Abstract
In the present study, a single Arcanobacterium (A.) pinnipediorum strain isolated from discharge of a jaw swelling of a grey seal pup (Halichoerus grypus) in England, UK, was identified. This strain was further characterized by phenotypical investigations, by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), by Fourier transform infrared spectroscopy (FT-IR), and genotypically by sequencing the 16S rRNA gene and the genes gap encoding glyceraldehyde 3-phosphate dehydrogenase, tuf encoding elongation factor tu, and rpoB encoding the β subunit of bacterial RNA polymerase. The present study gives a first detailed characterization of the species A. pinnipediorum from a grey seal in the UK. However, the route of infection of the grey seal with the bacterial pathogen remains unclear.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Arcanobacterium (A.) was described by Collins et al. (1982) as a group of facultative anaerobic, asporogenous Gram-positive rods. According to Yassin et al. (2011) genus Arcanobacterium consists of four species, namely, A. haemolyticum, A. phocae, A. pluranimalium, and A. hippocoleae. In the following years, A. canis (Hijazin et al. 2012b), A. phocisimile (Hijazin et al. 2013), A. pinnipediorum (Sammra et al. 2015), A. wilhelmae (Sammra et al. 2017), A. urinimassiliense (Diop et al. 2017), A. ihumii (Fall et al. 2019), and A. bovis (Sammra et al. 2020) were described as novel species of this genus. The type strain A. pinnipediorum DSM 2710 T was originally isolated in 2004 during a monitoring program from an anal swab of a female harbor seal and characterized phenotypically, by cell wall analysis and genotypically (Sammra et al. 2015, 2018). To the best of our knowledge, no other A. pinnipediorum strains have been described elsewhere. The present study was designed to characterize a single A. pinnipediorum strain, which represents the first isolation of this bacterial species in the UK and the first from a grey seal (Halichoerus grypus).
Materials and methods
The bacterial strains investigated in the present study included A. pinnipediorum 014418, type strain A. pinnipediorum DSM 28752 T, originally isolated in the German North Sea, the type strains A. phocae DSM 10002 T and A. phocisimile DSM 26142 T, and other type strains of genus Arcanobacterium (Hijazin et al. 2012b, 2013; Sammra et al. 2015). A. pinnipediorum 014418 was isolated in mixed culture from a swollen mandible discharge of an 8-month-old male grey seal pup in a rehabilitation centre in England. Upon submission, the seal had a high temperature, infected teeth, and gums with discharging pus around the teeth. The animal was treated with amoxicillin/clavulanic acid and clindamycin for 17 days and the discharge resolved. Six weeks later a swab was collected from the bilateral anterior mandible that was swelling and discharging leading to the isolation of A. pinnipediorum 014418. The animal was treated again with amoxicillin/clavulanic acid and clindamycin for 25 days. In addition, chin became a hard swelling but there was no discharge. The animal was released 10 days later.
The bacterial culturing of A. pinnipediorum 014418 was carried out on 5% sheep blood agar for 48 h at 37 °C under microaerobic conditions. The strain was investigated phenotypically by determination of hemolysis and by VITEK2-compact system (bioMérieux, Nürtingen, Germany) according to the instructions of the manufacturer. The A. pinnipediorum strain was additionally analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI TOF MS, Bruker Biotyper database 8.468, Bruker Daltonik, Bremen, Germany) (Sammra et al. 2018; Wickhorst et al. 2019) and by Fourier transform infrared spectroscopy (FT-IR, Bruker Tensor with HTS-XT, Bruker Optik, Ettlingen, Germany), (Nagib et al. 2014; Sammra et al. 2018).
The presence of A. phocae phocaelysin encoding gene phl was determined with a previously described loop-mediated isothermal amplification (LAMP) assay. This was performed using a real-time fluorometer (Genie II®, OptiGene, Horsham, UK) (Abdulmawjood et al. 2016; Alssahen et al. 2018).
A. pinnipediorum 014418 was additionally investigated by amplification and sequencing of 16S rRNA gene (Hassan et al. 2009; Sammra et al. 2014b, 2015). The DNA template was extracted from freshly cultivated bacterial colonies using the DNeasy Blood and Tissue Kit in accordance with the manufacturer’s instructions (Qiagen, Hilden, Germany). The nucleotide sequence of the bacterial 16S rRNA gene was amplified with forward primer 16S rRNA UNI-L (5′-AGA GTT TGA TCA TGG CTC AG-′3) and reverse primer 16S rRNA UNI-R (5′-GTG TGA CGG GCG GTG TGT AC-′3). The amplicon was used for sequencing with primer 16S rRNA-533F (5′-GTG CCA GCM GCC GCG GTA A-′3) and 16S rRNA-907-R (5′-CCG TCA ATT CMT TTG AGT TT-′3) by Eurofins Genomics GmbH (Ebersberg, Germany). The sequences were analyzed using FinchTV (version, 1.4.0), alignment, and phylogenetic analysis by the Clustal W method using DNASTAR Lasergene version 8.0.2 (DNASTAR Inc., Madison, USA). Furthermore, A. pinnipediorum 014418 was characterized by sequencing the genes gap encoding glyceraldehyde 3-phosphate dehydrogenase, tuf encoding elongation factor tu (Wickhorst et al. 2017, 2019; Sammra et al. 2018), and rpoB encoding the β subunit of bacterial RNA polymerase (Ülbegi-Mohyla et al. 2010; Sammra et al. 2013, 2018). The oligonucleotide primer sequences and PCR conditions of the target genes used in the present study are summarized in Table 1.
Results and discussion
A. pinnipediorum 014418 investigated in the present study displayed a relatively small zone of complete hemolysis on 5% sheep blood agar plates and could be classified biochemically using VITEK2-compact system. The biochemical properties of A. pinnipediorum 014418 mainly corresponded to properties of A. pinnipediorum DSM 28752 T. However, differences were observed in a positive D-galactose reaction and negative beta-D-fucosidase reaction of A. pinnipediorum 014418 (Table 2). Strain A. pinnipediorum 014418 could additionally be identified to the species level by MALDI-TOF MS analysis with the current Bruker database (MBT 8468 MSP library). A. pinnipediorum 014418 displayed a close relation to the type strain A. pinnipediorum DSM 28752 T and could be clearly separated from other species of genus Arcanobacterium. A typical dendrogram of the MALDI-TOF mass spectrum analysis of A. pinnipediorum 014418 and various other species of genus Arcanobacterium is shown in Fig. 1. MALDI-TOF MS has already been shown to be a rapid and reliable technique for identification of bacteria of genus Arcanobacterium and Trueperella (Hijazin et al. 2012a; Wickhorst et al. 2017), also including A. phocae (Alssahen et al. 2018), A. phocisimile (Sammra et al. 2014a), and A. pinnipediorum (Sammra et al. 2018) of seal origin. MALDI TOF mass-spectra of A. pinnipediorum 014418 and other species of genus Arcanobacterium are available by exchange via the MALDI-TOF user platform (https://www.maldi-up.ua-bw.de; Rau et al. 2016).
FT-IR spectroscopy has already been used as a tool for identification of a large number of clinically relevant pathogens (Kuhm et al. 2009; Samuels et al. 2009; Contzen et al. 2011; Grunert et al. 2013), also including T. pyogenes isolated from bovine mastitis (Nagib et al. 2014) and for characterization of A. pinnipediorum of seal origin (Sammra et al. 2018). The infrared spectra of A. pinnipediorum 014418 of the present study was analyzed by the method described by Nagib et al. (2014). Comparable to the MALDI-TOF MS analysis A. pinnipediorum 014418 of the present study yielded a close relation to A. pinnipediorum DSM 28752 T and to next closely related members A. phocae DSM 10002 T, A. phocisimile DSM 26142 T, and A. haemolyticum DSM 20595 T (Fig. 2).
Cluster analysis of infrared spectra of A. pinnipediorum 014418 investigated in the present study in comparison with type strain A. pinnipediorum DSM 28752 T and other species of genus Arcanobacterium. This analysis was performed by using the second derivatives of the spectra (n = 2 for each isolate) in the spectral range of 500 to 1400 cm−1. Ward’s algorithm was applied in OPUS (version 7.2, Bruker optic, Ettlingen)
The previously described loop mediated isothermal amplification (LAMP) assay for detection of phocaelysin encoding gene phl could be used successfully to identify A. phocae of various origins (Alssahen et al. 2018). However, A. pinnipediorum 014418 and comparatively investigated A. pinnipediorum DSM 28752 T and A. phocisimile DSM 26142 T yielded a negative phl-reaction indicating the high specificity of this assay for detection of A. phocae but not for other species of genus Arcanobacterium of seal origin (data not shown).
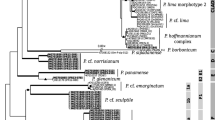
Strain A. pinnipediorum 014418 of the present study was identified genotypically by sequencing the 16S rRNA gene with a sequence similarity of 99.9% to 16S rRNA gene of type strain A. pinnipediorum DSM 28752 T (KJ596349) and a 16S rRNA gene sequence similarity ≤ 98.7% to other species of genus Arcanobacterium (Fig. 3).
Dendrogram analysis of 16S rRNA gene of A. pinnipediorum 014418 investigated in the present study in comparison with the type strain A. pinnipediorum DSM 28752 T and type strains of other species of genus Arcanobacterium obtained from NCBI GenBank using the Clustal W method of DNASTAR/Lasergene MegAlign program (version 8.0.2). *Accession numbers are given in brackets. T indicates type strains
According to previous studies A. phocae, A. phocisimile and A. pinnipediorum of seal origin could be further characterized genotypically by sequencing the genomic targets gap, tuf, and rpoB (Sammra et al. 2014a, 2018). Comparable to these studies, A. pinnipediorum 014418 showed sequence similarities of the genes gap, tuf, and rpoB of 97.1%, 98.7%, and 93.6%, respectively, to type strain A. pinnipediorum DSM 28752 T and sequence similarities of these genes of ≤ 91.2%, ≤ 94.0%, and ≤ 86.5%, respectively, to other species of genus Arcanobacterium. A dendrogram analysis of these target genes is shown in Fig. 4a, b, c.
Dendrogram analyses of the genes gap a, tuf b, and rpoB c of A. pinnipediorum 014418 investigated in the present study in comparison with the type strain A. pinnipediorum DSM 28752 T and type strains of other species of genus Arcanobacterium obtained from NCBI GenBank using the Clustal W method of DNASTAR/Lasergene MegAlign program (version 8.0.2). *Accession numbers are given in brackets
According to the results of the present study, A. pinnipediorum 014418 isolated from mandibular discharge of a grey seal pup in England (UK) could successfully be characterized biochemically, by MALDI-TOF MS, by FT-IR spectroscopy, and by sequencing the presented genomic targets. The usefulness of the determination of specific spectra by MALDI-TOF MS and FT-IR analyses and the various genotypic targets for identification of this species have to be further investigated with additional A. pinnipediorum strains isolated in other contexts. The biochemical differences observed between A. pinnipediorum 014418 and the type strain A. pinnipediorum DSM 28752 T might reflect an intra-specific variability caused by evolutionary processes. As already mentioned above, further phenotypical studies and whole genome sequence analyses of A. pinnipediorum of various origins will show the unique taxonomic position and the importance of this species in animal infections. However, strain A. pinnipediorum 014418 of the present study represented the first isolation of this species in the UK and the first isolation from a grey seal. A. pinnipediorum 014418 was isolated from a mixed culture with several other bacteria, including Enterococcus faecalis, Streptococcus lutetiensis, and a Gram negative cocco-bacillus that could not be identified further, indicating that the pathogenic importance of this species remains to be elucidated. It is perhaps of note, however, that A. phocae has often been isolated in mixed infections, including, as is the case in this report, Streptococcus spp., from seals in Scotland (G. Foster, unpublished findings). Furthermore, A. phocae has been recovered from local lesions including an infected jaw from a mouth lesion of a grey seal (Ramos et al. 1997), providing further similarities with the isolation of A. pinnipediorum 014418 from an infected jaw.
References
Abdulmawjood A, Wickhorst J, Hashim O, Sammra O, Hassan AA, Alssahen M, Lämmler C, Prenger-Berninghoff E, Klein G (2016) Application of a loop-mediated isothermal amplification (LAMP) assay for molecular identification of Trueperella pyogenes isolated from various origins. Mol Cell Probes 30:205–210
Alssahen M, Sammra O, Wickhorst J, Hassan AA, Lämmler C, Raunio-Saarnisto M, Prenger-Berninghoff E, Timke M, Becker A, Abdulmawjood A (2018) Identification of Arcanobacterium phocae isolated from fur animals by phenotypic properties, by MALDI-TOF MS analysis and by detection of phocaelysin encoding gene phl as probable novel target. Vet Microbiol 216:45–51
Collins MD, Jones D, Schofield GM (1982) Reclassification of Corynebacterium haemolyticum (MacLean, Liebow & Rosenberg) in the genus Arcanobacterium gen. nov. as Arcanobacterium haemolyticum nom.rev., comb.nov. J Gen Microbiol 28:1279–1281
Contzen M, Sting R, Blazey B, Rau J (2011) Corynebacterium ulcerans from diseased wild boars. Zoonoses Public Health 58:479–488
Diop K, Morand A, Dubus JC, Fournier EP, Raoult D, Fenollar F (2017) Arcanobacterium urinimassiliense sp. nov., a new bacterium isolated from the urogenital tract. New Microbe New Infect 18:15–17
Fall NS, Lo CI, Fournier PE, Sokhna C, Raoult D, Fenollar F, Lagier JC (2019) Arcanobacterium ihumii sp. nov., Varibaculum vaginae sp.nov. and Tessaracoccus timonensis sp. nov., isolated from vaginal swabs from healthy Senegalese women. New Microbe New Infect 31:100585
Grunert T, Wenning M, Barbagelata MS, Fricker M, Sordelli DO (2013) Rapid and reliable identification of Staphylococcus aureus capsular serotypes by means of artificial neural network-assisted Fourier transform infrared spectroscopy. J Clin Microbiol 51:2261–2266
Hassan AA, Ülbegi-Mohyla H, Kanbar T, Alber J, Lämmler C, Abdulmawjood A, Zschöck M, Weiss R (2009) Phenotypic and genotypic characterization of Arcanobacterium haemolyticum isolates from infections of horses. J Clin Microbiol 47:124–128
Hijazin M, Hassan AA, Alber J, Lämmler C, Timke M, Kostrzewa M, Prenger-Berninghoff E, Zschöck M (2012a) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for species identification of bacteria of genera Arcanobacterium and Trueperella. Vet Microbiol 157:243–245
Hijazin M, Prenger-Berninghoff E, Sammra O, Alber J, Lämmler C, Kämpfer P, Glaeser SP, Busse HJ, Hassan AA, Abdulmawjood A, Zschöck M et al (2012b) Arcanobacterium canis sp. nov., isolated from otitis externa of a dog, and emended description of the genus Arcanobacterium Collins, 1983 emend. Yassin et al., 2011. Int J Syt Evol Microbiol 62:2201–2205
Hijazin M, Sammra O, Ülbegi-Mohyla H, Nagib S, Alber J, Lämmler C, Kämpfer P, Glaeser SP, Busse HJ, Kassmannhuber J, Prenger-Berninghoff E, Weiss R, Siebert U, Hassan AA, Abdulmawjood A, Zschöck M (2013) Arcanobacterium phocisimile sp. nov., isolated from harbour seals. Int J Syst Evol Microbiol 63:2019–2024
Kuhm AE, Suter D, Felleisen R, Rau J (2009) Identification of Yersinia enterocolitica at the species and subspecies levels by Fourier transform infrared spectroscopy. Appl Environ Microbiol 75:5809–5813
Nagib S, Rau J, Sammra O, Lämmler C, Schlez K, Zschöck M, Prenger-Berninghoff E, Klein G, Abdulmawjood A (2014) Identification of Trueperella pyogenes isolated from bovine mastitis by Fourier transform infrared spectroscopy. PLoS One 9:e104654
Ramos C, Foster G, Collins MD (1997) Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov., Arcanobacter bernardiae comb. nov. and Arcanobacterium pyogenes comb. nov. J Clin Microbiol 47:46–53
Rau J, Eisenberg T, Männig A, Wind C, Lasch R, Sting R (2016) MALDI-UP – An internet platform for the exchange of mass spectra — user guide for http://maldi-up.ua-bw.de/. Aspects of food control and animal health 01/2016. http://ejournal.cvuas.de/issue201601.asp
Sammra O, Balbutskaya A, Zhang S, Hijazin M, Nagib S, Lämmler C, Abdulmawjood A, Prenger-Berninghoff E, Kostrzewa M, Timke M (2013) Further characteristics of Arcanobacterium canis, a novel species of genus Arcanobacterium. Vet Microbiol 167:619–622
Sammra O, Balbutskaya A, Hijazin M, Nagib S, Alber J, Lämmler C, Abdulmawjood A, Prenger-Berninghoff E, Timke M, Kostrzewa M, Siebert U (2014a) Further study on Arcanobacterium phocisimile a novel species of genus Arcanobacterium. J Vet Med ID 923592
Sammra O, Balbutskaya A, Nagib S, Alber J, Lämmler C, Abdulmawjood A, Timke M, Kostrzewa M, Prenger-Berninghoff E (2014b) Properties of an Arcanobacterium haemolyticum strain isolated from a donkey. Berl Münch Tierärztl Wochenschr 127:10–14
Sammra O, Balbutskaya A, Ülbegi-Mohyla H, Nagib S, Lämmler C, Kämpfer P, Glaeser SP, Golke J, Busse HJ, Prenger-Berninghoff E, Siebert U, Abdulmawjood A, Klein G (2015) Arcanobacterium pinnipediorum sp. nov., isolated from a harbour seal. Int J Syt Evol Microbiol 65:4539–4543
Sammra O, Foster G, Hassan AA, Alssahen M, Lämmler C, Glaeser SP, Kämpfer P, Busse H-J, Borowiak M, Malorny B, Ritchie CM, Prenger-Berninghoff E, Abdulmawjood A (2020) Arcanobacterium bovis sp. nov., isolated from the milk of a cow with mastitis. Int J Syst Evol Microbiol 70:4105–4110. https://doi.org/10.1099/ijsem.0.004230
Sammra O, Rau J, Wickhorst JP, Alssahen M, Hassan AA, Lämmler C, Kämpfer P, Glaeser SP, Busse HJ, Kleinhagauer T, Knauf-Witzens T, Prenger-Berninghoff E, Abdulmawjood A, Klein G (2017) Arcanobacterium wilhelmae sp. nov., isolated from the genital tract of a rhinoceros (Rhinoceros unicornis). Int J Syst Evol Microbiol 67:2093–2097
Sammra O, Rau J, Wickhorst JP, Alssahen M, Hassan AA, Lämmler C, Prenger-Berninghoff E, Abdulmawjood A (2018) Further characteristic of Arcanobacterium pinnipediorum DSM 28752T and Arcanobacterium wilhelmae DSM 102162T, two novel species of genus Arcanobacterium. Folia Microbiol 63:695–700
Samuels AC, Snyder AP, Emge DK, Amant D, Minter J (2009) Classification of select category A and B bacteria by Fourier transform infrared spectroscopy. Appl Spectrosc 63:14–24
Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, Hassan AA, Abdulmawjood A, Prenger-Berninghoff E, Weiss R, Zschöck M (2010) Identification of Arcanobacterium pyogenes isolated by post mortem examinations of a bearded dragon and a gecko by phenotypic and genotypic properties. J Vet Sci 11:265–267
Wickhorst JP, Hassan AA, Sheet OH, Eisenberg T, Sammra O, Alssahen M, Lämmler C, Prenger-Berninghoff E, Zschöck M, Timke M, Abdulmawjood A (2017) Trueperella pyogenes isolated from a brain abscess of an adult roebuck (Capreolus capreolus). Folia Microbiol 63:17–22
Wickhorst JP, Hassan AA, Sammra O, Alssahen M, Lämmler C, Prenger-Berninghoff E, Naggert M, Timke M, Rau J, Abdulmawjood A (2019) First report on the isolation of Trueperella abortisuis from companion animals. Res Vet Sci 125:465–467
Yassin AF, Hupfer H, Siering C, Schumann P (2011) Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al., 1982 emend. Lehnen et al., 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium. Int J Syst Evol Microbiol 61:1265–1274
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.A., C.L., G.F., A.A., T.E., A.H., E.P-B, and J.R. contributed to the design of the study and collected and analyzed the data. M.A. performed the initial examination of the isolates. M.A., C.L., A.A., and G.F. drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alssahen, M., Foster, G., Hassan, A.A. et al. First isolation of Arcanobacterium pinnipediorum from a grey seal pup (Halichoerus grypus) in the UK. Folia Microbiol 67, 291–297 (2022). https://doi.org/10.1007/s12223-021-00932-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-021-00932-7