Abstract
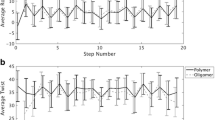
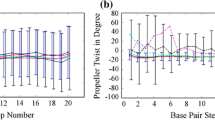
The three-dimensional structure of DNA contains various sequence-dependent structural information, which control many cellular processes in life, such as replication, transcription, DNA repair, etc. For the above functions, DNA double helices need to unwind or melt locally, which is different from terminal melting, as often seen in molecular dynamics (MD) simulations or even in many DNA crystal structures. We have carried out detailed MD simulations of DNA double helices of regular oligonucleotide fragments as well as in polymeric constructs with water and charge-neutralizing counter-ions at several different temperatures. We wanted to eliminate the end-effect or terminal melting propensity by employing MD simulation of DNA oligonucleotides in such a manner that gives rise to properties of polymeric DNA of infinite length. The polymeric construct is expected to allow us to see local melting at elevated temperatures. Comparative structural analysis of oligonucleotides and its corresponding virtual polymer at various temperatures ranging from 300 K to 400 K is discussed. The general behaviour, such as volume expansion coefficients of both the simulations show high similarity, indicating polymeric construct, does not give many artificial constraints. Local melting of a polymer, even at elevated temperature, may need a high nucleation energy that was not available in the short (7 ns) simulations. We expected to observe such nucleation followed by cooperative melting of the polymers in longer MD runs. Such simulations of different polymeric sequences would facilitate us to predict probable melting origins in a polymeric DNA.








Similar content being viewed by others
References
Albert B, Johnson A, Lewis J, Raff M, Roberts K and Walter P 2000 Molecular biology of the cell 4th edition (New York, Garland Science)
Allen MP and Tildesley DJ 1987 Computer simulations of liquids (New York, Oxford University Press)
Altan-Bonnet G, Libchaber A and Krichevsky O 2003 Bubble dynamics in double-stranded DNA. Phys. Rev. Lett. 90 138101
Bandyopadhyay D 2005 Different non-local interactions in stabilizing nucleic acid structures, PhD thesis, University of Calcutta, Kolkata
Bansal M, Bhattacharyya D and Ravi B 1995 NUPARM and NUCGEN: Software for analysis and generation of sequence dependent nucleic acid structures. Comput. Appl. Biosci. 11 281–287
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE 2000 The Protein Data Bank. Nucleic Acids Res. 28 235–242
Bhattacharyya D, Kundu S, Thakur AR and Majumdar R 1999 Sequence Directed flexibility of DNA and the role of cross-strand hydrogen bonds. J. Biomol. Struct. Dyn. 17 289–300
Blake RD and Delcourt SG 1998 Thermal stability of DNA. Nucleic Acids Res. 26 3323–3332
Blossey R and Carlon E 2003 Reparametrizing the loop entropy weights: Effect on DNA melting curves. Phys. Rev. E 68 061911
Breslauer KJ, Frank R, Blocker and Marky LA 1986 Predicting DNA duplex stability from base sequence. Proc. Natl. Acad. Sci. USA 83 3746–3750
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S and Karplus M 1983 CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4 187–217
Chan SS, Austin RH, Mukerji I and Spiro TG 1997 Temperature-dependent ultraviolet resonance Raman spectroscopy of the premelting state of dA.dT DNA. Biophys. J. 72 1512–1520
Chandrasekaran R and Arnott S 1996 The structure of B-DNA in oriented fibres. J. Biomol. Struct. Dyn. 13 1015–1027
Chowdhury S and Bansal M 2001 Modelling studies on neurodegenerative disease causing triplet repeat sequences d(GGC/GCC)n and d(CAG/CTG)n. J. Biosci. 26 649–665
Darden T, York D and Pedersen L 1993 Particle mesh Ewald-an NlogN method for Ewald sums in large systems. J. Chem. Phys. 9 10089–10092
Dickerson RE, Bansal M, Calladine CR, Diekmann S, Hunter WN, Kennard O, et al. 1989 Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 8 1–4
Feller SE, Zhang Y, Pastor RW and Brooks BR 1995 Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103 4613–4621
Foloppe N and MacKerell Jr AD 2000 All-atom empirical force field for nucleic acids: 1) Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21 86–104
Halder S and Bhattacharyya D 2010 Structural stability of tandemly occurring noncanonical basepairs within double helical fragments: molecular dynamics studies of functional RNA. J. Phys. Chem. B 114 14028–14040
Hoover WG 1985 Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 31 1695–1697
Izanloo C, Parsafar GA, Abroshan H and Akbarzadeh H 2011 Denaturation of Drew–Dickerson DNA in a high salt concentration medium: Molecular dynamics simulations. J. Comput. Chem. 32 3354–3361
Johansson E, Parkinson G and Neidle S 2000 A new crystal form for the dodecamer C-G-C-G-A-A-T-T-C-G-C-G: Symmetry effects on sequence-dependent DNA structure. J. Mol. Biol. 300 555–561
Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K and Schulten K 1999 NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151 283–312
Kannan S and Zacharias M 2009 Simulation of DNA double-strand dissociation and formation during replica-exchange molecular dynamics simulations. Phys. Chem. Chem. Phys. 11 10589–10595
Liu J, Malinina L, Huynh-Dinh T and Subirana JA 1998 The structure of the most studied DNA fragment changes under the influence of ions: a new packing of d(CGCGAATTCGCG). FEBS Lett. 438 211–214
MacKerell AD and Banavali NK 2000 All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 21 105–120
Mukherjee S, Bansal M and Bhattacharyya D 2006 Conformational specificity of non-canonical base pairs and higher order structures in nucleic acids: crystal structure database analysis. J. Comput. Aided Mol. Des. 20 629–645
Nelson HC, Finch JT, Luisi BF and Klug A 1987 The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature 330 221–226
Nelson M, Humphrey W, Gursoy A, Dalke A, Kalé L, Skeel R, Schulten K and Kufrin R. 1995 MDScope - A visual computing environment for structural biology. Comput. Phys. Commun. 91 111–134
Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, et al. 2001 A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 313 229–237
Panigrahi SK and Desiraju GR 2007 Strong and weak hydrogen bonds in drug-DNA complexes: a statistical analysis. J. Biosci. 32 677–691
Panigrahi S, Pal R and Bhattacharyya D 2011 Structure and energy of non-canonical basepairs: comparison of various computational chemistry methods with crystallographic ensembles. J. Biomol. Struct. Dyn. 29 424–596
Premilat S and Albiser G 1997 X-ray fibre diffraction study of an elevated temperature structure of Poly(dA).Poly(dT). J. Mol. Biol. 274 64–71
Samanta S, Mukherjee S, Chakrabarti, J and Bhattacharyya D 2009 Structural properties of polymeric DNA from molecular dynamics simulations. J. Chem. Phys. 130 115103–115114
SantaLucia J, Allawi Jr HT and Seneviratne PA 1996 Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35 3555–3562
Seibel GL, Singh UC and Kollman PA 1985 A molecular dynamics simulation of double-helical B-DNA including counterions and water. Proc. Natl. Acad. Sci. USA 82 6537–6540
Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL and Shlyakhtenko LS 2002 Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 27 53–65
Turner DH 1996 Thermodynamics of base pairing. Curr. Opin. Struct. Biol. 6 299–304
Urpi L, Tereshko V, Malinina L, Huynh-Dinh T and Subirana JA 1996 Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes. Nat. Struct. Biol. 3 325–328
van Erp TS, Cuesta-Lopez S and Peyrard M 2006 Bubbles and denaturation in DNA. Eur. Phys. J. E 20 421–434
Wartell RM and Benight AS 1985 Thermal denaturation of DNA molecules: A comparison of theory with experiment. Phys. Rep. 126 67–107
Wong KY and Pettitt BM 2008 The pathway of oligomeric DNA melting investigated by molecular dynamics simulations. Biophys. J. 95 5618–5626
Yakushevich LV 2001 Is DNA a nonlinear dynamical system where solitary conformational waves are possible? J. Biosci. 26 305–313
Zeng Y and Zocchi G 2006 Mismatches and bubbles in DNA. Biophys. J. 90 4522–4529
Author information
Authors and Affiliations
Corresponding author
Additional information
[Kundu S, Mukherjee S and Bhattacharyya D 2012 Effect of temperature on DNA double helix: An insight from molecular dynamics simulation. J. Biosci. 37 1–11] DOI
Rights and permissions
About this article
Cite this article
Kundu, S., Mukherjee, S. & Bhattacharyya, D. Effect of temperature on DNA double helix: An insight from molecular dynamics simulation. J Biosci 37, 445–455 (2012). https://doi.org/10.1007/s12038-012-9215-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-012-9215-5