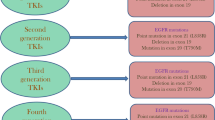
Abstract
As the largest cause of cancer-related deaths worldwide, pulmonary cancer is the most common form of the disease. Several genetic, epigenetic, and environmental factors come into play during the multi-step mechanism of tumorigenesis. The heterogeneity that makes discovering successful therapeutics for pulmonary cancer problematic is significantly influenced by the epigenetic landscape, including DNA methylation, chromatin architecture, histone modifications, and noncoding RNA control. Clinical activity of epigenetic-targeted medicines has been reported in hematological tumors, and these compounds may also have therapeutic effects in solid tumors. Over the course of the past few years, some researchers have successfully modified the expression of genes in cells using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated proteins) technique. The utilization of this technology allows for the induction of site-specific mutagenesis, epigenetic alterations, and the regulation of gene expression. This study will present an overview of the primary epigenetic alterations seen in pulmonary cancer, as well as a summary of therapeutic implications for targeting epigenetics in the management of pulmonary cancer, with a particular emphasis on the technique known as CRISPR/Cas9.


Similar content being viewed by others
Data availability
Not applicable.
References
Raso MG, Bota-Rabassedas N, Wistuba II. Pathology and Classification of SCLC. Cancers. 2021;13(4):820.
Tabib A, Khorgami MR, Meraji M, Omidi N, Mirmesdagh Y. Accuracy of doppler-derived indices in predicting pulmonary vascular resistance in children with pulmonary hypertension secondary to congenital heart disease with left-to-right shunting. Pediatr Cardiol. 2014;35(3):521–9.
Inamura K. Update on immunohistochemistry for the diagnosis of lung cancer. Cancers. 2018;10(3):72.
Liu Z, Su W, Ao J, Wang M, Jiang Q, He J, et al. Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology. Nat Commun. 2022;13(1):1–12.
Grossman DC, Curry SJ, Owens DK, Barry MJ, Davidson KW, Doubeni CA, et al. Screening for adolescent idiopathic scoliosis: US preventive services task force recommendation statement. JAMA. 2018;319(2):165–72.
Duan C, Deng H, Xiao S, Xie J, Li H, Zhao X, et al. Accelerate gas diffusion-weighted MRI for lung morphometry with deep learning. Eur Radiol. 2022;32(1):702–13.
Kadara H, Wistuba II. Field cancerization in non–small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc. 2012;9(2):38–42.
Su Y, Fang HB, Jiang F. An epigenetic classifier for early stage lung cancer. Clin Epigenetics. 2018;10(1):68.
Li H, Zhao X, Wang Y, Lou X, Chen S, Deng H, et al. Damaged lung gas exchange function of discharged COVID-19 patients detected by hyperpolarized 129Xe MRI. Sci Adv. 2021;7(1):eabc8180.
Jabbour SK, Lee KH, Frost N, Breder V, Kowalski DM, Pollock T, et al. Pembrolizumab plus concurrent chemoradiation therapy in patients with unresectable, locally advanced, stage III non-small cell lung cancer: the phase 2 keynote-799 nonrandomized trial. JAMA Oncol. 2021;7(9):1–9.
Vansteenkiste J, Crino L, Dooms C, Douillard J-Y, Faivre-Finn C, Lim E, et al. 2nd ESMO consensus conference on lung cancer early-stage non-small-cell lung cancer consensus on diagnosis treatment and follow-up. Ann Oncol. 2014;25(8):1462–74.
Hiddinga BI, Pauwels P, Janssens A, van Meerbeeck JP. O6-methylguanine-DNA methyltransferase (MGMT): a drugable target in lung cancer? Lung Cancer. 2017;107:91–9.
Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015;34(2):229–41.
Iranshahi N, Zafari P, Yari KH, Alizadeh E. The most common genes involved in epigenetics modifications among Iranian patients with breast cancer: a systematic review. Cell Mol Biol. 2016;62(12):116–22.
Liu S, Fabbri M, Gitlitz B, Laird-Offringa I. Epigenetic therapy in lung cancer. Front Oncol. 2013. https://doi.org/10.3389/fonc.2013.00135.
Liu H, Gao Y, Vafaei S, Gu X, Zhong X. The prognostic value of plasma cell-free DNA concentration in the prostate cancer: a systematic review and meta-analysis. Front Oncol. 2021;11: 599602.
Li Y, Yao C-F, Xu F-J, Qu Y-Y, Li J-T, Lin Y, et al. APC/CCDH1 synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nat Commun. 2019;10(1):1–16.
Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31(11):536–46.
Azadeh H, Alizadeh-Navaei R, Rezaiemanesh A, Rajabinejad M. Immune-related adverse events (irAEs) in ankylosing spondylitis (AS) patients treated with interleukin (IL)-17 inhibitors: a systematic review and meta-analysis. Inflammopharmacology. 2022;30(2):435–51.
Coiffier B, Pro B, Prince HM, Foss F, Sokol L, Greenwood M, et al. Results from a pivotal, open-label, phase II study of romidepsin in relapsed or refractory peripheral T-cell lymphoma after prior systemic therapy. J Clin Oncol. 2012;30(6):631–6.
Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–28.
Zou M, Yang Z, Fan Y, Gong L, Han Z, Ji L, et al. Gut microbiota on admission as predictive biomarker for acute necrotizing pancreatitis. Front Immunol. 2022. https://doi.org/10.3389/fimmu.2022.988326.
Vafaei S, Mirnejad R, Amirmozafari N. Determining the patterns of antimicrobial susceptibility and the distribution of blaCTX-M genes in strains of Acinetobacter Baumannii isolated from clinical samples. J Isfahan Med Sch. 2013;31(252):1443–51.
Rivera GA, Wakelee H. Lung cancer in never smokers. Lung Cancer Personal Med. 2016;893:43–57.
Zafari P, Yari K, Mostafaei S, Iranshahi N, Assar S, Fekri A, et al. Analysis of Helios gene expression and Foxp3 TSDR methylation in the newly diagnosed Rheumatoid Arthritis patients. Immunol Invest. 2018;47(6):632–42.
Turner BM. Epigenetic responses to environmental change and their evolutionary implications. Phil Trans R Soc B. 2009;364(1534):3403–18.
Iranshahi N, Assar S, Amiri SM, Zafari P, Fekri A, Taghadosi M. Decreased gene expression of epstein-barr virus-induced gene 3 (EBI-3) may contribute to the pathogenesis of rheumatoid arthritis. Immunol Invest. 2019;48(4):367–77.
Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9): a019505.
Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modificationsthe role of altered epigenetics in cancer development. Mol Cancer Ther. 2009;8(6):1409–20.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407.
Samimi Z, Kardideh B, Zafari P, Bahrehmand F, Roghani SA, Taghadosi M. The impaired gene expression of adenosine monophosphate-activated kinase (AMPK), a key metabolic enzyme in leukocytes of newly diagnosed rheumatoid arthritis patients. Mol Biol Rep. 2019;46(6):6353–60.
Shi Y-X, Wang Y, Li X, Zhang W, Zhou H-H, Yin J-Y, et al. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer. BMC Genomics. 2017;18(1):1–12.
Jia BY, Yang RH, Jiao WJ, Tian KH. Investigation of the effect of P14 promoter aberrant methylation on the biological function of human lung cancer cells. Thoracic Cancer. 2019;10(6):1388–94.
Tang Y, Xiao G, Chen Y, Deng Y. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29(8):725–35.
Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18(1):1–15.
Wang Y, Yu L, Wang T. MicroRNA-374b inhibits the tumor growth and promotes apoptosis in non-small cell lung cancer tissue through the p38/ERK signaling pathway by targeting JAM-2. J Thorac Dis. 2018;10(9):5489.
Shi Y-X, Sheng D-Q, Cheng L, Song X-Y. Current landscape of epigenetics in lung cancer: focus on the mechanism and application. J Oncol. 2019;2019:8107318.
Qu Y-Y, Zhao R, Zhang H-L, Zhou Q, Xu F-J, Zhang X, et al. Inactivation of the AMPK–GATA3–ECHS1 pathway induces fatty acid synthesis that promotes clear cell renal cell carcinoma growth. Cancer Res. 2020;80(2):319–33.
Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68(21):9005–14.
Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, et al. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer Interdiscip Int J Am Cancer Soc. 2006;107(5):1042–9.
Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275(46):35669–72.
Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22(12):4309–18.
Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, Carvalho AL, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNABiomarkers in non-small cell lung cancer. Clin Cancer Res. 2011;17(13):4494–503.
Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005;83(4):429–37.
Wang D, Zhao R, Qu Y-Y, Mei X-Y, Zhang X, Zhou Q, et al. Colonic lysine homocysteinylation induced by high-fat diet suppresses DNA damage repair. Cell Rep. 2018;25(2):398-412.e6.
Wu P-F, Kuo K-T, Kuo L-T, Lin Y-T, Lee W-C, Lu Y-S, et al. O6-Methylguanine-DNA methyltransferase expression and prognostic value in brain metastases of lung cancers. Lung Cancer. 2010;68(3):484–90.
Brabender J, Usadel H, Metzger R, Schneider PM, Park J, Salonga D, et al. Quantitative O 6-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res. 2003;9(1):223–7.
Li J, Poi MJ, Tsai M-D. Regulatory mechanisms of tumor suppressor P16INK4A and their relevance to cancer. Biochemistry. 2011;50(25):5566–82.
He X, Zhu Y, Yang L, Wang Z, Wang Z, Feng J, et al. MgFe-LDH nanoparticles: a promising leukemia inhibitory factor replacement for self-renewal and pluripotency maintenance in cultured mouse embryonic stem cells. Adv Sci. 2021;8(9):2003535.
Feng Y, Li F, Yan J, Guo X, Wang F, Shi H, et al. Pan-cancer analysis and experiments with cell lines reveal that the slightly elevated expression of DLGAP5 is involved in clear cell renal cell carcinoma progression. Life Sci. 2021;287: 120056.
Iwakawa R, Kohno T, Anami Y, Noguchi M, Suzuki K, Matsuno Y, et al. Association of p16 homozygous deletions with clinicopathologic characteristics and EGFR/KRAS/p53 mutations in lung adenocarcinoma. Clin Cancer Res. 2008;14(12):3746–53.
Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci. 1999;96(22):12754–9.
Yang W, Liu W, Li X, Yan J, He W. Turning chiral peptides into a racemic supraparticle to induce the self-degradation of MDM2. J Adv Res. 2022. https://doi.org/10.1016/j.jare.2022.05.009.
Xiao P, Chen J-r, Zhou F, Lu C-x, Yang Q, Tao G-h, et al. Methylation of P16 in exhaled breath condensate for diagnosis of non-small cell lung cancer. Lung Cancer. 2014;83(1):56–60.
Michie AM, McCaig AM, Nakagawa R, Vukovic M. Death-associated protein kinase (DAPK) and signal transduction: regulation in cancer. FEBS J. 2010;277(1):74–80.
Zhang Y, Wu J, Huang G, Xu S. Clinicopathological significance of DAPK promoter methylation in non-small-cell lung cancer: a systematic review and meta-analysis. Cancer Manag Res. 2018;10:6897.
Schrump DS. Targeting epigenetic mediators of gene expression in thoracic malignancies. Biochim Biophys Acta. 2012;1819(7):836–45.
Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18(6):682–9.
Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–43.
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95.
Nottke A, Colaiácovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Developmental. 2009;136(6):879–89.
Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res: Curr Rev. 2013;35(1):69.
Kim W, Kim R, Park G, Park J-W, Kim J-E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J Biol Chem. 2012;287(8):5588–99.
Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, et al. Loss of histone h4k20 trimethylation occurs in preneoplasia and influences prognosis of non–small cell lung cancer. Clin Cancer Res. 2008;14(22):7237–45.
Hosseini A, Minucci S. A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics. 2017;9(8):1123–42.
Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28(1):57–69.
Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–65.
Yavropoulou MP, Poulios C, Michalopoulos N, Gatzou A, Chrisafi S, Mantalovas S, et al. A role for circular non-coding RNAs in the pathogenesis of sporadic parathyroid adenomas and the impact of gender-specific epigenetic regulation. Cells. 2018;8(1):15.
Rajabinejad M, Asadi G, Ranjbar S, Varmaziar FR, Karimi M, Salari F, et al. The MALAT1-H19/miR-19b-3p axis can be a fingerprint for diabetic neuropathy. Immunol Lett. 2022;245:69–78.
Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045.
Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7(1):1–10.
An Y-x, Shang Y-j, Xu Z-w, Zhang Q-c, Wang Z, Xuan W-x, et al. STAT3-induced long noncoding RNA LINC00668 promotes migration and invasion of non-small cell lung cancer via the miR-193a/KLF7 axis. Biomed Pharmacother. 2019;116:109023.
Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X, et al. Silencing of LncRNA-HOTAIR decreases drug resistance of non-small cell lung cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun. 2018;497(4):1003–10.
Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol. 2017;12(2):179–200.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Can Res. 2005;65(21):9628–32.
Kang S-M, Lee H-J, Cho J-Y. MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 2013;335(2):487–94.
Zhang X, Liu L, Chen WC, Wang F, Cheng YR, Liu YM, et al. Gestational leucylation suppresses embryonic T-box transcription factor 5 signal and causes congenital heart disease. Adv Sci. 2022;9(15):2201034.
Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Investig. 2013;123(3):1241–61.
Braicu C, Zimta A-A, Harangus A, Iurca I, Irimie A, Coza O, et al. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers. 2019;11(5):605.
Di X, Jin X, Li R, Zhao M, Wang K. CircRNAs and lung cancer: biomarkers and master regulators. Life Sci. 2019;220:177–85.
Jiang M-M, Mai Z-T, Wan S-Z, Chi Y-M, Zhang X, Sun B-H, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–74.
Franic D, Dobrinic P, Korac P. Key achievements in gene therapy development and its promising progress with gene editing tools (ZFN, TALEN, CRISPR/CAS9). Mol Exp Biol Med. 2019;2(1):1–9.
Gaj T, Gersbach CA, Barbas CF III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405.
Razeghian E, Nasution MK, Rahman HS, Gardanova ZR, Abdelbasset WK, Aravindhan S, et al. A deep insight into CRISPR/Cas9 application in CAR-T cell-based tumor immunotherapies. Stem Cell Res Ther. 2021;12(1):1–17.
Liu M, Han X, Liu H, Chen D, Li Y, Hu W. The effects of CRISPR-Cas9 knockout of the TGF-β1 gene on antler cartilage cells in vitro. Cell Mol Biol Lett. 2019;24(1):1–12.
Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21.
Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ. Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol. 2018;13(2):389–96.
Levin WJ, Casey G, Ramos JC, Arboleda MJ, Reissmann PT, Slamon DJ. Tumor suppressor and immediate early transcription factor genes in non-small cell lung cancer. Chest. 1994;106(6):372S-S376.
Yu X, Wang W. Tumor suppressor microRNA-613 inhibits glioma cell proliferation, invasion and angiogenesis by targeting vascular endothelial growth factor A. Mol Med Rep. 2017;16(5):6729–35.
Wu J, Tang B, Tang Y. Allele-specific genome targeting in the development of precision medicine. Theranostics. 2020;10(7):3118.
Neri M, Cesario A, Granone P, Dominioni L, Puntoni R, D’Angelillo RM, et al. Prognostic role of K-Ras mutations in non-small cell lung cancer: still an issue for open debate. Lung Cancer. 2006;53(3):393–5.
Koo T, Yoon A-R, Cho H-Y, Bae S, Yun C-O, Kim J-S. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45(13):7897–908.
Zhang X, Qu Y-Y, Liu L, Qiao Y-N, Geng H-R, Lin Y, et al. Homocysteine inhibits pro-insulin receptor cleavage and causes insulin resistance via protein cysteine-homocysteinylation. Cell Rep. 2021;37(2): 109821.
Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein H-U, Rohde C, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23(1):69–78.
Luo J. CRISPR/Cas9: from genome engineering to cancer drug discovery. Trends Cancer. 2016;2(6):313–24.
Shen Y, Chen F, Liang Y. MicroRNA-133a inhibits the proliferation of non-small cell lung cancer by targeting YES1. Oncol Lett. 2019;18(6):6759–65.
Zhu H, Lu Q, Lu Q, Shen X, Yu L. Matrine regulates proliferation, apoptosis, cell cycle, migration, and invasion of non-small cell lung cancer cells through the circFUT8/miR-944/YES1 axis. Cancer Manag Res. 2021;13:3429.
Garmendia I, Pajares MJ, Hermida-Prado F, Ajona D, Bértolo C, Sainz C, et al. YES1 drives lung cancer growth and progression and predicts sensitivity to dasatinib. Am J Respir Crit Care Med. 2019;200(7):888–99.
Bilal E, Alexe G, Yao M, Cong L, Kulkarni A, Ginjala V, et al. Identification of the YES1 kinase as a therapeutic target in basal-like breast cancers. Genes Cancer. 2010;1(10):1063–73.
Zhou Y, Wang C, Ding J, Chen Y, Sun Y, Cheng Z. miR-133a targets YES1 to reduce cisplatin resistance in ovarian cancer by regulating cell autophagy. Cancer Cell Int. 2022;22(1):1–13.
Zang Q, Xu L, Li J, Jia H. GATA6 activated long non-coding RNA PCAT1 maintains stemness of non-small cell lung cancer by mediating FRK. J BUON. 2020;25(5):2371–81.
Grunblatt E, Wu N, Zhang H, Liu X, Norton JP, Ohol Y, et al. MYCN drives chemoresistance in small cell lung cancer while USP7 inhibition can restore chemosensitivity. Genes Dev. 2020;34(17–18):1210–26.
Zhang L, Yang Y, Chai L, Bu H, Yang Y, Huang H, et al. FRK plays an oncogenic role in non-small cell lung cancer by enhancing the stemness phenotype via induction of metabolic reprogramming. Int J Cancer. 2020;146(1):208–22.
Nau MM, Brooks BJ Jr, Carney DN, Gazdar AF, Battey JF, Sausville EA, et al. Human small-cell lung cancers show amplification and expression of the N-myc gene. Proc Natl Acad Sci. 1986;83(4):1092–6.
Gao Y, Chen S, Vafaei S, Zhong X. Tumor-infiltrating immune cell signature predicts the prognosis and chemosensitivity of patients with pancreatic ductal adenocarcinoma. Front Oncol. 2020;10: 557638.
Czech-Sioli M, Siebels S, Radau S, Zahedi RP, Schmidt C, Dobner T, et al. The ubiquitin-specific protease Usp7, a novel Merkel cell polyomavirus large T-antigen interaction partner, modulates viral DNA replication. J Virol. 2020;94(5):e01638-e1719.
Nair J, Nair A, Veerappan S, Sen D. Translatable gene therapy for lung cancer using Crispr CAS9—an exploratory review. Cancer Gene Ther. 2020;27(3):116–24.
Bidnur S, Savdie R, Black P. Inhibiting immune checkpoints for the treatment of bladder cancer. Bladder Cancer. 2016;2(1):15–25.
Thomas R, Al-Khadairi G, Roelands J, Hendrickx W, Dermime S, Bedognetti D, et al. NY-ESO-1 based immunotherapy of cancer: current perspectives. Front Immunol. 2018;9:947.
Miliotou AN, Papadopoulou LC. CAR T-cell therapy: a new era in cancer immunotherapy. Curr Pharm Biotechnol. 2018;19(1):5–18.
Su S, Hu B, Shao J, Shen B, Du J, Du Y, et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep. 2016;6(1):1–14.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
AK contributed to the idea design, literature search, and writing the manuscript. HK drafted the work.
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical approval
It is not applicable.
Informed consent
It is not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kazemizadeh, H., Kashefizadeh, A. CRISPR-Cas9-mediated gene therapy in lung cancer. Clin Transl Oncol 25, 1156–1166 (2023). https://doi.org/10.1007/s12094-022-03039-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-03039-8