Abstract
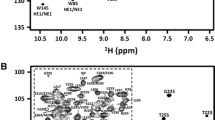
The N-terminal half of La protein consists of two concatenated motifs: La motif (LAM) and the N-terminal RNA recognition motif (RRM1) both of which are responsible for poly(U) RNA binding. Here, we present the backbone and side-chain assignments of the 1H, 13C and 15N resonances of the 191-residue LAM–RRM1 region of the La protein from the lower eukaryote Dictyostelium discoideum and its secondary structure prediction.

Similar content being viewed by others
Abbreviations
- NMR:
-
Nuclear magnetic resonance
- HSQC:
-
Heteronuclear single quantum coherence
- LAM:
-
La motif
- RRM:
-
RNA recognition motif
- RRM1:
-
N-terminal RNA recognition motif
References
Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 14:323–329
Apostolidi M, Vourtsis DJ, Chasapis CT, Stathopoulos C, Bentrop D, Spyroulias GA (2014) 1H, 15N, 13C assignment and secondary structure determination of two domains of La protein from D. discoideum. Biomol NMR Assign 8:47–51
Argyriou A, Chasapis CT, Apostolidi M, Konstantinidou P, Stathopoulos C, Bentrop D, Spyroulias GA (2014) Backbone and side chain NMR assignment, along with the secondary structure determination of RRM2 domain of La protein from a lower eukaryote exhibiting identical structural organization with its human homolog. Biomol NMR Assign. doi:10.1007/s12104-014-9578-7
Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Bayfield MA, Maraia RJ (2009) Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol 16:430–437
Bayfield MA, Yang R, Maraia RJ (2010) Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 1799:365–378
Bousquet-Antonelli C, Deragon JM (2009) A comprehensive analysis of the La-motif protein superfamily. RNA 15:750–764
Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ (1997) Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell 88:707–715
Ford LP, Shay JW, Wright WE (2001) The La antigen associates with the human telomerase ribonucleoprotein and influences telomere length in vivo. RNA 7:1068–1075
Horke S, Reumann K, Schulze C, Grosse F, Heise T (2004) The La motif and the RNA recognition motifs of human La autoantigen contribute individually to RNA recognition and subcellular localization. J Biol Chem 279:50302–50309
Keller R (2004) The computer aided resonance assignment tutorial CH-6410. Cantina Verlag, Goldau
Kucera NJ, Hodsdon ME, Wolin SL (2011) An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci USA 108:1308–1313
Liang C, Xiong K, Szulwach KE, Zhang Y, Wang Z, Peng J, Fu M, Jin P, Suzuki HI, Liu Q (2013) Sjogren syndrome antigen B (SSB)/La promotes global microRNA expression by binding microRNA precursors through stem-loop recognition. J Biol Chem 288:723–736
Maraia RJ, Bayfield MA (2006) The La protein-RNA complex surfaces. Mol Cell 21:149–152
Maraia RJ, Intine RVA (2001) Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol Cell Biol 21:367–379
Martino L, Pennell S, Kelly G, Bui TT, Kotik-Kogan O, Smerdon SJ, Drake AF, Curry S, Conte MR (2012) Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res 3:1381–1394
Sanfelice D, Kelly G, Curry S, Conte MR (2008) NMR assignment of the N-terminal region of human La free and in complex with RNA. Biomol NMR Assign 2:107–109
Schubert M, Labudde D, Oschkinat H, Schmieder P (2002) A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR 24:149–154
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
van Venrooij WJ, Pruijn GJ (1995) Ribonucleoprotein complexes as autoantigens. Curr Opin Immunol 7:819–824
Wolin SL, Cedervall T (2002) The La protein. Annu Rev Biochem 71:375–403
Acknowledgments
The authors thank Prof. D. Drainas for kindly providing the cDNA library. ‘‘SEE-DRUG’’ Grant (EU FP7 REGPOT CT-2011-285950; www.seedrug.upatras.gr) and ‘‘K. Karatheodori’’ Grant 2010 (D.164; UPAT Research Committee to C.S.) are acknowledged for financial support. This work was additionally supported in part and implemented under the ‘‘ARISTEIA’’ action of the ‘‘OPERATIONAL PROGRAMME EDUCATION AND LIFELONG LEARNING’’ and is co-funded by the European Social Fund (ESF) and National Resources (MIS 1225, No. D608). D.B. thanks Dr. B. Fakler (University of Freiburg) for continued support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Christos T. Chasapis and Aikaterini I. Argyriou have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Chasapis, C.T., Argyriou, A.I., Apostolidi, M. et al. 1H, 13C and 15N backbone and side-chain resonance assignment of the LAM–RRM1 N-terminal module of La protein from Dictyostelium discoideum . Biomol NMR Assign 9, 303–307 (2015). https://doi.org/10.1007/s12104-015-9597-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-015-9597-z