Abstract
Objectives
Multiple b-value gas diffusion-weighted MRI (DW-MRI) enables non-invasive and quantitative assessment of lung morphometry, but its long acquisition time is not well-tolerated by patients. We aimed to accelerate multiple b-value gas DW-MRI for lung morphometry using deep learning.
Methods
A deep cascade of residual dense network (DC-RDN) was developed to reconstruct high-quality DW images from highly undersampled k-space data. Hyperpolarized 129Xe lung ventilation images were acquired from 101 participants and were retrospectively collected to generate synthetic DW-MRI data to train the DC-RDN. Afterwards, the performance of the DC-RDN was evaluated on retrospectively and prospectively undersampled multiple b-value 129Xe MRI datasets.
Results
Each slice with size of 64 × 64 × 5 could be reconstructed within 7.2 ms. For the retrospective test data, the DC-RDN showed significant improvement on all quantitative metrics compared with the conventional reconstruction methods (p < 0.05). The apparent diffusion coefficient (ADC) and morphometry parameters were not significantly different between the fully sampled and DC-RDN reconstructed images (p > 0.05). For the prospectively accelerated acquisition, the required breath-holding time was reduced from 17.8 to 4.7 s with an acceleration factor of 4. Meanwhile, the prospectively reconstructed results showed good agreement with the fully sampled images, with a mean difference of −0.72% and −0.74% regarding global mean ADC and mean linear intercept (Lm) values.
Conclusions
DC-RDN is effective in accelerating multiple b-value gas DW-MRI while maintaining accurate estimation of lung microstructural morphometry, facilitating the clinical potential of studying lung diseases with hyperpolarized DW-MRI.
Key Points
• The deep cascade of residual dense network allowed fast and high-quality reconstruction of multiple b-value gas diffusion-weighted MRI at an acceleration factor of 4.
• The apparent diffusion coefficient and morphometry parameters were not significantly different between the fully sampled images and the reconstructed results (p > 0.05).
• The required breath-holding time was reduced from 17.8 to 4.7 s and each slice with size of 64 × 64 × 5 could be reconstructed within 7.2 ms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyperpolarized gas (3He or 129Xe) MRI provides a unique strategy for high-resolution functional and structural imaging of the lung [1, 2]. In particular, hyperpolarized gas diffusion-weighted MRI (DW-MRI) has been demonstrated to be sensitive to microstructural changes associated with some lung diseases, such as chronic obstructive lung disease (COPD) [3,4,5] and idiopathic pulmonary fibrosis [6, 7]. Furthermore, multiple b-value gas DW-MRI, together with theoretical diffusion models (e.g., cylinder model [8, 9] or stretched exponential model [10]), enables non-invasive and quantitative estimation of lung morphometry parameters at the alveolar level [11]. However, such technique suffers from long scan times due to multiple b-value acquisitions and long diffusion gradient time [12], which imposes great burdens on subjects, especially for children and patients with lung diseases.
Compressed sensing MRI has been applied to accelerate hyperpolarized gas DW-MRI acquisition [11,12,13,14,15,16], because it can reduce scan time by undersampling k-space without additional hardware requirement [17,18,19]. However, compressed sensing MRI generally uses fixed sparsifying transforms, which may not capture complex image characteristics associated with biological tissues [20, 21]. Moreover, the selection of hyper-parameters (e.g., update rates and weighting parameters) is tedious and non-trivial, and inappropriate hyper-parameters can result in excessively smooth or residual artifacts in reconstructed results [20, 22]. Therefore, effective approaches are highly desirable for accelerating multiple b-value gas DW-MRI.
Recently, deep learning methods have been successfully used for reconstructing undersampled MRI data [20,21,22,23], providing superior performance in both reconstruction quality and speed. Such methods typically use convolutional neural networks to learn an end-to-end mapping from undersampled images or k-space data to reference images [24,25,26], and implicitly explore regularization prior from training data [27]. In particular, deep learning has also been used to fast and accurately reconstruct hyperpolarized gas ventilation MRI from highly undersampled k-space data [28]. Nevertheless, using deep learning to accelerate multiple b-value gas DW-MRI has not been explored so far. One reason is that large amounts of high-quality fully sampled (FS) images are required in most of deep learning–based reconstruction methods. As mentioned previously, it is a difficult task for multiple b-value gas DW-MRI. The other reason is that DW-MRI data contain quantitative microstructural information, which should be well-preserved during the reconstruction process.
The purpose of this work was to develop a novel deep learning framework for efficiently accelerating multiple b-value gas DW-MRI and evaluate its performance in terms of reconstruction quality and the effects on morphometry measurements by using both retrospectively and prospectively undersampled multiple b-value 129Xe DW-MRI data.
Materials and methods
Network architecture
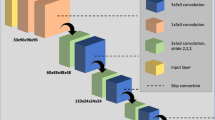
The proposed deep cascade of residual dense network (DC-RDN) for accelerating multiple b-value gas DW-MRI is illustrated in Fig. 1, which forms a cascading network by using reconstruction modules interleaved with data consistency (DC) layers several times [29]. We adopt residual dense blocks (RDBs) as the reconstruction modules, because the RDB can not only reuse abundant features from different levels but also substantially reduce the number of parameters [30, 31]. Furthermore, the DC layer is used to ensure the consistency between the reconstruction of each cascade and the measured k-space data, which is important to preserve quantitative microstructural information during reconstruction [13]. Additionally, because DW images have highly correlated edges and structural information along the diffusion dimension, the undersampled DW images with different b-values are stacked as a multi-channel input and then jointly reconstructed, which has potential to achieve more favorable reconstruction quality [32,33,34]. The details of the network architecture are described in Supplementary material.
Illustration of the proposed DC-RDN model for reconstructing multiple b-value gas DW-MRI from highly undersampled k-space data. DC, data consistency; DC-RDN, deep cascade of residual dense network; DW-MRI, diffusion-weighted MRI; IFT, inverse Fourier transform; kd, diffusion direction; kx, frequency-encoding direction; ky, phase-encoding direction; RDB, residual dense block; xd-1, the input of the d-th RDB; xd,r, the reconstruction of the d-th RDB
Training data simulation
Since it is difficult to acquire large amounts of FS DW-MRI data, we generated synthetic multiple b-value DW-MRI data from hyperpolarized 129Xe lung ventilation images. All MRI experiments were performed under the approval of the local Institutional Review Board, and written informed consents were obtained from all study participants.
The hyperpolarized 129Xe lung ventilation images were acquired from 101 participants (68 males and 33 females; age, 47.6 ± 17.2 years) between June 2016 and September 2019. Imaging was performed with a 1.5-T MRI scanner (Avanto; Siemens Healthineers). The imaging parameters were as follows: matrix size = 96 × 84, number of slices = 24, repetition time/echo time = 4.2/1.9 ms. After removing the slices with low signal-to-noise ratio (SNR), a total of 1404 ventilation images were obtained.
If the ventilation images are considered as the signal without diffusion-sensitizing gradients, the signal of non-zero b-value will be generated through the cylinder model with pseudorandom values of acinar duct radius (R), acinar lumen radius (r), and the measured b-values (see details in Supplementary material). Examples of synthetic images are presented in Supplementary Fig. 1. The synthetic images served as the references in the training process. The undersampled data were generated by retrospectively undersampling the reference k-space data according to a variable-density Cartesian random undersampling matrix at an acceleration factor of 4.
Implementation and training
The DC-RDN was implemented using TensorFlow package (version R2.0; https://tensorflow.google.org). All the training and testing were performed on a desktop computer with an Intel Xeon® Gold 6128 quad-core CPU, 64-GB RAM, and an NVIDIA RTX 2080Ti GPU.
During the training process, the network weights were initialized using the He initialization [35] and optimized using the Adam algorithm [36] with a fixed learning rate of 0.0002, β1 = 0.9, β2 = 0.999, and batch size of 8. The mean-squared error between the DC-RDN reconstructed results and the FS images was chosen as the loss function. Total training time was about 4 h for 200 epochs. Once the training process is completed, the parameters of DC-RDN are fixed, which can be adopted to effectively transform new undersampled DW-MRI data to the corresponding reconstruction results directly.
Testing data
Fully sampled multiple b-value 129Xe DW-MRI datasets were acquired from 5 healthy volunteers (age, 25.6 ± 2.9 years) using a 2D gradient echo diffusion sequence [11]. For each subject, 4 coronal slices were acquired with 30-mm thickness and 6-mm gap using a gas mixture of 500 mL 129Xe and 500 mL N2. Additional imaging parameters were: 5 b-values (b = 0, 10, 20, 30, 40 s/cm2), matrix size = 64 × 64, files of view = 384 × 384 mm2, repetition time/echo time = 13.8/10.9 ms, diffusion time = 5 ms, flip angle = 5°, scan time = 17.8 s.
Because some COPD patients cannot tolerate the long breath-holding time, two-fold undersampled multiple b-value 129Xe DW-MRI data were acquired from 5 COPD patients (age, 57.2 ± 6.8 years) and then reconstructed using a conventional compressed sensing method [11]. The reconstructed results were also treated as “FS” images in the following evaluation. These FS datasets were retrospectively undersampled to evaluate the reconstruction performance of the DC-RDN.
To demonstrate the performance of DC-RDN for in vivo imaging, FS and four-fold undersampled data were prospectively acquired from 5 healthy subjects (age, 24.8 ± 2.3 years). For the FS acquisition, the flip angle was set to 5°, whereas a higher flip angle of 9° was used for the undersampled acquisition to preserve signal-to-noise ratio [18]. The acceleration factor of 4 reduced the scan time from 17.8 to 4.7 s.
Evaluation
The DC-RDN was compared with two CS-MRI methods, i.e., total variation (TV) and signal decay into the reconstruction (SIDER) methods [15], and a deep learning method, i.e., U-Net [37], on the retrospectively undersampled datasets. TV and SIDER methods were implemented using the source codes provided in https://github.com/HGGM-LIM/compressed-sensing-diffusion-lung-MRI [15]. The original U-Net architecture was modified to be applicable to the reconstruction of multiple b-value gas DW-MRI in this study. The U-Net took the undersampled images with different b-values as a multi-channel input, and was operated on the TensorFlow using the framework specifications in Hyun et al [37].
The reconstructed images were evaluated using 3 quantitative metrics: mean absolute error (MAE), structure similarity (SSIM) [38], and MAEADC [13]. The MAEADC was to evaluate the MAE between the FS and reconstructed ADC maps. ADC maps were derived from the first two b-value images (b = 0 and 10 s/cm2) through a pixel-by-pixel mono-exponential fitting [13]. Additionally, qualitative assessment of the image quality was performed by two experienced readers using a 4-point ordinal scale [20, 39] (see details in Supplementary material).
The lung regions were manually segmented from the b = 0 image, which served as the region of interest for the calculations of MAE, SSIM, and MAEADC, as well as the lung morphometry analysis. Lung morphometry maps, including R, r, alveolar sleeve depth (h), mean linear intercept (Lm), and surface-to-volume ratio (SVR), were derived from the multiple b-value fit to the cylinder model [11].
Statistical analysis
Paired two-tailed Student’s t-tests were performed to compare the quantitative results between the different reconstruction methods. Comparisons in terms of image quality scores, averaged over the two readers, were performed using Wilcoxon signed-rank tests. Bland-Altman plots were adopted to assess the agreement of ADC and lung morphometry parameters between the FS and DC-RDN reconstructed images on a slice-by-slice level. All statistical analyses were performed by using SPSS (version 20.0; IBM), and p < 0.05 was considered statistically significant.
Results
Comparisons with baseline methods
Figure 2 shows the FS and reconstructed DW images with b = 0 for a healthy volunteer and a COPD patient. It can be seen that the zero-filling images present severe artifacts and blurred lung structures. Through the TV and SIDER, the artifacts are removed and the fine details are recovered to some extent, but noticeable smooth patterns are produced in the lung regions. Nevertheless, through the deep learning–based methods, such artifacts are successfully eliminated and most of the lung structures are restored. In the DC-RDN results, the sharpness and details of the lung are better preserved than the U-Net, especially in the regions indicated by the green arrows in Fig. 2f1 and f2. These qualitative comparisons are also supported by the quantitative metrics. The FS images with b = 10, 20, 30, and 40 cm2/s and the corresponding reconstructed results are provided in Supplementary Figs. 2 and 3.
Reconstructed DW images with b = 0 obtained using the zero-filling, TV, SIDER, U-Net, and DC-RDN at an acceleration factor of 4. a1 One slice of FS image from a 23-year-old male healthy volunteer. a2 One slice of FS image from a 62-year-old male patient with COPD. b1, b2; c1, c2; d1, d2; e1, e2; and f1, f2 Reconstructed results through the zero-filling, TV, SIDER, U-Net, and DC-RDN, respectively. g1–k1 and g2–k2 Corresponding differences between the FS images and reconstructed results. The green arrows indicate fine details of the lung, which are well preserved by the DC-RDN. Color bar for the difference images is shown at the bottom. COPD, chronic obstructive lung disease; DW, diffusion-weighted; FS, fully sampled; MAE, mean absolute error; SIDER, signal decay into the reconstruction; SSIM, structure similarity; TV, total variation
For DW images with b = 0, Fig. 3a and b display the mean MAE and SSIM values obtained using different methods for the total, healthy, and COPD test data. The healthy and COPD test data include 5 healthy volunteers and 5 COPD patients, respectively, and the total test data is the combination of the healthy and COPD test data. When compared with the two CS-MRI methods, DC-RDN achieves significantly lower MAE and higher SSIM values (the mean MAE is 2.5% and the mean SSIM is 0.9569). DC-RDN also has significantly lower MAE values and similar SSIM values compared to the U-Net. For DW images with b = 10, 20, 30, and 40 s/cm2, the mean MAE and SSIM values are provided in Supplementary Figs. 4 and 5.
Quantitative comparisons between different reconstruction methods at an acceleration factor of 4. a, b Average MAE and SSIM values of b = 0 DW images obtained using the zero-filling, TV, SIDER, U-Net, and DC-RDN for the total, healthy, and COPD test datasets. c Average MAEADC through different reconstruction algorithms. * denotes p < 0.05; ** denotes p < 0.001
Supplementary Fig. 6 summarizes the results of image quality scores for the total, healthy, and COPD test data. It can be seen that both the U-Net and DC-RDN methods get comparable scores to the FS images in terms of sharpness, SNR, aliasing artifacts, and overall image quality. In addition, DC-RDN achieves significantly better image quality than the two CS-MRI methods in terms of all the scoring criteria.
For a healthy volunteer and COPD patient, Supplementary Fig. 7 shows ADC maps derived from the reconstructed images through different algorithms. It can be seen that the zero-filling and TV lead to large errors in the mean slice ADC values, and the SIDER results in certain smoothing effects in the derived ADC maps. In contrast, the U-Net and DC-RDN methods can provide a favorable overall appearance in the derived ADC maps, and the DC-RDN owns the smallest MAEADC values.
For all the test data, Fig. 3c summarizes the average MAEADC obtained using the zero-filling, TV, SIDER, U-Net, and DC-RDN. DC-RDN achieves significantly lower MAEADC values than other methods for the total, healthy, and COPD test data (the mean MAEADC is 0.0056 cm2/s, 0.0052 cm2/s, and 0.0060 cm2/s, respectively).
Lung morphometry analysis
Figure 4 shows the lung morphometry maps derived from the FS and DC-RDN reconstructed images for a healthy volunteer (a1, b1) and a COPD patient (a2, b2), respectively. It can be found that all the lung morphometry parameters derived from the reconstructed results visually resemble those from the FS images, and there exists a slight bias in the mean slice values for each morphometry parameter.
Lung morphometry maps derived from the FS and DC-RDN reconstructed images. a1, b1 and a2, b2 Lung morphometry maps of R, r, h, Lm, and SVR in a 26-year-old healthy male volunteer and a 58-year-old male patient with COPD, respectively. The mean slice value of each morphometry parameter is listed under each image. Color bar for the morphometry maps is shown at the bottom. The color-bar scale is μm for the R, r, h, and Lm and cm−1 for the SVR. h, alveolar sleeve depth; Lm, mean linear intercept; R, acinar duct radius; r, acinar lumen radius; SVR, surface-to-volume ratio
Figure 5 shows the Bland-Altman plots of mean ADC and morphometry parameters derived from the FS and DC-RDN reconstructed images on a slice-by-slice basis. Two clusters of data points are observed in the plots, corresponding to the healthy and COPD groups in the test data. For each clustering data, there are good agreements between the FS and DC-RDN results for the six parameters. The mean percentage differences of ADC, R, r, h, Lm, and SVR values are −0.29% (95% confidence interval (CI): −1.84% to 1.25%), 0.19% (95% CI: −1.29% to 1.66%), 0.06% (95% CI: −1.69% to 1.81%), 0.35% (95% CI: −2.19% to 2.89%), 0.17% (95% CI: −2.04% to 2.38%), and 0.17% (95% CI: −1.73% to 2.07%), respectively. Meanwhile, there are no significant differences between the FS and DC-RDN results for such slice parameters (p > 0.05).
Bland-Altman analyses of the mean parameters derived from the FS and DC-RDN reconstructed images on the slice-by-slice basis in 5 healthy volunteers and 5 COPD patients. a–f Bland-Altman plots of ADC, R, r, h, Lm, and SVR values estimated from the FS and DC-RDN reconstructed images, respectively. Solid line indicates the mean difference and dotted lines indicate the 95% confidence interval
Table 1 provides the global mean values of ADC and morphometry parameters derived from the FS and DC-RDN reconstructed images for the 5 healthy volunteers and 5 COPD patients. For each participant, the global mean values of ADC, R, r, h, Lm, and SVR derived from the DC-RDN are nearly identical with the references. While for all the participants, the corresponding mean differences are −0.25%, 0.20%, 0.09%, 0.50%, 0.14%, and 0.10%, respectively. Moreover, for both the FS and DC-RDN reconstructed images, the global mean values of ADC and morphometry parameters obtained from the healthy volunteers are significantly different from those of the COPD patients (p < 0.001).
In vivo
Figure 6 shows the FS and reconstructed multiple b-value 129Xe DW images for a healthy volunteer at an acceleration factor of 4. It can be seen that Fig. 6b suffers from severe oscillatory artifacts and obscured lung structures. After the DC-RDN, such artifacts are successfully removed, and most of the structural details are restored. Figure 6d and e show morphometry maps derived from Fig. 6a and c, respectively. Figure 6e demonstrates good agreement with Fig. 6d, and the mean slice values of lung morphometry parameters are similar between them, confirming good preservation of quantitative microstructural information during DC-RDN reconstruction.
Prospective reconstruction results for a 23-year-old healthy male volunteer at an acceleration factor of 4. a–c Reconstruction images with 5 b-values obtained using the FS, zero-filling, and DC-RDN, respectively. d, e Morphometry maps estimated from the corresponding FS images and DC-RDN reconstructed results, respectively. Color bar for the morphometry maps is shown at the bottom. The color-bar scale is μm for the R, r, h, and Lm and cm−1 for the SVR. The mean slice values of morphometry parameters are listed under each image
Table 2 displays the global mean values of ADC, R, r, h, Lm, and SVR for the 5 healthy volunteers. The respective mean percentage difference is −0.72%, −0.81%, 0.46%, −1.52%, −0.74%, and 1.41% between the FS and DC-RDN reconstructed images. Through the Bland-Altman analysis on a slice-by-slice basis, the mean percentage differences for the six parameters are −0.57% (95% CI: −4.75% to 3.61%), −0.58% (95% CI: −3.93% to 2.78%), 0.68% (95% CI: −6.27% to 7.63%), −1.46% (95% CI: −8.13% to 5.21%), −0.28% (95% CI: −5.35% to 4.79%), and 1.17% (95% CI: −3.33% to 5.67%), respectively.
Discussion
In this work, a novel DC-RDN model is proposed to reconstruct highly accelerated multiple b-value gas DW-MRI for lung morphometry. Our results show that DC-RDN can effectively eliminate aliasing artifacts as well as restore detailed structures of the lung at an acceleration factor of 4. Meanwhile, DC-RDN outperforms the baseline methods regarding MAE, MAEADC, and SSIM terms, as well as image quality scores. For the healthy and COPD test data, the DC-RDN reconstructed images are able to accurately estimate ADC and lung morphometry parameters, which are not significantly different from the parameters derived from the FS images. Furthermore, good agreements in ADC and lung morphometry parameters are observed between the FS and prospectively undersampled datasets, suggesting the clinical potential of the DC-RDN in real accelerated multiple b-value gas DW-MRI.
Since SIDER explores the prior knowledge of diffusion signal decay, it can provide better preservation of fine details of the lung than TV (Fig. 2). Nevertheless, the reconstruction of SIDER needs pre-estimated model parameters provided by the TV reconstructed results, resulting in long reconstruction time [12]. Although TV and SIDER can remove aliasing artifacts to some extent, they lead to over-smooth appearances in lung regions owing to the high acceleration factor. In contrast, the deep learning–based methods can successfully remove aliasing artifacts and provide comparable visual quality to the reference images. Furthermore, DC-RDN can achieve better performance for recovering fine structures of the lung and lower reconstruction errors (i.e., MAE and MAEADC) than the U-Net.
At an acceleration factor of 4, DC-RDN has a negligible influence on the estimation of ADC and lung morphometry parameters. Moreover, the global mean values of ADC and morphometry parameters from the healthy and COPD test data are consistent with those from previous studies [4, 11]. Compared with the healthy volunteers, higher mean values of R, r, and Lm while lower mean values of h and SVR are observed in the COPD patients. This agrees with previous studies [4, 10, 11]. For the DC-RDN reconstructed results, the ADC and morphometry parameters from the COPD patients are significantly different from those of the healthy volunteers. This indicates that DC-RDN has potential to detect changes in lung microstructure associated with COPD.
For the prospectively undersampled acquisition, the required breath-holding time is reduced from 17.8 to 4.7 s for multiple b-value 129Xe DW-MRI. This could be easily tolerable for subjects even with pulmonary diseases. Alternatively, the time savings could be used to increase the image resolution and/or the number of slices. Meanwhile, DC-RDN provides good qualitative and quantitative agreements with the FS datasets. As shown in Table 2, a small bias in global mean values of ADC and morphometry parameters is obtained between the FS and prospectively undersampled datasets. The small bias can be negligible when compared with the large differences in lung microstructure between healthy and COPD subjects [11, 13].
Successful application of deep learning relies on large and high-quality training data, which may not be feasible for some medical imaging problems [40]. This is especially true for multiple b-value gas DW-MRI due to the limitations of non-renewable gas hyperpolarization and breath-holding duration [12]. Training on simulated data is one of the strategies to eliminate this issue and permit generating large amounts of training data with diverse experimental conditions [41, 42]. Since DW signal decay of 129Xe in lungs can be well described by the cylinder model [9], and ventilation images have similar characteristics with b = 0 images, we synthetize DW-MRI training data from hyperpolarized 129Xe ventilation images in this work. Experimental results demonstrate that DC-RDN trained on the simulated data can generalize well to both the retrospectively and prospectively undersampled multiple b-value 129Xe DW-MRI datasets.
Our study has several limitations. First, the synthetic training data may be different from real multiple b-value gas DW images due to the simulation strategy (e.g., different levels of noise and/or artifacts). The deviations between the training and testing can be alleviated by fine-tuning the trained DC-RDN with a small number of real DW-MRI data [24, 43]. Second, the hyper-parameters used in the DC-RDN structure and training process are empirically selected in this work. This cannot ensure optimal reconstruction performance. In the future, the influence of the hyper-parameters should be extensively evaluated to optimize the network performance. Third, the feasibility of DC-RDN on prospective acquisition is only tested on 5 healthy volunteers. However, it would be more important to validate DC-RDN using pathological cases, such as patients with COPD or interstitial lung disease. Therefore, additional evaluation on larger prospective datasets with various diseases will be performed to further demonstrate the generalization of DC-RDN in future studies.
In conclusion, a DC-RDN model is developed to accelerate multiple b-value 129Xe DW-MRI for lung morphometry. The DC-RDN is able to provide both high-quality reconstructed images and good preservation of quantitative microstructural information, which has clinical potential for studying lung diseases, not only COPD but also pneumonia, e.g., coronavirus disease 2019 (COVID-19) [44,45,46].
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- CI:
-
Confidence interval
- COPD:
-
Chronic obstructive lung disease
- DC:
-
Data consistency
- DC-RDN:
-
Deep cascade of residual dense network
- DW-MRI:
-
Diffusion-weighted MRI
- FS:
-
Fully sampled
- h :
-
Alveolar sleeve depth
- L m :
-
Mean linear intercept
- MAE:
-
Mean absolute error
- R :
-
Acinar duct radius
- r :
-
Acinar lumen radius
- RDB:
-
Residual dense block
- SIDER:
-
Signal decay into the reconstruction
- SNR:
-
Signal-to-noise ratio
- SSIM:
-
Structure similarity
- SVR :
-
Surface-to-volume ratio
- TV:
-
Total variation
References
Mugler JP III, Altes TA (2013) Hyperpolarized 129Xe MRI of the human lung. J Magn Reson Imaging 37(2):313–331
Doganay O, Matin T, Chen M et al (2019) Time-series hyperpolarized xenon-129 MRI of lobar lung ventilation of COPD in comparison to V/Q-SPECT/CT and CT. Eur Radiol 29:4058–4067
Kaushik SS, Cleveland ZI, Cofer GP et al (2011) Diffusion-weighted hyperpolarized 129Xe MRI in healthy volunteers and subjects with chronic obstructive pulmonary disease. Magn Reson Med 65:1154–1165
Ouriadov A, Farag A, Kirby M, McCormack DG, Parraga G, Santyr GE (2013) Lung morphometry using hyperpolarized 129Xe apparent diffusion coefficient anisotropy in chronic obstructive pulmonary disease. Magn Reson Med 70:1699–1706
Tafti S, Garrison WJ, Mugler JP III et al (2020) Emphysema index based on hyperpolarized 3He or 129Xe diffusion MRI: performance and comparison with quantitative CT and pulmonary function tests. Radiology 297:201–210
Thomen RP, Quirk JD, Roach D et al (2017) Direct comparison of 129Xe diffusion measurements with quantitative histology in human lungs. Magn Reson Med 77:265–272
Chan HF, Weatherley ND, Johns CS et al (2019) Airway microstructure in idiopathic pulmonary fibrosis: assessment at hyperpolarized 3He diffusion-weighted MRI. Radiology 291:223–229
Sukstanskii AL, Yablonskiy DA (2008) In vivo lung morphometry with hyperpolarized 3He diffusion MRI: theoretical background. J Magn Reson 190:200–210
Sukstanskii AL, Yablonskiy DA (2012) Lung morphometry with hyperpolarized 129Xe: theoretical background. Magn Reson Med 67:856–866
Chan HF, Collier GJ, Weatherley ND, Wild JM (2019) Comparison of in vivo lung morphometry models from 3D multiple b-value 3He and 129Xe diffusion-weighted MRI. Magn Reson Med 81:2959–2971
Zhang H, Xie J, Xiao S et al (2018) Lung morphometry using hyperpolarized 129Xe multi-b diffusion MRI with compressed sensing in healthy subjects and patients with COPD. Med Phys 45:3097–3108
Westcott A, Guo F, Parraga G, Ouriadov A (2019) Rapid single-breath hyperpolarized noble gas MRI-based biomarkers of airspace enlargement. J Magn Reson Imaging 49:1713–1722
Chan HF, Stewart NJ, Parra-Robles J, Collier GJ, Wild JM (2017) Whole lung morphometry with 3D multiple b-value hyperpolarized gas MRI and compressed sensing. Magn Reson Med 77:1916–1925
Chan HF, Stewart NJ, Norquay G, Collier GJ, Wild JM (2018) 3D diffusion-weighted 129Xe MRI for whole lung morphometry. Magn Reson Med 79:2986–2995
Abascal JFPJ, Desco M, Parra-Robles J (2018) Incorporation of prior knowledge of signal behavior into the reconstruction to accelerate the acquisition of diffusion MRI data. IEEE Trans Med Imaging 37:547–556
Ouriadov A, Guo F, McCormack DG, Parraga G (2019) Accelerated 129Xe MRI morphometry of terminal airspace enlargement: feasibility in volunteers and those with alpha-1 antitrypsin deficiency. Magn Reson Med 84:416–426
Lustig M, Donoho D, Pauly JM (2007) Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med 58:1182–1195
Ajraoui S, Lee KJ, Deppe MH, Parnell SR, Parra-Robles J, Wild JM (2010) Compressed sensing in hyperpolarized 3He lung MRI. Magn Reson Med 63:1059–1069
Delattre BMA, Boudabbous S, Hansen C, Neroladaki A, Hachulla AL, Vargas MI (2020) Compressed sensing MRI of different organs: ready for clinical daily practice? Eur Radiol 30:308–319
Hammernik K, Klatzer T, Kobler E et al (2018) Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 79:3055–3071
Yang G, Yu S, Dong H et al (2018) DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE Trans Med Imaging 37:1310–1321
Yang Y, Sun J, Li H et al (2020) ADMM-CSNet: a deep learning approach for image compressive sensing. IEEE Trans Pattern Anal Mach Intell 42:521–538
Chen F, Taviani V, Malkiel I et al (2018) Variable-density single-shot fast spin-echo MRI with deep learning reconstruction by using variational networks. Radiology 289:366–373
Han Y, Yoo J, Kim HH, Shin HJ, Sung K, Ye JC (2018) Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn Reson Med 80:1189–1205
Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA (2019) Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning–proof of concept in congenital heart disease. Magn Reson Med 81:1143–1156
Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS (2018) Image reconstruction by domain-transform manifold learning. Nature 555:487–492
Qin C, Schlemper J, Caballero J, Price AN, Hajnal JV, Rueckert D (2019) Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE Trans Med Imag 38:280–290
Duan C, Deng H, Xiao S et al (2019) Fast and accurate reconstruction of human lung gas MRI with deep learning. Magn Reson Med 82:2273–2285
Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D (2018) A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 37:491–503
Zhang Y, Tian Y, Kong Y, Zhong B, Fu Y (2018) Residual dense network for image super-resolution. Proceedings of the IEEE conference on computer vision and pattern recognition 2472-2481.
Huang G, Liu Z, Maaten Lvd, Weinberger KQ (2017) Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition 2261-2269.
Sun L, Fan Z, Fu X, Huang Y, Ding X, Paisley J (2019) A deep information sharing network for multi-contrast compressed sensing MRI reconstruction. IEEE Trans Image Process 28:6141–6153
Xiang L, Chen Y, Chang W et al (2019) Deep learning based multi-modal fusion for fast MR reconstruction. IEEE Trans Biomed Eng 66:2105–2114
Wang H, Zheng R, Dai F, Wang Q, Wang C (2019) High-field MR diffusion-weighted image denoising using a joint denoising convolutional neural network. J Magn Reson Imag 50:1937–1947
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. Proceedings of the IEEE international conference on computer vision 1026-1034.
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. https://arxiv.org/abs/1412.6980. .
Hyun CM, Kim HP, Lee SM et al (2018) Deep learning for undersampled MRI reconstruction. Phys Med Biol 63:135007
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13:600–612
Yaman B, Hosseini SAH, Moeller S et al (2020) Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data. Magn Reson Med 84:3172–3191
Gong K, Catana C, Qi J, Li Q (2019) PET image reconstruction using deep image prior. IEEE Trans Med Imaging 38:1655–1665
Cai C, Wang C, Zeng Y et al (2018) Single-shot T2 mapping using overlapping-echo detachment planar imaging and a deep convolutional neural network. Magn Reson Med 80:2202–2214
Cohen O, Zhu B, Rosen MS (2018) MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magn Reson Med 80:885–894
Knoll F, Hammernik K, Kobler E, Pock T, Recht MP, Sodickson DK (2019) Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn Reson Med 81:116–128
Guan W, Ni Z, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720
Revel MP, Parkar AP, Prosch H et al (2020) COVID-19 patients and the radiology department–advice from the European Society of Radiology (ESR) and the European Society of Thoracic Imaging (ESTI). Eur Radiol 30:4903–4909
Li H, Zhao X, Wang Y et al (2021) Damaged lung gas-exchange function of discharged COVID-19 patients detected by hyperpolarized 129Xe MRI. Sci Adv. https://doi.org/10.1126/sciadv.abc8180
Funding
This study has received funding from the National Key R&D Program of China (2018YFA0704000), National Natural Science Foundation of China (81625011, 91859206, 81771917, 81825012, 81730048, 21921004), and Key Research Program of Frontier Sciences, CAS (ZDBS-LY-JSC004). Xin Zhou acknowledges the support from the Tencent Foundation through the XPLORER PRIZE.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Xin Zhou.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
•Prospective and retrospective
•Observational
•Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 2325 kb)
Rights and permissions
About this article
Cite this article
Duan, C., Deng, H., Xiao, S. et al. Accelerate gas diffusion-weighted MRI for lung morphometry with deep learning. Eur Radiol 32, 702–713 (2022). https://doi.org/10.1007/s00330-021-08126-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08126-y