Abstract
Human LARP4A belongs to a superfamily of RNA binding proteins called La-related proteins (LARPs). Whilst being a positive regulator of protein synthesis and a promoter of mRNA stability, LARP4A also controls cell morphology and motility in human breast and prostate cancer cells. All LARPs share a characteristic RNA binding unit named the La–module, which despite a high level of primary structure conservation exhibits a great versatility in RNA target selection. Human LARP4A La–module is the most divergent compared with other LARPs and its RNA recognition properties have only recently started to be revealed. Given the key role of LARP4A protein in cancer cell biology, we have initiated a complete NMR characterisation of its La-module and here we report the assignment of 1H, 15N and 13C resonances resulting from our studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
Human LARP4A is an RNA binding protein (RBP) involved in mRNA stabilisation and translation enhancement, 3′UTR polyA lengthening and miRNA processing (Maraia et al. 2017; Mattijssen et al. 2017; Nussbacher and Yeo 2018; Yang et al. 2011). As it localises to stress granules, membraneless structures associated with mRNA turnover and protection of mRNA during stress conditions, LARP4A has been suggested to play a role in the stress response (Gilbertson et al. 2018; Yang et al. 2011). LARP4A also regulates cancer cell morphology and motility: its siRNA-mediated depletion has been shown to increase cell migration and invasion, whereas its overexpression promotes cell circularity in breast and prostate cancers (Seetharaman et al. 2016).
LARP4A binds to the 3′polyA tail of mRNAs, and associates to translating ribosomes and protein partners including RACK1 (Receptor for Activated C Kinase) and PolyA binding protein (PABP) (Maraia et al. 2017; Yang et al. 2011). How the cellular functions of LARP4A in RNA and tumour biology are mediated by its molecular associations to RNA targets and/or other proteins remains unclear.
LARP4A possesses a La–module, a unique RNA binding unit conserved across all the members of the La-related proteins (LARPs) superfamily and consisting of two domains, the La motif (LaM) and an RNA recognition motif (RRM1) (Bousquet-Antonelli and Deragon 2009; Maraia et al. 2017). Despite sequence conservation, the RNA targets and functions of the La–modules in different LARPs are quite diverse, but the molecular bases of this versatility remain poorly understood (Maraia et al. 2017). The La–module of the human La protein has been extensively studied at the molecular level and its interactions with the 3′UUUOH tail of the nascent RNA polymerase III transcripts well characterised: the LaM and RRM1 act in synergy to accommodate the 3′UUUOH target, with the LaM establishing the majority of the intermolecular contacts with the RNA (Alfano et al. 2004; Kotik-Kogan et al. 2008; Teplova et al. 2006). An analogous mechanism has been reported for human LARP7 and LARP6 (Maraia et al. 2017; Martino et al. 2015; Uchikawa et al. 2015).
The La–motifs (LaM) of LARPs exhibit a high degree of sequence conservation across the superfamily, particularly in six key residues identified in human La as prime mediators of RNA recognition, namely Q20, Y23, Y24, D33, F35, F55 (human La protein numbering). Intriguingly, in human LARP4A Y24 and F55 are replaced by Cys and Met respectively (Merret et al. 2013). Moreover, primary sequence analysis suggests that LARP4A lacks the otherwise conserved wing 2 loop at the C-terminus of the LaM and contains a short inter domain linker between the LaM and the RRM (Maraia et al. 2017; Martino et al. 2015; unpublished). These distinctive characteristics, divergent from other LARPs, may impact on the RNA binding properties of LARP4A. We therefore set out to unveil the structure and the determinants of RNA recognition of LARP4A, to understand its cellular functions and roles in cancer biology. Here, we report the chemical shift assignments of the backbone and side-chain resonances of LARP4A La–module.
Methods and experiments
Protein expression and purification
LARP4A La–module, spanning residues 111–287, was cloned in a pET-Duet1 vector (Novagen) with a hexa-Histidine tag at the N-terminus, followed by a TEV protease cleavage site. The recombinant protein was expressed in Escherichia coli Rosetta II cells (Novagen) and uniformly labelled with 15N or 15N/13C in minimal media containing 15NH4Cl (1 g/L) and 13C glucose (2 g/L). The cells were grown to an OD value of 0.6 and induced at 18 °C with 1 mM IPTG (Isopropyl β-d-1-thiogalactopyranoside) overnight. The harvested cells were resuspended in a buffer containing 50 mM Tris pH 8, 300 mM NaCl, 10 mM imidazole, 5% glycerol, one Complete protease inhibitor cocktail tablet (Roche), 2 mM phenylmethylsulfonyl fluoride and lysozyme. After sonication and clarification, the lysate was loaded on a 5 mL His-Trap (GE Healthcare) affinity column and the His-tagged protein was eluted with a gradient from 0 to 300 mM of imidazole. The protein was dialyzed into a buffer comprising 50 mM Tris pH 7.25, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT and digested with TEV protease at 4 °C overnight. To isolate the un-tagged protein from the protease, tags and non-digested protein, the mixture was applied onto a Nickel affinity column (Generon). To eliminate any nucleic acid from the sample, the protein was further purified using a 5 mL Hi-Trap heparin column (GE Healthcare) and eluted with a KCl gradient from 0 to 1 M. The pure protein was dialysed into a buffer containing 20 mM Tris pH 7.25, 100 mM KCl, 0.2 mM EDTA and 1 mM DTT.
NMR spectroscopy
The 15N and 15N,13C-labeled samples of LARP4A La–module (111–287) were concentrated to 600 µM in 20 mM Tris pH 7.25, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT in 99.8% D2O or 10%D2O/90%H2O as appropriate. All the NMR experiments were performed at 25 °C on Bruker Avance III or NEO NMR spectrometers operating at 700, 800 and 950 MHz, equipped with triple resonance cryoprobes. NMR data were processed with Topspin 3.5pl7 software (Bruker) and NMRPipe/NMRDraw (Delaglio et al. 1995). Assignment was performed with CcpNmr Analysis (Vranken et al. 2005) and/or CARA/NEASY (Bartels et al. 1995) softwares. For the assignment of the backbone resonances a set of experiments including 1H-15N HSQC, HNCO, HNCA, HN(CO)CA, HNCACB and CBCA(CO)NH was used. The side-chain resonance assignments were determined using 1H-15N HSQC, 1H/15N- and 1H/13C-edited NOESY-HSQC and HCCH-TOCSY spectra (Fesik et al. 1988).
Extent of assignment and data deposition
The chemical shift assignment for LARP4A La–module has been deposited in the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/), accession number 27666.
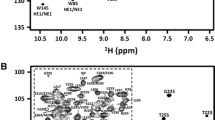
Human LARP4A La–module displays a well-resolved 1H-15N HSQC indicating that the protein is folded (Fig. 1a). It comprises 177 residues, with 4 glycine and 8 proline residues. An almost complete backbone assignment (93%) was achieved, identifying 93% of NH (156/168), 87% of Hα (154/177), 97% of Cα (171/177) and 95% of Cβ (165/173) resonances unambiguously. For side chains, 87% of the aliphatic and 48% of the aromatic side-chains (1H and 13C resonances beyond the Cγ position) were assigned. Resonances of Asn111, Ser132, His234, Asn275, Thr276 and the linker residues (His196–Arg198) could not be assigned. Non-native residues derived from the vector sequence after the His-tag cleavage, a serine and a valine preceding Asn111, were not assigned.
LARP4A La–module NH amide assignment and secondary structure. a1H-15N HSQC spectrum of human LARP4A recorded at 800 MHz and 25 °C. Amide group peaks are labelled with the residue type and numbered corresponding to the protein sequence. A closer view of the central part of the spectrum is shown for clarity. b TALOS+ prediction of secondary structure elements for LARP4A La–module. The secondary structure probabilities (red, α-helices; blue, β-strands), plotted against the residue number, are based on backbone HN, N, C′, Cα, and Cβ chemical shifts. Residues for which backbone amide resonance assignments are missing are indicated by asterisks
An analysis of the backbone chemical shifts performed with TALOS+ (Shen et al. 2009) revealed that LARP4A La–module contains eight α-helices and seven β-strands distributed between the LaM and RRM1 (Fig. 1b). LARP4A LaM displays the same secondary structure topology found in other LaMs previously described (Alfano et al. 2004; Martino et al. 2015): α1(115–129)–α1′(133–137)–α2(139–145)–β1(151–153)–α3(155–158)–α4(161–165)–α5(170–178)–β2(183–185)–β3(190–193). Likewise, the RRM1 harbours the canonical topology for this class of domains (Clery et al. 2008): β1(200–204)–α1(212–218)–β2(229–233)–β3(236–242)–α2(247–259) β4(267–274).
The backbone and side-chains chemical shifts of the isolated LaM (111–196) and RRM1 (196–287) were also analysed, revealing that they remained largely unchanged in the context of the La–module, and suggesting that the two domains do not adopt a rigid orientation relative to one other in solution.
References
Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11:323–329
Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Bousquet-Antonelli C, Deragon JM (2009) A comprehensive analysis of the La-motif protein superfamily. RNA 15:750–764
Clery A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18:290–298
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Fesik SW, Luly JR, Erickson JW, Abad-Zapatero C (1988) Isotope-edited proton NMR study on the structure of a pepsin/inhibitor complex. Biochemistry 27:8297–8301
Gilbertson S, Federspiel JD, Hartenian E, Cristea IM, Glaunsinger B (2018) Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. eLife https://doi.org/10.7554/eLife.37663
Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S (2008) Structural analysis reveals conformational plasticity in the recognition of RNA 3’ ends by the human La protein. Structure 16:852–862
Maraia RJ, Mattijssen S, Cruz-Gallardo I, Conte MR (2017) The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA. https://doi.org/10.1002/wrna.1430
Martino L, Pennell S, Kelly G, Busi B, Brown P, Atkinson RA, Salisbury NJH, Ooi ZH, See KW, Smerdon SJ, Alfano C, Bui TTT, Conte MR (2015) Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res 43:645–660
Mattijssen S, Arimbasseri AG, Iben JR, Gaidamakov S, Lee J, Hafner M, Maraia RJ (2017) LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. eLife https://doi.org/10.7554/eLife.28889
Merret R, Martino L, Bousquet-Antonelli C, Fneich S, Descombin J, Billey E, Conte MR, Deragon JM (2013) The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA 19:36–50
Nussbacher JK, Yeo GW (2018) Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol Cell 69:1005–1016.e7
Seetharaman S, Flemyng E, Shen J, Conte MR, Ridley AJ (2016) The RNA-binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton 73:680–690
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ (2006) Structural basis for recognition and sequestration of UUUOH 3’ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Molecular cell 21:75–85
Uchikawa E, Natchiar KS, Han X, Proux F, Roblin P, Zhang E, Durand A, Klaholz BP, Dock-Bregeon AC (2015) Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res 43:3373–3388
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Yang R, Gaidamakov SA, Xie J, Lee J, Martino L, Kozlov G, Crawford AK, Russo AN, Conte MR, Gehring K, Maraia RJ (2011) La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol Cell Biol 31:542–556
Acknowledgements
ICG and MRC acknowledge support from a Newton Royal Society Fellowship ref. NF140482 and a H2020 actions: Marie Sklodowska Curie Fellowship ref. 655341; LM acknowledges support by an EMBO Long Term Fellowship ref. ALT400-2010. RT and SDT were visiting PhD students from the Department of Pharmacy, University of Naples “Federico II”. We thank the Centre for Biomolecular Spectroscopy for access to biophysical infrastructure. The Centre was funded by the Wellcome Trust and British Heart Foundation (Grant Nos. 202767/Z/16/Z and IG/16/2/32273 respectively). This work was supported by the Francis Crick Institute through provision of access to the MRC Biomedical NMR Centre. The Francis Crick Institute receives its core funding from Cancer Research UK (Grant No. FC001029), the UK Medical Research Council (FC001029), and the Wellcome Trust (Grant No. FC001029).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cruz-Gallardo, I., Martino, L., Trotta, R. et al. Resonance assignment of human LARP4A La module. Biomol NMR Assign 13, 169–172 (2019). https://doi.org/10.1007/s12104-019-09871-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-019-09871-4