apoptosis with regard to endocytosis in TRAIL-sensitive cells. DR5 activation by TRAIL induces cleavage of adaptor protein (AP)2α, AP1/2β, and clathrin heavy chain (CHC), which are machinery of clathrin-dependent endocytosis, and attenuates DR5 endocytosis, leading to amplification of TRAIL-induced apoptosis signaling. In contrast, clathrin-dependent endocytosis is critical for TRAIL-induced apoptosis via lysosomal membrane permeabilization in hepatocellular carcinoma (Akazawa et al. 2009). TRAIL triggers endocytosis of DR5, and DR5 with trafficking to lysosomal membrane induces release of cathepsin and apoptosis. Inhibition of endocytosis by dominant negative dynamin blocks TRAIL-induced apoptosis via inhibition of endocytosis (Akazawa et al. 2009). Therefore, so far, the role of endocytosis in DRs-mediated apoptosis is unclear.
Autophagosome
The role of autophagy in cell death is controversial, and expression of DR5 by autophagy is also dependent of stimulators. Gefitinib and ginsenoside compound K increase DR5 expression and inhibition of autophagy reduces DR5 expression in human colon cancer cells (Chen et al. 2016a, b). In contrast, telmisartan, a drug for hypertension, causes induction of DR5 via inhibition of autophagy (Rasheduzzaman et al. 2018). However, there is no direct mechanism how autophagy controls the expression of DR5. Therefore, we only discuss about direct regulation of DR5 expression by autophagy. Di et al. found that the sensitivity to TRAIL negatively correlated with LC3 II in multiple breast carcinomas, and DR5 is localized in autophagosomes in TRAIL-resistance cells. When autophagy is inhibited by knock-down of ATG7, beclin-1, or LC3, DR5 expression on cell surface is increased, leading to induction of TRAIL-induced apoptosis. However, inhibitor of lysosomal activity had no effect on DR5 surface expression (Di et al. 2013). Recently, although it is not related with TRAIL-induced apoptosis, HBx induces DR5 protein degradation via activation of autophagy. The underlying mechanisms are that DR5 is recruited to phagophores by direct interaction with HBx and autophagy induction by HBx (Shin et al. 2016). It is certain that DR5 is controlled by autophagy-lysosome pathway, but it need further study to identify the accurate mechanism. We summarized the regulation of cell death by modulation of DR5 localization (Fig. 2).
Nucleus
DR5 has two nuclear localization signals (NLS), and nuclear localization of DR5 is correlated with TRAIL resistance. Kojima et al. reported that HeLa and HepG2 cells highly express DR5 at nucleus, and both cells are resistant to TRAIL-mediated apoptosis. In contrast, levels of DR5 nuclear expression are low in DU145 cells, which are sensitive to TRAIL. Nuclear translocation of DR5 is mediated by importin β1 via the recognition of NLS sequences, and knock-down importin β1 abrogates DR5 expression at the nucleus (Kojima et al. 2011).
Conclusion
Because of TRAIL induces apoptosis preferentially in cancer cells, target for DRs-mediated signaling is promising anti-cancer strategy. For this reason, various approaches have been conducted to strengthen DRs-mediated apoptosis signals. Here, we describe the modulatory mechanisms of DR5 activation and expression (Fig. 1), and importance of DR5 localization in the cells (Fig. 2). Understanding of these mechanisms could contribute to the improvement of the anti-cancer effect using recombinant TRAIL or antagonistic monoclonal antibodies, as well as DR-specific TRAIL variant and combination treatment.
References
Akazawa Y, Mott JL, Bronk SF, Werneburg NW, Kahraman A, Guicciardi ME, Meng XW, Kohno S, Shah VH, Kaufmann SH, Mcniven MA, Gores GJ (2009) Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology 136:2365–2376
Akpinar B, Bracht EV, Reijnders D, Safarikova B, Jelinkova I, Grandien A, Vaculova AH, Zhivotovsky B, Olsson M (2015) 5-Fluorouracil-induced RNA stress engages a TRAIL-DISC-dependent apoptosis axis facilitated by p53. Oncotarget 6:43679–43697
Austin CD, Lawrence DA, Peden AA, Varfolomeev EE, Totpal K, De Mazière AM, Klumperman J, Arnott D, Pham V, Scheller RH, Ashkenazi A (2006) Death-receptor activation halts clathrin-dependent endocytosis. Proc Natl Acad Sci 103:10283–10288
Baeuerle PA, Baltimore D (1996) NF-kappa B: ten years after. Cell 87:13–20
Baritaki S, Huerta-Yepez S, Sakai T, Spandidos DA, Bonavida B (2007a) Chemotherapeutic drugs sensitize cancer cells to TRAIL-mediated apoptosis: up-regulation of DR5 and inhibition of Yin Yang 1. Mol Cancer Ther 6:1387–1399
Baritaki S, Katsman A, Chatterjee D, Yeung KC, Spandidos DA, Bonavida B (2007b) Regulation of tumor cell sensitivity to TRAIL-induced apoptosis by the metastatic suppressor Raf kinase inhibitor protein via Yin Yang 1 inhibition and death receptor 5 up-regulation. J Immunol 179:5441–5453
Baritaki S, Suzuki E, Umezawa K, Spandidos DA, Berenson J, Daniels TR, Penichet ML, Jazirehi AR, Palladino M, Bonavida B (2008) Inhibition of Yin Yang 1-dependent repressor activity of DR5 transcription and expression by the novel proteasome inhibitor NPI-0052 contributes to its TRAIL-enhanced apoptosis in cancer cells. J Immunol 180:6199–6210
Berry WL, Janknecht R (2013) KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 73:2936–2942
Bollinger CR, Teichgraber V, Gulbins E (2005) Ceramide-enriched membrane domains. Biochim Biophys Acta 1746:284–294
Bonavida B (2007) Rituximab-induced inhibition of antiapoptotic cell survival pathways: implications in chemo/immunoresistance, rituximab unresponsiveness, prognostic and novel therapeutic interventions. Oncogene 26:3629–3636
Burton TR, Henson ES, Azad MB, Brown M, Eisenstat DD, Gibson SB (2013) BNIP3 acts as transcriptional repressor of death receptor-5 expression and prevents TRAIL-induced cell death in gliomas. Cell Death Dis 4:e587–e587
Byun HS, Zhou W, Park I, Kang K, Lee SR, Piao X, Bong Park J, Kyu Kwon T, Na M, Min Hur G (2018) C-27-carboxylated oleanane triterpenoids up-regulate TRAIL DISC assembly via p38 MAPK and CHOP-mediated DR5 expression in human glioblastoma cells. Biochem Pharmacol 158:243–260
Carter BZ, Mak DH, Schober WD, Dietrich MF, Pinilla C, Vassilev LT, Reed JC, Andreeff M (2008) Triptolide sensitizes AML cells to TRAIL-induced apoptosis via decrease of XIAP and p53-mediated increase of DR5. Blood 111:3742–3750
Chen X, Kandasamy K, Srivastava RK (2003) Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res 63:1059–1066
Chen JJ, Chou CW, Chang YF, Chen CC (2008) Proteasome inhibitors enhance TRAIL-induced apoptosis through the intronic regulation of DR5: involvement of NF-kappa B and reactive oxygen species-mediated p53 activation. J Immunol 180:8030–8039
Chen CY, Yiin SJ, Hsu JL, Wang WC, Lin SC, Chern CL (2012a) Isoobtusilactone A sensitizes human hepatoma Hep G2 cells to TRAIL-induced apoptosis via ROS and CHOP-mediated up-regulation of DR5. J Agric Food Chem 60:3533–3539
Chen JJ, Shen HC, Rivera Rosado LA, Zhang Y, Di X, Zhang B (2012b) Mislocalization of death receptors correlates with cellular resistance to their cognate ligands in human breast cancer cells. Oncotarget 3:833–842
Chen L, Meng Y, Guo X, Sheng X, Tai G, Zhang F, Cheng H, Zhou Y (2016a) Gefitinib enhances human colon cancer cells to TRAIL-induced apoptosis of via autophagy- and JNK-mediated death receptors upregulation. Apoptosis 21:1291–1301
Chen L, Meng Y, Sun Q, Zhang Z, Guo X, Sheng X, Tai G, Cheng H, Zhou Y (2016b) Ginsenoside compound K sensitizes human colon cancer cells to TRAIL-induced apoptosis via autophagy-dependent and -independent DR5 upregulation. Cell Death Dis 7:e2334
Chen M, Wang X, Zha D, Cai F, Zhang W, He Y, Huang Q, Zhuang H, Hua ZC (2016c) Apigenin potentiates TRAIL therapy of non-small cell lung cancer via upregulating DR4/DR5 expression in a p53-dependent manner. Sci Rep 6:35468
Chen P, Hu T, Liang Y, Li P, Chen X, Zhang J, Ma Y, Hao Q, Wang J, Zhang P, Zhang Y, Zhao H, Yang S, Yu J, Jeong LS, Qi H, Yang M, Hoffman RM, Dong Z, Jia L (2016d) Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin Cancer Res 22:4145–4157
D’arcy P, Linder S (2012) Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol 44:1729–1738
Das S, Tripathi N, Siddharth S, Nayak A, Nayak D, Sethy C, Bharatam PV, Kundu CN (2017) Etoposide and doxorubicin enhance the sensitivity of triple negative breast cancers through modulation of TRAIL-DR5 axis. Apoptosis 22:1205–1224
Deng Z, Yan H, Hu J, Zhang S, Peng P, Liu Q, Guo D (2012) Hepatitis C virus sensitizes host cells to TRAIL-induced apoptosis by up-regulating DR4 and DR5 via a MEK1-dependent pathway. PLoS ONE 7:e37700
Di X, Zhang G, Zhang Y, Takeda K, Rivera Rosado LA, Zhang B (2013) Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget 4:1349–1364
Dilshara MG, Jayasooriya R, Molagoda IMN, Jeong JW, Lee S, Park SR, Kim GY, Choi YH (2018) Silibinin sensitizes TRAIL-mediated apoptosis by upregulating DR5 through ROS-induced endoplasmic reticulum stress-Ca(2+)-CaMKII-Sp1 pathway. Oncotarget 9:10324–10342
Donia M, Maksimovic-Ivanic D, Mijatovic S, Mojic M, Miljkovic D, Timotijevic G, Fagone P, Caponnetto S, Al-Abed Y, Mccubrey J, Stosic-Grujicic S, Nicoletti F (2011) In vitro and in vivo anticancer action of Saquinavir-NO, a novel nitric oxide-derivative of the protease inhibitor saquinavir, on hormone resistant prostate cancer cells. Cell Cycle 10:492–499
Dumitru CA, Gulbins E (2006) TRAIL activates acid sphingomyelinase via a redox mechanism and releases ceramide to trigger apoptosis. Oncogene 25:5612–5625
Dumitru CA, Carpinteiro A, Trarbach T, Hengge UR, Gulbins E (2007) Doxorubicin enhances TRAIL-induced cell death via ceramide-enriched membrane platforms. Apoptosis 12:1533–1541
Eckhardt I, Roesler S, Fulda S (2013) Identification of DR5 as a critical, NF-kappaB-regulated mediator of Smac-induced apoptosis. Cell Death Dis 4:e936
Edagawa M, Kawauchi J, Hirata M, Goshima H, Inoue M, Okamoto T, Murakami A, Maehara Y, Kitajima S (2014) Role of activating transcription factor 3 (ATF3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J Biol Chem 289:21544–21561
Fassl A, Tagscherer KE, Richter J, De-Castro Arce J, Savini C, Rosl F, Roth W (2015) Inhibition of Notch1 signaling overcomes resistance to the death ligand Trail by specificity protein 1-dependent upregulation of death receptor 5. Cell Death Dis 6:e1921
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102
Gajate C, Mollinedo F (2007) Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood 109:711–719
Glab JA, Doerflinger M, Nedeva C, Jose I, Mbogo GW, Paton JC, Paton AW, Kueh AJ, Herold MJ, Huang DC, Segal D, Brumatti G, Puthalakath H (2017) DR5 and caspase-8 are dispensable in ER stress-induced apoptosis. Cell Death Differ 24:944–950
Gopalan A, Yu W, Sanders BG, Kline K (2013) Simvastatin inhibition of mevalonate pathway induces apoptosis in human breast cancer cells via activation of JNK/CHOP/DR5 signaling pathway. Cancer Lett 329:9–16
Grassme H, Riethmuller J, Gulbins E (2007) Biological aspects of ceramide-enriched membrane domains. Prog Lipid Res 46:161–170
Guo F, Sigua C, Tao J, Bali P, George P, Li Y, Wittmann S, Moscinski L, Atadja P, Bhalla K (2004) Cotreatment with histone deacetylase inhibitor LAQ824 enhances Apo-2L/tumor necrosis factor-related apoptosis inducing ligand-induced death inducing signaling complex activity and apoptosis of human acute leukemia. Cells. 64:2580–2589
Guo X, Meng Y, Sheng X, Guan Y, Zhang F, Han Z, Kang Y, Tai G, Zhou Y, Cheng H (2017) Tunicamycin enhances human colon cancer cells to TRAIL-induced apoptosis by JNK-CHOP-mediated DR5 upregulation and the inhibition of the EGFR pathway. Anticancer Drugs 28:66–74
He Q, Lee DI, Rong R, Yu M, Luo X, Klein M, El-Deiry WS, Huang Y, Hussain A, Sheikh MS (2002) Endoplasmic reticulum calcium pool depletion-induced apoptosis is coupled with activation of the death receptor 5 pathway. Oncogene 21:2623–2633
He Q, Huang Y, Sheikh MS (2004) Proteasome inhibitor MG132 upregulates death receptor 5 and cooperates with Apo2L/TRAIL to induce apoptosis in Bax-proficient and -deficient cells. Oncogene 23:2554–2558
He Q, Luo X, Jin W, Huang Y, Reddy MV, Reddy EP, Sheikh MS (2008) Celecoxib and a novel COX-2 inhibitor ON09310 upregulate death receptor 5 expression via GADD153/CHOP. Oncogene 27:2656–2660
He K, Zheng X, Li M, Zhang L, Yu J (2016) mTOR inhibitors induce apoptosis in colon cancer cells via CHOP-dependent DR5 induction on 4E-BP1 dephosphorylation. Oncogene 35:148–157
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Higuchi H, Grambihler A, Canbay A, Bronk SF, Gores GJ (2004) Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J Biol Chem 279:51–60
Hori T, Kondo T, Kanamori M, Tabuchi Y, Ogawa R, Zhao QL, Ahmed K, Yasuda T, Seki S, Suzuki K, Kimura T (2010) Nutlin-3 enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through up-regulation of death receptor 5 (DR5) in human sarcoma HOS cells and human colon cancer HCT116 cells. Cancer Lett 287:98–108
Huerta-Yepez S, Vega M, Escoto-Chavez SE, Murdock B, Sakai T, Baritaki S, Bonavida B (2009) Nitric oxide sensitizes tumor cells to TRAIL-induced apoptosis via inhibition of the DR5 transcription repressor Yin Yang 1. Nitric Oxide 20:39–52
Jung KJ, Min KJ, Bae JH, Kwon TK (2015) Carnosic acid sensitized TRAIL-mediated apoptosis through down-regulation of c-FLIP and Bcl-2 expression at the post translational levels and CHOP-dependent up-regulation of DR5, Bim, and PUMA expression in human carcinoma caki cells. Oncotarget 6:1556–1568
Kandasamy K, Kraft AS (2008) Proteasome inhibitor PS-341 (VELCADE) induces stabilization of the TRAIL receptor DR5 mRNA through the 3′-untranslated region. Mol Cancer Ther 7:1091–1100
Kang CH, Moon DO, Choi YH, Choi IW, Moon SK, Kim WJ, Kim GY (2011) Piceatannol enhances TRAIL-induced apoptosis in human leukemia THP-1 cells through Sp1- and ERK-dependent DR5 up-regulation. Toxicol In Vitro 25:605–612
Kim YH, Park JW, Lee JY, Kwon TK (2004) Sodium butyrate sensitizes TRAIL-mediated apoptosis by induction of transcription from the DR5 gene promoter through Sp1 sites in colon cancer cells. Carcinogenesis 25:1813–1820
Kim JY, Kim EH, Park SS, Lim JH, Kwon TK, Choi KS (2008) Quercetin sensitizes human hepatoma cells to TRAIL-induced apoptosis via Sp1-mediated DR5 up-regulation and proteasome-mediated c-FLIPS down-regulation. J Cell Biochem 105:1386–1398
Kim MO, Moon DO, Kang CH, Kwon TK, Choi YH, Kim GY (2010) beta-Ionone enhances TRAIL-induced apoptosis in hepatocellular carcinoma cells through Sp1-dependent upregulation of DR5 and downregulation of NF-kappaB activity. Mol Cancer Ther 9:833–843
Kim IY, Kang YJ, Yoon MJ, Kim EH, Kim SU, Kwon TK, Kim IA, Choi KS (2011) Amiodarone sensitizes human glioma cells but not astrocytes to TRAIL-induced apoptosis via CHOP-mediated DR5 upregulation. Neuro Oncol 13:267–279
Kim SY, Kim JH, Song JJ (2013) c-Cbl shRNA-expressing adenovirus sensitizes TRAIL-induced apoptosis in prostate cancer DU-145 through increases of DR4/5. Cancer Gene Ther 20:82
Kohlhaas SL, Craxton A, Sun XM, Pinkoski MJ, Cohen GM (2007) Receptor-mediated endocytosis is not required for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. J Biol Chem 282:12831–12841
Kojima Y, Nakayama M, Nishina T, Nakano H, Koyanagi M, Takeda K, Okumura K, Yagita H (2011) Importin β1 protein-mediated nuclear localization of death receptor 5 (DR5) limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced cell death of human tumor cells. J Biol Chem 286:43383–43393
Kong F, You H, Zhao J, Liu W, Hu L, Luo W, Hu W, Tang R, Zheng K (2015) The enhanced expression of death receptor 5 (DR5) mediated by HBV X protein through NF-kappaB pathway is associated with cell apoptosis induced by (TNF-alpha related apoptosis inducing ligand) TRAIL in hepatoma cells. Virol J 12:192
Kouhara J, Yoshida T, Nakata S, Horinaka M, Wakada M, Ueda Y, Yamagishi H, Sakai T (2007) Fenretinide up-regulates DR5/TRAIL-R2 expression via the induction of the transcription factor CHOP and combined treatment with fenretinide and TRAIL induces synergistic apoptosis in colon cancer cell lines. Int J Oncol 30:679–687
Kuribayashi K, Finnberg N, Jeffers JR, Zambetti GP, El-Deiry WS (2011) The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo. Cell Cycle 10:2380–2389
Kurita S, Higuchi H, Saito Y, Nakamoto N, Takaishi H, Tada S, Saito H, Gores GJ, Hibi T (2010) DNMT1 and DNMT3b silencing sensitizes human hepatoma cells to TRAIL-mediated apoptosis via up-regulation of TRAIL-R2/DR5 and caspase-8. Cancer Sci 101:1431–1439
Lee TJ, Um HJ, Min Do S, Park JW, Choi KS, Kwon TK (2009) Withaferin A sensitizes TRAIL-induced apoptosis through reactive oxygen species-mediated up-regulation of death receptor 5 and down-regulation of c-FLIP. Free Radic Biol Med 46:1639–1649
Lee JY, Jung KH, Morgan MJ, Kang YR, Lee HS, Koo GB, Hong SS, Kwon SW, Kim YS (2013) Sensitization of TRAIL-induced cell death by 20(S)-ginsenoside Rg3 via CHOP-mediated DR5 upregulation in human hepatocellular carcinoma cells. Mol Cancer Ther 12:274–285
Lee CF, Yang JS, Tsai FJ, Chiang NN, Lu CC, Huang YS, Chen C, Chen FA (2016) Kaempferol induces ATM/p53-mediated death receptor and mitochondrial apoptosis in human umbilical vein endothelial cells. Int J Oncol 48:2007–2014
Lee YS, Lee DH, Jeong SY, Park SH, Oh SC, Park YS, Yu J, Choudry HA, Bartlett DL, Lee YJ (2018) Ferroptosis-inducing agents enhance TRAIL-induced apoptosis through upregulation of death receptor 5. J Cell Biochem 120:928–939
Li KK, Li F, Li QS, Yang K, Jin B (2013) DNA methylation as a target of epigenetic therapeutics in cancer. Anticancer Agents Med Chem 13:242–247
Liang Y, Xu W, Liu S, Chi J, Zhang J, Sui A, Wang L, Liang Z, Li D, Chen Y, Niu H (2018) N-acetyl-glucosamine sensitizes non-small cell lung cancer cells to TRAIL-induced apoptosis by activating death receptor 5. Cell Physiol Biochem 45:2054–2070
Lim SC, Han SI (2017) MDL-12330A potentiates TRAIL-induced apoptosis in gastric cancer cells through CHOP-mediated DR5 upregulation. Korean J Physiol Pharmacol 21:397–405
Lim JH, Cho J-Y, Park YB, Park J-W, Kwon TK (2006) ESE-3 transcription factor is involved in the expression of death receptor (DR)-5 through putative Ets sites. Biochem Biophys Res Commun 350:736–741
Lim JH, Park JW, Choi KS, Park YB, Kwon TK (2009) Rottlerin induces apoptosis via death receptor 5 (DR5) upregulation through CHOP-dependent and PKC delta-independent mechanism in human malignant tumor cells. Carcinogenesis 30:729–736
Lim SC, Duong HQ, Choi JE, Lee TB, Kang JH, Oh SH, Han SI (2011) Lipid raft-dependent death receptor 5 (DR5) expression and activation are critical for ursodeoxycholic acid-induced apoptosis in gastric cancer cells. Carcinogenesis 32:723–731
Lin T, Chen Y, Ding Z, Luo G, Liu J, Shen J (2013) Novel insights into the synergistic interaction of a thioredoxin reductase inhibitor and TRAIL: the activation of the ASK1-ERK-Sp1 pathway. PLoS ONE 8:e63966
Liu X, Yue P, Chen S, Hu L, Lonial S, Khuri FR, Sun SY (2007) The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res 67:4981–4988
Lowe SW, Ruley HE, Jacks T, Housman DE (1993) p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74:957–967
Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A (2014) Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345:98–101
Macfarlane M, Ahmad M, Srinivasula SM, Fernandes-Alnemri T, Cohen GM, Alnemri ES (1997) Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J Biol Chem 272:25417–25420
Mahalingam D, Szegezdi E, Keane M, De Jong S, Samali A (2009) TRAIL receptor signalling and modulation: are we on the right TRAIL? Cancer Treat Rev 35:280–288
Maldonado ME, Bousserouel S, Gosse F, Lobstein A, Raul F (2010) Implication of NF-kappaB and p53 in the expression of TRAIL-death receptors and apoptosis by apple procyanidins in human metastatic SW620 cells. Biomedica 30:577–586
Martin S, Phillips DC, Szekely-Szucs K, Elghazi L, Desmots F, Houghton JA (2005) Cyclooxygenase-2 inhibition sensitizes human colon carcinoma cells to TRAIL-induced apoptosis through clustering of DR5 and concentrating death-inducing signaling complex components into ceramide-enriched caveolae. Cancer Res 65:11447–11458
Martinez-Paniagua MA, Baritaki S, Huerta-Yepez S, Ortiz-Navarrete VF, Gonzalez-Bonilla C, Bonavida B, Vega MI (2011) Mcl-1 and YY1 inhibition and induction of DR5 by the BH3-mimetic Obatoclax (GX15-070) contribute in the sensitization of B-NHL cells to TRAIL apoptosis. Cell Cycle 10:2792–2805
Meijer A, Kruyt FA, Van Der Zee AG, Hollema H, Le P, Ten Hoor KA, Groothuis GM, Quax WJ, De Vries EG, De Jong S (2013) Nutlin-3 preferentially sensitises wild-type p53-expressing cancer cells to DR5-selective TRAIL over rhTRAIL. Br J Cancer 109:2685–2695
Meng XW, Koh BD, Zhang JS, Flatten KS, Schneider PA, Billadeau DD, Hess AD, Smith BD, Karp JE, Kaufmann SH (2014) Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression. J Biol Chem 289:20543–20558
Mert U, Sanlioglu ADJC, Sciences ML (2017) Intracellular localization of DR5 and related regulatory pathways as a mechanism of resistance to TRAIL in cancer. Cell Mol Life Sci 74:245–255
Micheau O (2018) Regulation of TNF-related apoptosis-inducing ligand signaling by glycosylation. Int J Mol Sci 19:715
Min Y, Shi J, Zhang Y, Liu S, Liu Y, Zheng D (2009) Death receptor 5-recruited raft components contributes to the sensitivity of Jurkat leukemia cell lines to TRAIL-induced cell death. IUBMB Life 61:261–267
Min KJ, Jang JH, Lee JT, Choi KS, Kwon TK (2012) Glucocorticoid receptor antagonist sensitizes TRAIL-induced apoptosis in renal carcinoma cells through up-regulation of DR5 and down-regulation of c-FLIP(L) and Bcl-2. J Mol Med (Berl) 90:309–319
Min KJ, Nam JO, Kwon TK (2017) Fisetin induces apoptosis through p53-mediated up-regulation of DR5 expression in human renal carcinoma caki cells. Molecules 22:1285
Moon DO, Kim MO, Choi YH, Kim GY (2010) Butein sensitizes human hepatoma cells to TRAIL-induced apoptosis via extracellular signal-regulated kinase/Sp1-dependent DR5 upregulation and NF-kappaB inactivation. Mol Cancer Ther 9:1583–1595
Moon DO, Park SY, Choi YH, Ahn JS, Kim GY (2011) Guggulsterone sensitizes hepatoma cells to TRAIL-induced apoptosis through the induction of CHOP-dependent DR5: involvement of ROS-dependent ER-stress. Biochem Pharmacol 82:1641–1650
Moon DO, Kang CH, Kang SH, Choi YH, Hyun JW, Chang WY, Kang HK, Koh YS, Maeng YH, Kim YR, Kim GY (2012) Capsaicin sensitizes TRAIL-induced apoptosis through Sp1-mediated DR5 up-regulation: involvement of Ca(2+) influx. Toxicol Appl Pharmacol 259:87–95
Moon DO, Asami Y, Long H, Jang JH, Bae EY, Kim BY, Choi YH, Kang CH, Ahn JS, Kim GY (2013) Verrucarin A sensitizes TRAIL-induced apoptosis via the upregulation of DR5 in an eIF2alpha/CHOP-dependent manner. Toxicol In Vitro 27:257–263
Nakata S, Yoshida T, Horinaka M, Shiraishi T, Wakada M, Sakai T (2004) Histone deacetylase inhibitors upregulate death receptor 5/TRAIL-R2 and sensitize apoptosis induced by TRAIL/APO2-L in human malignant tumor cells. Oncogene 23:6261
Nakata S, Yoshida T, Shiraishi T, Horinaka M, Kouhara J, Wakada M, Sakai T (2006) 15-Deoxy-Delta 12,14-prostaglandin J(2) induces death receptor 5 expression through mRNA stabilization independently of PPARgamma and potentiates TRAIL-induced apoptosis. Mol Cancer Ther 5:1827–1835
Newbold A, Falkenberg KJ, Prince HM, Johnstone RW (2016) How do tumor cells respond to HDAC inhibition? FEBS J 283:4032–4046
Oh YT, Deng L, Deng J, Sun SY (2017) The proteasome deubiquitinase inhibitor b-AP15 enhances DR5 activation-induced apoptosis through stabilizing DR5. Sci Rep 7:8027
Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11:381–389
Park EJ, Min KJ, Choi KS, Kubatka P, Kruzliak P, Kim DE, Kwon TK (2016) Chloroquine enhances TRAIL-mediated apoptosis through up-regulation of DR5 by stabilization of mRNA and protein in cancer cells. Sci Rep 6:22921
Pennati M, Sbarra S, De Cesare M, Lopergolo A, Locatelli SL, Campi E, Daidone MG, Carlo-Stella C, Gianni AM, Zaffaroni N (2015) YM155 sensitizes triple-negative breast cancer to membrane-bound TRAIL through p38 MAPK- and CHOP-mediated DR5 upregulation. Int J Cancer 136:299–309
Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B, Winter JM, Weber MC, Londin ER, Rigoutsos I, Yeo CJ, Gorospe M, Witkiewicz AK, Sachs JN, Brody JR (2012) HuR’s post-transcriptional regulation of Death Receptor 5 in pancreatic cancer cells. Cancer Biol Ther 13:946–955
Rasheduzzaman M, Moon JH, Lee JH, Nazim UM, Park SY (2018) Telmisartan generates ROS-dependent upregulation of death receptor 5 to sensitize TRAIL in lung cancer via inhibition of autophagy flux. Int J Biochem Cell Biol 102:20–30
Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C, Fuchs EJ, Bedi A (2001) Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat Cell Biol 3:409–416
Schlegel CR, Fonseca AV, Stocker S, Georgiou ML, Misterek MB, Munro CE, Carmo CR, Seckl MJ, Costa-Pereira AP (2014) DAPK2 is a novel modulator of TRAIL-induced apoptosis. Cell Death Differ 21:1780–1791
Schuler S, Fritsche P, Diersch S, Arlt A, Schmid RM, Saur D, Schneider G (2010) HDAC2 attenuates TRAIL-induced apoptosis of pancreatic cancer cells. Mol Cancer 9:80
Sheikh MS, Burns TF, Huang Y, Wu GS, Amundson S, Brooks KS, Fornace AJ Jr, El-Deiry WS (1998) p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res 58:1593–1598
Shin GC, Kang HS, Lee AR, Kim KH (2016) Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy 12:2451–2466
Shiraishi T, Yoshida T, Nakata S, Horinaka M, Wakada M, Mizutani Y, Miki T, Sakai T (2005) Tunicamycin enhances tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human prostate cancer cells. Cancer Res 65:6364–6370
Son YG, Kim EH, Kim JY, Kim SU, Kwon TK, Yoon AR, Yun CO, Choi KS (2007) Silibinin sensitizes human glioma cells to TRAIL-mediated apoptosis via DR5 up-regulation and down-regulation of c-FLIP and survivin. Cancer Res 67:8274–8284
Song JJ, Szczepanski MJ, Kim SY, Kim JH, An JY, Kwon YT, Alcala MA Jr, Bartlett DL, Lee YJ (2010) c-Cbl-mediated degradation of TRAIL receptors is responsible for the development of the early phase of TRAIL resistance. Cell Signal 22:553–563
Stern HM, Padilla M, Wagner K, Amler L, Ashkenazi A (2010) Development of immunohistochemistry assays to assess GALNT14 and FUT3/6 in clinical trials of dulanermin and drozitumab. Clin Cancer Res 16:1587–1596
Su RY, Chi KH, Huang DY, Tai MH, Lin WW (2008) 15-deoxy-Delta 12,14-prostaglandin J2 up-regulates death receptor 5 gene expression in HCT116 cells: involvement of reactive oxygen species and C/EBP homologous transcription factor gene transcription. Mol Cancer Ther 7:3429–3440
Sun SC (2011) Non-canonical NF-kappaB signaling pathway. Cell Res 21:71–85
Sun SY, Liu X, Zou W, Yue P, Marcus AI, Khuri FR (2007) The farnesyltransferase inhibitor lonafarnib induces CCAAT/enhancer-binding protein homologous protein-dependent expression of death receptor 5, leading to induction of apoptosis in human cancer cells. J Biol Chem 282:18800–18809
Sun M, Zhang J, Liu S, Liu Y, Zheng D (2008) Sp1 is involved in 8-chloro-adenosine-upregulated death receptor 5 expression in human hepatoma cells. Oncol Rep 19:177–185
Sun YY, Xiao L, Wang D, Ji YC, Yang YP, Ma R, Chen XH (2017) Triptolide inhibits viability and induces apoptosis in liver cancer cells through activation of the tumor suppressor gene p53. Int J Oncol 50:847–852
Surget S, Chiron D, Gomez-Bougie P, Descamps G, Menoret E, Bataille R, Moreau P, Le Gouill S, Amiot M, Pellat-Deceunynck C (2012) Cell death via DR5, but not DR4, is regulated by p53 in myeloma cells. Cancer Res 72:4562–4573
Takimoto R, El-Deiry WS (2000) Wild-type p53 transactivates the KILLER/DR5 gene through an intronic sequence-specific DNA-binding site. Oncogene 19:1735–1743
Thien CB, Langdon WY (2005) c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J 391:153–166
Thomas MJ, Seto E (1999) Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197–208
Tian X, Ye J, Alonso-Basanta M, Hahn SM, Koumenis C, Dorsey JF (2011) Modulation of CCAAT/enhancer binding protein homologous protein (CHOP)-dependent DR5 expression by nelfinavir sensitizes glioblastoma multiforme cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Biol Chem 286:29408–29416
Truneh A, Sharma S, Silverman C, Khandekar S, Reddy MP, Deen KC, Mclaughlin MM, Srinivasula SM, Livi GP, Marshall LA, Alnemri ES, Williams WV, Doyle ML (2000) Temperature-sensitive differential affinity of TRAIL for its receptors. DR5 is the highest affinity receptor. J Biol Chem 275:23319–23325
Tur V, Sloot AM, Reis CR, Szegezdi E, Cool RH, Samali A, Serrano L, Quax WJ (2008) DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J Biol Chem 283:20560–20568
Urso L, Cavallari I, Silic-Benussi M, Biasini L, Zago G, Calabrese F, Conte PF, Ciminale V, Pasello G (2017) Synergistic targeting of malignant pleural mesothelioma cells by MDM2 inhibitors and TRAIL agonists. Oncotarget 8:44232–44241
Van Der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, Serrano L, Quax WJ (2006) Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci USA 103:8634–8639
Van Noesel MM, Van Bezouw S, Salomons GS, Voute PA, Pieters R, Baylin SB, Herman JG, Versteeg R (2002) Tumor-specific down-regulation of the tumor necrosis factor-related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res 62:2157–2161
Van Roosmalen IM, Quax WJ, FaE Kruyt (2014) Two death-inducing human TRAIL receptors to target in cancer: similar or distinct regulation and function? Biochem Pharmacol 91:447–456
Vizcaino C, Mansilla S, Portugal J (2015) Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol Ther 152:111–124
Voelkel-Johnson C, Hannun YA, El-Zawahry A (2005) Resistance to TRAIL is associated with defects in ceramide signaling that can be overcome by exogenous C6-ceramide without requiring down-regulation of cellular FLICE inhibitory protein. Mol Cancer Ther 4:1320–1327
Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, Von Goetz M, Yee SF, Totpal K, Huw L, Katta V, Cavet G, Hymowitz SG, Amler L, Ashkenazi A (2007) Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 13:1070–1077
Wang S, El-Deiry WS (2003) Requirement of p53 targets in chemosensitization of colonic carcinoma to death ligand therapy. Proc Natl Acad Sci USA 100:15095–15100
Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC (2007) NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol 27:4374–4387
Wang J, Wang H, Wang LY, Cai D, Duan Z, Zhang Y, Chen P, Zou JX, Xu J, Chen X, Kung HJ, Chen HW (2016) Silencing the epigenetic silencer KDM4A for TRAIL and DR5 simultaneous induction and antitumor therapy. Cell Death Differ 23:1886–1896
Wang S-S, Feng L, Hu B-G, Lu Y-F, Wang W-M, Guo W, Suen C-W, Jiao B-H, Pang J-X, Fu W-M, Zhang J-F (2017) miR-133a promotes TRAIL resistance in glioblastoma via suppressing death receptor 5 and activating NF-κB signaling. Molecular therapy. Nucleic Acids 8:482–492
White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, Voelkel-Johnson C (2009) Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene 28:1132–1141
Wu GS, Burns TF, Mcdonald ER 3rd, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, El-Deiry WS (1997) KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 17:141–143
Xiaowen H, Yi S (2012) Triptolide sensitizes TRAIL-induced apoptosis in prostate cancer cells via p53-mediated DR5 up-regulation. Mol Biol Rep 39:8763–8770
Xu L, Qu X, Zhang Y, Hu X, Yang X, Hou K, Teng Y, Zhang J, Sada K, Liu Y (2009) Oxaliplatin enhances TRAIL-induced apoptosis in gastric cancer cells by CBL-regulated death receptor redistribution in lipid rafts. FEBS Lett 583:943–948
Yamaguchi H, Wang HG (2004) CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem 279:45495–45502
Yan S, Qu X, Xu C, Zhu Z, Zhang L, Xu L, Song N, Teng Y, Liu Y (2012) Down-regulation of Cbl-b by bufalin results in up-regulation of DR4/DR5 and sensitization of TRAIL-induced apoptosis in breast cancer cells. J Cancer Res Clin Oncol 138:1279–1289
Yao Z, Chen A, Li X, Zhu Z, Jiang X (2017) Hsp90 inhibitor sensitizes TRAIL-mediated apoptosis via chop-dependent DR5 upregulation in colon cancer cells. Am J Transl Res 9:4945–4953
Yoshida T, Maeda A, Tani N, Sakai T (2001) Promoter structure and transcription initiation sites of the human death receptor 5/TRAIL-R2 gene1. FEBS Lett 507:381–385
Yoshida T, Shiraishi T, Nakata S, Horinaka M, Wakada M, Mizutani Y, Miki T, Sakai T (2005) Proteasome inhibitor MG132 induces death receptor 5 through CCAAT/enhancer-binding protein homologous protein. Cancer Res 65:5662–5667
Yuan K, Yong S, Xu F, Zhou T, Mcdonald JM, Chen Y (2015) Calmodulin antagonists promote TRA-8 therapy of resistant pancreatic cancer. Oncotarget 6:25308–25319
Yuan D, Xu J, Wang J, Pan Y, Fu J, Bai Y, Zhang J, Shao C (2016) Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 7:32707–32722
Yue D, Sun X (2018) Ixazomib promotes CHOP-dependent DR5 induction and apoptosis in colorectal cancer cells. Cancer Biol Ther 1:1–11. https://doi.org/10.1080/15384047.2018.1529095
Zhang Y, Zhang B (2008) TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol Cancer Res 6:1861–1871
Zhang XY, Zhang XD, Borrow JM, Nguyen T, Hersey P (2004) Translational control of tumor necrosis factor-related apoptosis-inducing ligand death receptor expression in melanoma cells. J Biol Chem 279:10606–10614
Acknowledgements
This work was supported by NRF grant funded by the Korea Government (MSIP) (Grant Nos. 2014R1A5A2010008 and NRF-2016R1A2B2013393).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Min, Kj., Woo, S.M., Shahriyar, S.A. et al. Elucidation for modulation of death receptor (DR) 5 to strengthen apoptotic signals in cancer cells. Arch. Pharm. Res. 42, 88–100 (2019). https://doi.org/10.1007/s12272-018-01103-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-018-01103-y