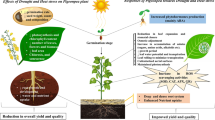
Abstract
The abiotic stress factors associated with climate change frequently enhance the severity of plant diseases, which have a detrimental impact on the growth and productivity of the various crops including legumes. After common beans, the chickpea (Cicer spp.) is the second most cultivated legume crop all over the world. They are susceptible to decreased productivity caused by the detrimental effects of several fungal and bacterial infections, which are regulated by environmental conditions. To understand crop growth, it is crucial to study how plants respond to infections in the presence/fluctuations of abiotic factors. However, to cope with these environmental changes, plants have developed a variety of specific signaling mechanisms for intracellular communications, leading to the initiation of complex defense systems of signal perception and signal transduction to induce/enhance defense responses. Various transcription factors (TFs), along with their cofactors and cis-regulatory elements, play a crucial role in plant defense mechanisms. Transcriptional control by TFs has a vital role in building plant defense mechanisms and related activities in response to viral and bacterial infections. However, the molecular mechanisms including the role of transcription factors (TFs) behind environmental cues are still little understood in chickpea. Therefore, the objective of this review is to outline the potential functions of key stress-responsive transcription factors (TFs), such as WRKY, bHLH, bZIP, AP2/ERF, and MYB gene families, in regulating defense-related genes and facilitating communication across the network of stress-responses during adverse conditions. Furthermore, understanding the function of transcription factors (TFs) could be advantageous in enhancing crop tolerance to develop stress-resistant chickpea cultivars utilizing advanced biotechnological techniques.





Similar content being viewed by others
References
Agarwal G, Garg V, Kudapa H, Doddamani D, Pazhamala LT, Khan AW, Thudi M, Lee SH, Varshney RK (2016) Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol J 14(7):1563–1577. https://doi.org/10.1111/pbi.12520
Agarwal P, Reddy MP, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38(6):3883–3896. https://doi.org/10.1007/s11033-010-0504-5
Ahmad S, Wei X, Sheng Z, Hu P, Tang S (2020) CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations and future prospects. Brief Funct Genomics 19(1):26–39. https://doi.org/10.1093/bfgp/elz041
Akbari A, Ismaili A, Amirbakhtiar N, Pouresmael M, Shobbar ZS (2023) Genome-wide transcriptional profiling provides clues to molecular mechanisms underlying cold tolerance in chickpea. Sci Rep 13(1):6279
Ali A, Javed T, Zaheer U, Zhou JR, Huang MT, Fu HY, Gao SJ (2021) Genome-wide identification and expression profiling of the bHLH transcription factor gene family in Saccharum spontaneum under bacterial pathogen stimuli. Tropical Plant Biology 14(3):283–294
Ali Z, Sarwat SS, Karim I, Faridi R, Jaskani MJ, Khan AA (2016) Functions of plant’s bZIP transcription factors. Pak J Agric Sci 53(2)
Alves MS, Dadalto SP, Gonçalves AB, De Souza GB, Barros VA, Fietto LG (2014) Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2(1):85–106. https://doi.org/10.3390/proteomes2010085
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19(3):307–321. https://doi.org/10.1007/s12298-013-0179-1
Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M (2013) Co-expression of at bHLH17 and at WRKY 28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res 22(2):327–341. https://doi.org/10.1007/s11248-012-9645-8
Babitha KC, Vemanna RS, Nataraja KN, Udayakumar M (2015) Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 10(9):e0137098. https://doi.org/10.1371/journal.pone.0137098
Bai F, Stratmann JW, Matton DP (2022) A complete MAPK cascade, a calmodulin, and a protein phosphatase act downstream of CRK receptor kinases and regulate rabidopsis innate immunity. bioRxiv 2022–03. https://doi.org/10.1101/2022.03.27.486008
Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10(10):771. https://doi.org/10.3390/genes10100771
Bajaj D, Upadhyaya HD, Das S, Kumar V, Gowda CLL, Sharma S, Tyagi AK, Parida SK (2016) Identification of candidate genes for dissecting complex branch number trait in chickpea. Plant Sci 245:61–70
Bajpai R, Kumar G, Sarma BK (2021) Comparative expression analysis and characterization of the ethylene response factor in Cajanus cajan under the influence of Fusarium udum, NaCl and Pseudomonas fluorescens OKC. Environ Exp Bot 186:104428
Bakshi M, Oelmüller R (2014) WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav 9(2):e27700. https://doi.org/10.4161/psb.27700
Bano N, Patel P, Chakrabarty D, Bag SK (2021) Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol Mol Biol Plants 27(8):1747–1764
Bertolini E, Verelst W, Horner DS, Gianfranceschi L, Piccolo V, Inzé D, Pè ME, Mica E (2013) Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant 6(2):423–443. https://doi.org/10.1093/mp/sss160
Bharath P, Gahir S, Raghavendra AS (2021) Abscisic acid-induced stomatal closure: An important component of plant defense against abiotic and biotic stress. Front Plant Sci 12:615114
Bian S, Jin D, Li R, Xie X, Gao G, Sun W, Li Y, Zhai L, Li X (2017) Genome-wide analysis of CCA1-like proteins in soybean and functional characterization of GmMYB138a. Int J Mol Sci 18(10):2040. https://doi.org/10.3390/ijms18102040
Bian Z, Gao H, Wang C (2020) NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops. Int J Mol Sci 22(1):81. https://doi.org/10.3390/ijms22010081
Borrego-Benjumea A, Carter A, Tucker JR, Yao Z, Xu W, Badea A (2020) Genome-wide analysis of gene expression provides new insights into waterlogging responses in barley (Hordeum vulgare L.). Plants 9(2):240. https://doi.org/10.3390/plants9020240
Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13(8):1002–1014
Cai H, Huang Y, Chen F, Liu L, Chai M, Zhang M, Zhang M, Yan M, Aslam M, He Q, Qin Y (2021) ERECTA signaling regulates plant immune responses via chromatin-mediated promotion of WRKY33 binding to target genes. New Phytol 230(2):737–756. https://doi.org/10.1111/nph.17200
Cenci A, Rouard M (2017) Evolutionary analyses of GRAS transcription factors in angiosperms. Front Plant Sci 8:273. https://doi.org/10.3389/fpls.2017.00273
Chakraborty J, Ghosh P, Sen S, Das S (2018) Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci 276:250–267
Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Rep 31(1):87–97. https://doi.org/10.1007/s11105-012-0481-z
Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y, Zhang L (2017) The WRKY transcription factor family in model plants and crops. Crit Rev Plant Sci 36(5–6):311–335. https://doi.org/10.1080/07352689.2018.1441103
Chen Y, Yang Z, Xiao Y, Wang P, Wang Y, Ge X, Li F (2018) Genome-wide analysis of the NF-YB gene family in Gossypium hirsutum L. and characterization of the role of GhDNF-YB22 in embryogenesis. Int J Mol Sci 19(2):483. https://doi.org/10.3390/ijms19020483
Cheng L, Li S, Hussain J, Xu X, Yin J, Zhang Y, Chen X, Li L (2013) Isolation and functional characterization of a salt responsive transcriptional factor, LrbZIP from lotus root (Nelumbo nucifera Gaertn). Mol Biol Rep 40(6):4033–4045. https://doi.org/10.1007/s11033-012-2481-3
Cohen SP, Leach JE (2019) Abiotic and biotic stresses induce a core transcriptome response in rice. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-42731-8
Corrêa LGG, Riaño-Pachón DM, Schrago CG, Vicentini dos Santos R, Mueller-Roeber B, Vincentz M (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3(8):e2944. https://doi.org/10.1371/journal.pone.0002944
Dang F, Wang Y, She J, Lei Y, Liu Z, Eulgem T, Lai Y, Lin J, Yu L, Lei D, He S (2014) Overexpression of CaWRKY27, a subgroup Iie WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol Plant 150(3):397–411. https://doi.org/10.1111/ppl.12093
Das A, Pramanik K, Sharma R, Gantait S, Banerjee J (2019) In-silico study of biotic and abiotic stress-related transcription factor binding sites in the promoter regions of rice germin-like protein genes. PLoS ONE 14(2):e0211887. https://doi.org/10.1111/ppl.12093
Deokar AA, Kondawar V, Kohli D, Aslam M, Jain PK, Karuppayil SM, Varshney RK, Srinivasan R (2015) The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct Integr Genomics 15:27–46
Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88(5):685–694. https://doi.org/10.1016/S0092-8674(00)81911-X
Dong Y, Wang C, Han X, Tang S, Liu S, Xia X, Yin W (2014) A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem Biophys Res Commun 450(1):453–458. https://doi.org/10.1016/j.bbrc.2014.05.139
Erpen L, Devi HS, Grosser JW, Dutt M (2018) Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult (PCTOC) 132(1):1–25. https://doi.org/10.1007/s11240-017-1320-6
Fan Y, Lai D, Yang H, Xue G, He A, Chen L, Feng L, Ruan J, Xiang D, Yan J, Cheng J (2021) Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genomics 22:1–18
Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66(1):94–116. https://doi.org/10.1111/j.1365-313X.2010.04459.x
Feng K, Hao JN, Liu JX, Huang W, Wang GL, Xu ZS, Huang Y, Xiong AS (2019) Genome-wide identification, classification, and expression analysis of TCP transcription factors in carrot. Can J Plant Sci 99(4):525–535. https://doi.org/10.1139/cjps-2018-0232
Foyer CH, Rasool B, Davey JW, Hancock RD (2016) Cross-tolerance to biotic and abiotic stresses in plants: a focus on resistance to aphid infestation. J Exp Bot 67(7):2025–2037. https://doi.org/10.1093/jxb/erw079
Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci 111(6):2367–2372. https://doi.org/10.1073/pnas.1316278111
Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M (2015) Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep 5(1):1–7. https://doi.org/10.1038/srep07773
Gao F, Robe K, Gaymard F, Izquierdo E, Dubos C (2019) The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Front Plant Sci 10:6. https://doi.org/10.3389/fpls.2019.00006
Gao H, Wang Y, Xu P, Zhang Z (2018) Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front Plant Sci 9:997. https://doi.org/10.3389/fpls.2018.00997
Godiard L, Lepage A, Moreau S, Laporte D, Verdenaud M, Timmers T, Gamas P (2011) MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol 191(2):391–404
Gu C, Guo ZH, Hao PP, Wang GM, Jin ZM, Zhang SL (2017) Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot Stud 58(1):1–8. https://doi.org/10.1186/s40529-016-0159-1
Gu L, Ma Q, Zhang C, Wang C, Wei H, Wang H, Yu S (2019) The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front Plant Sci 10:1352. https://doi.org/10.3389/fpls.2019.01352
Gu L, Wei H, Wang H, Su J, Yu S (2018) Characterization and functional analysis of GhWRKY42, a group Iid WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genet 19(1):1–14. https://doi.org/10.1186/s12863-018-0653-4
Guo B, Wei Y, Xu R, Lin S, Luan H, Lv C, Zhang X, Song X, Xu R (2016) Genome-wide analysis of APETALA2/ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS ONE 11(9):e0161322. https://doi.org/10.1371/journal.pone.0161322
Gupta S, Mishra VK, Kumari S, Chand R, Varadwaj PK (2019) Deciphering genome-wide WRKY gene family of Triticum aestivum L. and their functional role in response to Abiotic stress. Genes Genomics 41(1):79–94. https://doi.org/10.1007/s13258-018-0742-9
Ha CV, Nasr Esfahani M, Watanabe Y, Tran UT, Sulieman S, Mochida K, Tran LSP (2014) Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLoS ONE 9(12):e114107. https://doi.org/10.1371/journal.pone.0114107
Häffner E, Konietzki S, Diederichsen E (2015) Keeping control: the role of senescence and development in plant pathogenesis and defense. Plants 4(3):449–488. https://doi.org/10.3390/plants4030449
Han G, Lu C, Guo J, Qiao Z, Sui N, Qiu N, Wang B (2020) C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front Plant Sci 11:115. https://doi.org/10.3389/fpls.2020.00115
Hao Y, Zong X, Ren P, Qian Y, Fu A (2021) Basic Helix-Loop-Helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int J Mol Sci 22(13):7152. https://doi.org/10.3390/ijms22137152
Haque E, Taniguchi H, Hassan MM, Bhowmik P, Karim MR, Śmiech M, Zhao K, Rahman M, Islam T (2018) Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: recent progress, prospects, and challenges. Front Plant Sci 9:617. https://doi.org/10.3389/fpls.2018.00617
He L, Shi X, Wang Y, Guo Y, Yang K, Wang Y (2017) Arabidopsis ANAC069 binds to C [A/G] CG [T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol 93(4–5):369–387. https://doi.org/10.1007/s11103-016-0567-3
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20(5):735–747. https://doi.org/10.1093/molbev/msg088
Hrmova M, Hussain SS (2021) Plant transcription factors involved in drought and associated stresses. Int J Mol Sci 22(11):5662. https://doi.org/10.3390/ijms22115662
Hsu FC, Chou MY, Chou SJ, Li YR, Peng HP, Shih MC (2013) Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 25(7):2699–2713. https://doi.org/10.1105/tpc.113.114447
Huang Y, Li T, Xu ZS, Wang F, Xiong AS (2017) Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol Biochem 120:61–74. https://doi.org/10.1016/j.plaphy.2017.09.020
Hung YH, Slotkin RK (2021) The initiation of RNA interference (RNAi) in plants. Curr Opin Plant Biol 61:102014. https://doi.org/10.1016/j.pbi.2021.102014
Huang Q, Wang Y, Li B, Chang J, Chen M, Li K, He G (2015) TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol 15(1):1–15. https://doi.org/10.1186/s12870-015-0644-9
Huang C, Yu J, Cai Q, Chen Y, Li Y, Ren Y, Miao Y (2020) Triple-localized WHIRLY2 influences leaf senescence and silique development via carbon allocation. Plant Physiol 184(3):1348–1362. https://doi.org/10.1104/pp.20.00832
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet MGG 244(6):563–571. https://doi.org/10.1007/BF00282746
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74(5):715–729
Jalmi SK, Bhagat PK, Verma D, Noryang S, Tayyeba S, Singh K, Sharma D, Sinha AK (2018) Traversing the links between heavy metal stress and plant signaling. Front Plant Sci 9:12. https://doi.org/10.3389/fpls.2018.00012
Jamil S, Shahzad R, Kanwal S, Yasmeen E, Rahman SU, Iqbal MZ (2020) DNA fingerprinting and population structure of date palm varieties grown in Punjab Pakistan using simple sequence repeat markers. Int J Agric Biol 23(5):943–950. https://doi.org/10.17957/IJAB/15.1373
Javed T, Shabbir R, Ali A, Afzal I, Zaheer U, Gao SJ (2020) Transcription factors in plant stress responses: challenges and potential for sugarcane improvement. Plants 9(4):491. https://doi.org/10.3390/plants9040491
Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J (2017) WRKY transcription factors in plant responses to stresses. J Integr Plant Biol 59(2):86–101. https://doi.org/10.1111/jipb.12513
Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G (2016) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw982
Jofuku KD, Den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6(9):1211–1225. https://doi.org/10.1105/tpc.6.9.1211
Kaur S, Samota MK, Choudhary M, Choudhary M, Pandey AK, Sharma A, Thakur J (2022) How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol Mol Biol Plants 28(2):485–504. https://doi.org/10.1007/s12298-022-01146-y
Kavas M, Kizildogan A, Gökdemir G, Baloglu MC (2015) Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J 14:1187. https://doi.org/10.17179/excli2015-600
Ke YZ, Wu YW, Zhou HJ, Chen P, Wang MM, Liu MM, Li PF, Yang J, Li JN, Du H (2020) Genome-wide survey of the bHLH super gene family in Brassica napus. BMC Plant Biol 20:1–16
Khan SA, Li MZ, Wang SM, Yin HJ (2018) Revisiting the role of plant transcription factors in the battle against abiotic stress. Int J Mol Sci 19(6):1634. https://doi.org/10.3390/ijms19061634
Khandal H, Gupta SK, Dwivedi V, Mandal D, Sharma NK, Vishwakarma NK, Pal L, Choudhary M, Francis A, Malakar P, Chattopadhyay D (2020) Root-specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol J 18(11):2225–2240. https://doi.org/10.1111/pbi.13378
Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61:561–591. https://doi.org/10.1146/annurev-arplant-042809-112226
Kiranmai K, Lokanadha Rao G, Pandurangaiah M, Nareshkumar A, Amaranatha Reddy V, Lokesh U, Venkatesh B, Anthony Johnson AM, Sudhakar C (2018) A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front Plant Sci 9:346. https://doi.org/10.3389/fpls.2018.00346
Konda AK, Farmer R, Soren KR, PS S, Setti A (2018) Structural modelling and molecular dynamics of a multi-stress responsive WRKY TF-DNA complex towards elucidating its role in stress signalling mechanisms in chickpea. J Biomol Struct Dyn 36(9):2279–2291. https://doi.org/10.1080/07391102.2017.1349690
Konda AK, Sabale PR, Soren KR, Subramaniam SP, Singh P, Rathod S, Chaturvedi SK, Singh NP (2019) Systems biology approaches reveal a multi-stress responsive WRKY transcription factor and stress associated gene co-expression networks in chickpea. Curr Bioinform 14(7):591–601. https://doi.org/10.2174/1574893614666190204152500
Kujur ALICE, Bajaj DEEPAK, Saxena MS, Tripathi SHAILESH, Upadhyaya HD, Gowda CLL, Singh SUBE, Jain MUKESH, Tyagi AK, Parida SK (2013) Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res 20(4):355–374
Kumar G, Bajpai R, Sarkar A, Mishra RK, Kumar Gupta V, Singh HB, Sarma BK (2019) Identification, characterization and expression profiles of Fusarium udum stress-responsive WRKY transcription factors in Cajanus cajan under the influence of NaCl stress and Pseudomonas fluorescens OKC. Sci Rep 9:14344. https://doi.org/10.1038/s41598-019-50696-x
Kumar K, Srivastava V, Purayannur S, Kaladhar VC, Cheruvu PJ, Verma PK (2016) WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene (s). DNA Res 23(3):225–239
Lan Thi Hoang X, Du Nhi NH, Binh Anh Thu N, Phuong Thao N, Phan Tran LS (2017) Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr Genomics 18(6):483–497. https://doi.org/10.2174/1389202918666170227150057
Le Hir R, Castelain M, Chakraborti D, Moritz T, Dinant S, Bellini C (2017) At bHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol Plant 160(3):312–327. https://doi.org/10.1111/ppl.12549
Le TN, Schumann U, Smith NA, Tiwari S, Au PCK, Zhu QH, Taylor JM, Kazan K, Llewellyn DJ, Zhang R, Wang MB (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol 15(9):1–18. https://doi.org/10.1186/s13059-014-0458-3
Lee SB, Kim H, Kim RJ, Suh MC (2014) Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep 33(9):1535–1546
Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK (2006) Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224(5):1209–1225
Li J, Lin K, Zhang S, Wu J, Fang Y, Wang Y (2021) Genome-wide analysis of myeloblastosis-related genes in Brassica napus L. and positive modulation of osmotic tolerance by BnMRD107. Front Plant Sci 12:978
Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233(6):1237–1252
Li S, Liu S, Zhang Q, Cui M, Zhao M, Li N, Wang S, Wu R, Zhang L, Cao Y, Wang L (2022) The interaction of ABA and ROS in plant growth and stress resistances. Front Plant Sci 13:1050132. https://doi.org/10.3389/fpls.2022.1050132
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Zhang D (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141(4):1167–1184
Li X, Fan S, Hu W, Liu G, Wei Y, He C, Shi H (2017) Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Front Plant Sci 8:2110. https://doi.org/10.3389/fpls.2017.02110
Li X, Guo C, Gu J, Duan W, Zhao M, Ma C, Du X, Lu W, Xiao K (2014) RETRACTED: overexpression of VP, a vacuolar H+-pyrophosphatase gene in wheat (Triticum aestivum L.), improves tobacco plant growth under Pi and N deprivation, high salinity, and drought. J Exp Bot 65(2):683–696
Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol 170(4):2478–2493
Li Z, Thomas TL (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10(3):383–398
Li Z, Hua X, Zhong W, Yuan Y, Wang Y, Wang Z, Ming R, Zhang J (2020) Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol 61(3):616–630
Liang J, Fang Y, An C, Yao Y, Wang X, Zhang W, Liu R, Wang L, Aslam M, Cheng Y, Zheng P (2023b) Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int J Biol Macromol 225:389–403
Liang Y, Heyman J, Lu R, De Veylder L (2023a) Evolution of wound-activated regeneration pathways in the plant kingdom. Eur J Cell Biol 102(2):151291
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3(1):1–11
Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22(9):2930–2942
Liu X, Huang J, Parameswaran S, Ito T, Seubert B, Auer M, Rymaszewski A, Jia G, Owen HA, Zhao D (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151(3):1401–1411
Liu GS, Li HL, Grierson D, Fu DQ (2022a) NAC transcription factor family regulation of fruit ripening and quality: a review. Cells 11(3):525. https://doi.org/10.3390/cells11030525
Liu Q, Liu Y, Tang Y, Chen J, Ding W (2017) Overexpression of NtWRKY50 increases resistance to Ralstonia solanacearum and alters salicylic acid and jasmonic acid production in tobacco. Front Plant Sci 8:1710
Liu S, Liu Y, Liu C, Zhang F, Wei J, Li B (2022c) Genome-wide characterization and expression analysis of GeBP family genes in soybean. Plants 11(14):1848
Liu M, Ma Z, Sun W, Huang L, Wu Q, Tang Z, Bu T, Li C, Chen H (2019) Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genomics 20(1):1–16
Liu JH, Peng T, Dai W (2014a) Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol Biol Rep 32:303–317
Liu H, Song S, Zhang H, Li Y, Niu L, Zhang J, Wang W (2022b) Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int J Mol Sci 23(23):14824
Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX (2014b) bHLH 122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201(4):1192–1204
López-Galiano MJ, González-Hernández AI, Crespo-Salvador O, Rausell C, Real MD, Escamilla M, Camañes G, García-Agustín P, González-Bosch C, García-Robles I (2018) Epigenetic regulation of the expression of WRKY75 transcription factor in response to biotic and abiotic stresses in Solanaceae plants. Plant Cell Rep 37:167–176
Lu X, Zhang H, Hu J, Nie G, Khan I, Feng G, Zhang X, Wang X, Huang L (2022) Genome-wide identification and characterization of bHLH family genes from orchardgrass and the functional characterization of DgbHLH46 and DgbHLH128 in drought and salt tolerance. Funct Integr Genomics 1–14. https://doi.org/10.1007/s10142-022-00890-4
Ludwig SR, Habera LF, Dellaporta SL, Wessler S (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci 86(18):7092–7096
Mabuchi K, Maki H, Itaya T, Suzuki T, Nomoto M, Sakaoka SS, Morikami A, Higashiyama T, Tada Y, Busch W, Tsukagoshi H (2018) MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc Natl Acad Sci 115(20):E4710–E4719
Maiti RK, Satya P (2014) Research advances in major cereal crops for adaptation to abiotic stresses. GM Crops & Food 5(4):259–279
Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23(4):1639–1653
Meher J, Lenka S, Sarkar A, Sarma BK (2023a) Transcriptional regulation of OsWRKY genes in response to individual and overlapped challenges of Magnaporthe oryzae and drought in indica genotypes of rice. Environ Exp Bot 207:105221. https://doi.org/10.1016/j.envexpbot.2023.105221
Meher J, Sarkar A, Sarma BK (2023b) Binding of stress-responsive OsWRKY proteins through WRKYGQK heptapeptide residue with the promoter region of two rice blast disease resistance genes Pi2 and Pi54 is important for development of blast resistance. 3 Biotech 13(9):294. https://doi.org/10.1007/s13205-023-03711-y
Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63(2):179–188
Michael AK, Stoos L, Crosby P, Eggers N, Nie XY, Makasheva KK, Minnich M, Healy KL, Weiss J, Kempf G, Thomä NH (2023) Cooperation between bHLH transcription factors and histones for DNA access. Nature 619(7969):385–393
Molina C, Rotter B, Horres R, Udupa SM, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G, Winter P (2008) SuperSAGE: the drought stress-responsive transcriptome 880 of chickpea roots. BMC Genomics 9:553
Monsur MB, Shao G, Lv Y, Ahmad S, Wei X, Hu P, Tang S (2020) Base editing: the ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants. Genes 11(4):466
Muthamilarasan M, Khandelwal R, Yadav CB, Bonthala VS, Khan Y, Prasad M (2014) Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 9(10):e109920. https://doi.org/10.1371/journal.pone.0109920
Nan H, Lin Y, Wang X, Gao L (2021) Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Hortic Plant J 7(5):469–478
Ng DW, Abeysinghe JK, Kamali M (2018) Regulating the regulators: the control of transcription factors in plant defense signaling. Int J Mol Sci 19(12):3737
Ng DW, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ (2011) Cis-and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23(5):1729–1740
Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, Giraud E (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25(9):3450–3471
Nguyen D, Rieu I, Mariani C, van Dam NM (2016) How plants handle multiple stresses: hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol Biol 91(6):727–740
Niu X, Luo T, Zhao H, Su Y, Ji W, Li H (2020) Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 740:144514
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465(1–2):30–44
Oladosu Y, Rafii MY, Arolu F, Chukwu SC, Muhammad I, Kareem I, Salisu MA, Arolu IW (2020) Submergence tolerance in rice: review of mechanism, breeding and future prospects. Sustainability 12(4):1632
Pandey B, Gupta OP, Pandey DM, Sharma I, Sharma P (2013) Identification of new stress-induced microRNA and their targets in wheat using computational approach. Plant Signal Behav 8(5):e23932
Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15(7):1538–1551
Parveen S, Pandey A, Jameel N, Chakraborty S, Chakraborty N (2018) Transcriptional regulation of chickpea ferritin CaFer1 influences its role in iron homeostasis and stress response. J Plant Physiol 222:9–16
Peng H, Cheng HY, Chen C, Yu XW, Yang JN, Gao WR, Shi QH, Zhang H, Li JG, Ma H (2009) A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J Plant Physiol 166(17):1934–1945
Peng W, Yang Y, Xu J, Peng E, Dai S, Dai L, Wang Y, Yi T, Wang B, Li D, Song N (2021) TALE transcription factors in sweet orange (Citrus sinensis): Genome-wide identification, characterization, and expression in response to biotic and abiotic stresses. Front Plant Sci 12
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Prasad KV, Xing D, Reddy AS (2018) Vascular plant one-zinc-finger (VOZ) transcription factors are positive regulators of salt tolerance in Arabidopsis. Int J Mol Sci 19(12):3731
Qin Y, Tian Y, Liu X (2015) A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun 464(2):428–433
Rai N, Kumari S, Singh S, Saha P, Pandey-Rai S (2023) Genome-wide identification of bZIP transcription factor family in Artemisia annua, its transcriptional profiling and regulatory role in phenylpropanoid metabolism under different light conditions. Physiol Mol Biol Plants 29(7):905–925. https://doi.org/10.1007/s12298-023-01338-0
Rajput R, Tyagi S, Naik J, Pucker B, Stracke R, Pandey A (2022) The R2R3-MYB gene family in Cicer arietinum: genome-wide identification and expression analysis leads to functional characterization of proanthocyanidin biosynthesis regulators in the seed coat. Planta 256(4):67
Ramalingam A, Kudapa H, Pazhamala LT, Garg V, Varshney RK (2015) Gene expression and yeast two-hybrid studies of 1R-MYB transcription factor mediating drought stress response in chickpea (Cicer arietinum L.). Front Plant Sci 6:1117
Rambani A, Hu Y, Piya S, Long M, Rice JH, Pantalone V, Hewezi T (2020) Identification of differentially methylated miRNA genes during compatible and incompatible interactions between soybean and soybean cyst nematode. Mol Plant Microbe Interact 33(11):1340–1352. https://doi.org/10.1094/MPMI-07-20-0196-R
Roorkiwal M, Bharadwaj C, Barmukh R, Dixit GP, Thudi M, Gaur PM, Chaturvedi SK, Fikre A, Hamwieh A, Kumar S, Varshney RK (2020) Integrating genomics for chickpea improvement: achievements and opportunities. Theor Appl Genet 133:1703–1720
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15(5):247–258
Sahil, Keshan R, Patra A, Mehta S, Abdelmotelb KF, Lavale SA, Chaudhary M, Aggarwal SK, Chattopadhyay A (2021) Expression and regulation of stress-responsive genes in plants under harsh environmental conditions. Harsh Environ Plant Resilience: Mol Funct Aspects 25–44
Saxena S, Pal L, Naik J, Singh Y, Verma PK, Chattopadhyay D, Pandey A (2023) The R2R3-MYB-SG7 transcription factor CaMYB39 orchestrates surface phenylpropanoid metabolism and pathogen resistance in chickpea. New Phytol 238(2):798–816
Schuster-Böckler B, Schultz J, Rahmann S (2004) HMM Logos for visualization of protein families. BMC Bioinformatics 5(1):1–8
Shahzad R, Shakra Jamil SA, Nisar A, Amina Z, Saleem S, Iqbal MZ, Atif MR, Wang X (2021) Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: Current and future perspectives. Saudi J Biol Sci 28(4):2323
Shao H, Wang H, Tang X (2015) NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci 6:902
Sharma V, Goel P, Kumar S, Singh AK (2019) An apple transcription factor, MdDREB76, confers salt and drought tolerance in transgenic tobacco by activating the expression of stress-responsive genes. Plant Cell Rep 38(2):221–241
Shen XJ, Wang YY, Zhang YX, Guo W, Jiao YQ, Zhou XA (2018) Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int J Mol Sci 19(12):3958
Shimray PW, Bajaj D, Srivastava R, Daware A, Upadhyaya HD, Kumar R, Bharadwaj C, Tyagi AK, Parida SK (2017) Identifying transcription factor genes associated with yield traits in chickpea. Plant Mol Biol Report 35:562–574. https://doi.org/10.1007/s11105-017-1044-0
Shriti S, Paul S, Das S (2023) Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against Fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway. Protoplasma 260(2):589–605
Shriti S, Das S (2023) Chickpea R2R3 transcription factor CaMYB78 enhances abiotic stress tolerance in tobacco. J Plant Growth Regul 1–17. https://doi.org/10.1007/s00344-023-10916-1
Siddiq M, Uebersax MA, Siddiq F (2022) Global production, trade, processing and nutritional profile of dry beans and other pulses. Dry Beans Pulses: Prod Process Nutr 1–28
Song H, Liu Y, Dong G, Zhang M, Wang Y, Xin J, Su Y, Sun H, Yang M, Yang M (2022) Genome-wide characterization and comprehensive analysis of NAC transcription factor family in Nelumbo nucifera. Front Genet 13:901838. https://doi.org/10.3389/fgene.2022.901838
Stagnari F, Maggio A, Galieni A, Pisante M (2017) Multiple benefits of legumes for agriculture sustainability: an overview. Chem Biol Technol Agric 4(1):1–13
Su LT, Li JW, Liu DQ, Zhai Y, Zhang HJ, Li XW, Zhang QL, Wang Y, Wang QY (2014) A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538(1):46–55
Sukumari Nath V, Kumar Mishra A, Kumar A, Matoušek J, Jakše J (2019) Revisiting the role of transcription factors in coordinating the defense response against citrus bark cracking viroid infection in commercial hop (Humulus Lupulus L.). Viruses 11(5):419
Tak H, Mhatre M (2013) Cloning and molecular characterization of a putative bZIP transcription factor VvbZIP23 from Vitis vinifera. Protoplasma 250(1):333–345
Tanaka K, Mudgil Y, Tunc-Ozdemir M (2023) Abiotic stress and plant immunity–a challenge in climate change. Front Plant Sci 14:1197435. https://doi.org/10.3389/fpls.2023.1197435
Tian Y, Pu X, Yu H, Ji A, Gao R, Hu Y, Xu Z, Wang H (2020) Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Crocin Biosynthesis in Gardenia jasminoides Ellis (Rubiaceae). BioMed Res Int. https://doi.org/10.1155/2020/2903861
Tiwari RK, Lal MK, Kumar R, Mangal V, Altaf MA, Sharma S, Singh B, Kumar M (2021) Insight into melatonin-mediated response and signaling in the regulation of plant defense under biotic stress. Plant Mol Biol 1–15
Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16(9):2481–2498
Tsuda K, Somssich IE (2015) Transcriptional networks in plant immunity. New Phytol 206(3):932–947
van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ (2008) A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146(4):1983–1995
Varshney RK, Ribaut JM, Buckler ES, Tuberosa R, Rafalski JA, Langridge P (2012) Can genomics boost productivity of orphan crops? Nat Biotechnol 30(12):1172–1176. https://doi.org/10.1038/nbt.2440
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, Millan T, Cook DR (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31(3):240–246. https://doi.org/10.1038/nbt.2491
Verma V, Srivastava AK, Gough C, Campanaro A, Srivastava M, Morrell R, Joyce J, Bailey M, Zhang C, Krysan PJ, Sadanandom A (2021) SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc Natl Acad Sci 118(10):e2021351118. https://doi.org/10.1111/jipb.12973
Wang Z, Cheng K, Wan L, Yan L, Jiang H, Liu S, Lei Y, Liao B (2015b) Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1–15. https://doi.org/10.1186/s12864-015-2258-x
Wang K, Liu H, Mei Q, Yang J, Ma F, Mao K (2023) Characteristics of bHLH transcription factors and their roles in the abiotic stress responses of horticultural crops. Sci Hortic 310:111710
Wang MM, Liu MM, Ran F, Guo PC, Ke YZ, Wu YW, Wen J, Li PF, Li JN, Du H (2018c) Global analysis of WOX transcription factor gene family in Brassica napus reveals their stress-and hormone-responsive patterns. Int J Mol Sci 19(11):3470
Wang L, Liu F, Zhang X, Wang W, Sun T, Chen Y, Dai M, Yu S, Xu L, Su Y, Que Y (2018b) Expression characteristics and functional analysis of the ScWRKY3 gene from sugarcane. Int J Mol Sci 19(12):4059
Wang CT, Ru JN, Liu YW, Yang JF, Li M, Xu ZS, Fu JD (2018a) The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int J Mol Sci 19(9):2580
Wang J, Tao F, An F, Zou Y, Tian W, Chen X, Hu X (2017a) Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol Plant Pathol 18(5):649–661
Wang H, Wang H, Shao H, Tang X (2016b) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front Plant Sci 7:67
Wan L, Wu Y, Huang J, Dai X, Lei Y, Yan L, Jiang H, Liao B (2014) Identification of ERF genes in peanuts and functional analysis of AhERF008 and AhERF019 in abiotic stress response. Funct Integr Genomics 14(3):467–477
Wang L, Xiang L, Hong J, Xie Z, Li B (2019) Genome-wide analysis of bHLH transcription factor family reveals their involvement in biotic and abiotic stress responses in wheat (Triticum aestivum L.). 3 Biotech 9:1–12
Wang L, Zheng L, Zhang C, Wang Y, Lu M, Gao C (2015a) ThWRKY4 from Tamarix hispida can form homodimers and heterodimers and is involved in abiotic stress responses. Int J Mol Sci 16(11):27097–27106
Wang F, Zhu H, Kong W, Peng R, Liu Q, Yao Q (2016a) The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 244(1):59–73
Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F (2017b) Drought tolerance conferred in soybean (Glycine max L.) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol 58(10):1764–1776
Waqas M, Azhar MT, Rana IA, Azeem F, Ali MA, Nawaz MA, Chuang G, Atif RM (2019) Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum L.) reveal their role in abiotic stress-responses. Genes Genomics 41(4):467–481
Wei KAIFA, Chen JUAN, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D (2012a) Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19(6):463–476
Wei KF, Chen J, Chen YF, Wu LJ, Xie DX (2012b) Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res 19(2):153–164
Wei W, Liang DW, Bian XH, Shen M, Xiao JH, Zhang WK, Ma B, Lin Q, Lv J, Chen X, Zhang JS (2019) GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J 100(2):384–398
Weiste C, Pedrotti L, Selvanayagam J, Muralidhara P, Fröschel C, Novák O, Ljung K, Hanson J, Dröge-Laser W (2017) The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet 13(2):e1006607
Xiao P, Feng JW, Zhu XT, Gao J (2021) Evolution analyses of CAMTA transcription factor in plants and its enhancing effect on cold-tolerance. Front Plant Sci 12
Xiong Y, Lin D, Ma S, Wang C, Lin S (2022) Genome-wide identification of the calcium-dependent protein kinase gene family in Fragaria vesca and expression analysis under different biotic stresses. Eur J Plant Pathol 164(2):283–298. https://doi.org/10.1007/s10658-022-02560-4
Xu Z, Wang C, Xue F, Zhang H, Ji W (2015) Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Biochem 96:356–363
Yadav BS, Mani A (2019) Analysis of bHLH coding genes of Cicer arietinum during heavy metal stress using biological network. Physiol Mol Biol Plants 25:113–121. https://doi.org/10.1007/s12298-018-0625-1
Yadav S, Yadava YK, Meena S, Singh L, Kansal R, Grover M, MS N, Bharadwaj C, Paul V, Gaikwad K, Jain PK (2023) The SPL transcription factor genes are potential targets for epigenetic regulation in response to drought stress in chickpea (C. arietinum L.). Mol Biol Rep 1–9
Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S (2013) DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci 18(5):267–276
Yang JH, Lee KH, Du Q, Yang S, Yuan B, Qi L, Wang H (2020) A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol 226(1):59–74
Yang O, Popova OV, Süthoff U, Lüking I, Dietz KJ, Golldack D (2009) The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436(1–2):45–55
Yang G, Zhang W, Liu Z, Yi-Maer AY, Zhai M, Xu Z (2017) Both Jr WRKY 2 and Jr WRKY 7 of Juglans regia mediate responses to abiotic stresses and abscisic acid through formation of homodimers and interaction. Plant Biol 19(2):268–278
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Li-Jia Q (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60(1):107–124
Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120(2):249–259
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada KH, Shimono M, Sugano S, Takatsuji H, Nishizawa Y (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64(16):5085–5097
You J, Chan Z (2015) ROS regulation during abiotic stress responses in crop plants. Front Plant Sci 6:1092
Younis A, Siddique MI, Kim CK, Lim KB (2014) RNA interference (RNAi) induced gene silencing: a promising approach of hi-tech plant breeding. Int J Biol Sci 10(10):1150. https://doi.org/10.7150/ijbs.10452
Yu X, Liu Y, Wang S, Tao Y, Wang Z, Shu Y, Peng H, Mijiti A, Wang Z, Zhang H, Ma H (2016) CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep 35:613–627. https://doi.org/10.1007/s00299-015-1907-5
Yuan X, Wang H, Cai J, Li D, Song F (2019) NAC transcription factors in plant immunity. Phytopathol Res 1(1):1–13. https://doi.org/10.1186/s42483-018-0008-0
Zargar SM, Zargar MY (Eds.) (2018) Abiotic stress-mediated sensing and signaling in plants: An omics perspective. Springer
Zhang C, McGee RJ, Vandemark GJ, Sankaran S (2021) Crop performance evaluation of chickpea and dry pea breeding lines across seasons and locations using phenomics data. Front Plant Sci 12:640259. https://doi.org/10.3389/fpls.2021.640259
Zhang G, Chen M, Chen X, Xu Z, Guan S, Li LC, Ma Y (2008) Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot 59(15):4095–4107
Zhang J, Liu B, Li J, Zhang L, Wang Y, Zheng H, Chen J (2015) Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics 16(1):1–19
Zhang Y, Li D, Wang Y, Zhou R, Wang L, Zhang Y, Gong H, You J, Zhang X (2018b) Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS ONE 13(6):e0199262
Zhang T, Zhao Y, Wang Y, Liu Z, Gao C (2018a) Comprehensive analysis of MYB gene family and their expressions under abiotic stresses and hormone treatments in Tamarix hispida. Front Plant Sci 9:1303
Zhao M, Song A, Li P, Chen S, Jiang J, Chen F (2014) A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci Rep 4(1):1–6
Zhao SP, Song XY, Guo LL, Zhang XZ, Zheng WJ (2020) Genome-wide analysis of the shi-related sequence family and functional identification of GmSRS18 involving in drought and salt stresses in soybean. Int J Mol Sci 21(5):1810
Zhao Q, Xiang X, Liu D, Yang A, Wang Y (2018) Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front Plant Sci 9:381
Zhao SP, Xu ZS, Zheng WJ, Zhao W, Wang YX, Yu TF, Chen M, Zhou YB, Min DH, Ma YZ, Zhang XH (2017) Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front Plant Sci 8:905
Zhao Y, Zhang YY, Liu H, Zhang XS, Ni R, Wang PY, Gao S, Lou HX, Cheng AX (2019) Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol 19(1):1–13
Zhou J, Wang X, He Y, Sang T, Wang P, Dai S, Zhang S, Meng X (2020) Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell 32(8):2621–2638. https://doi.org/10.1105/tpc.19.00971
Zhou Q, Zhang S, Chen F, Liu B, Wu L, Li F, Zhang J, Bao M, Liu G (2018) Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genomics 19(1):1–18. https://doi.org/10.1186/s12864-018-4537-9
Zhou Y, Zheng R, Peng Y, Chen J, Zhu X, Xie K, Su Q, Huang R, Zhan S, Peng D, Liu ZJ (2023) Bioinformatic assessment and expression profiles of the AP2/ERF superfamily in the melastoma dodecandrum genome. Int J Mol Sci 24(22):16362. https://doi.org/10.3390/ijms242216362
Acknowledgements
The authors are grateful to the Department of Botany, Institute of Science and Department of Mycology & Plant Pathology, Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, India for providing the necessary facilities for preparing the manuscript. NR, SPR, and BKS are thankful to BHU (IoE) and the UGC Non-NET BHU Fellowship for the financial support of the present work.
Author information
Authors and Affiliations
Contributions
NR, BKS, and SPR conceptualized the article. NR wrote the manuscript, designed the tables, and made the diagrams. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Communicated by: Liwu Zhang
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rai, N., Sarma, B.K. & Rai, S.P. Transcriptional Regulation of Biotic and Abiotic Stress Responses: Challenges and Potential Mechanism for Stress Tolerance and Chickpea Improvement. Tropical Plant Biol. (2024). https://doi.org/10.1007/s12042-024-09354-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12042-024-09354-4