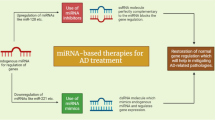
Abstract
Disruptions in multiple neurobiological pathways and neuromolecular processes have been widely implicated in the etiopathology of Alzheimer’s disease (AD), a complex, progressive, and ultimately lethal neurological disorder whose current incidence, both domestically and globally, is reaching epidemic proportions. While only a few percent of all AD cases appear to have a strong genetic or familial component, the major form of this disease, known as idiopathic or sporadic AD, displays a multi-factorial pathology and represents one of the most complex and perplexing neurological disorders known. More effective and innovative pharmacological strategies for the successful intervention and management of AD might be expected: (i) to arise from strategic-treatments that simultaneously address multiple interrelated AD targets that are directed at the initiation, development, and/or propagation of this disease and (ii) those that target the “neuropathological core” of the AD process at early or upstream stages of AD. This “Perspectives paper” will review current research involving microRNA (miRNA)-mediated, messenger RNA (mRNA)-targeted gene expression pathways in sporadic AD and address the potential implementation of evolving anti-microRNA (AM) strategies in the amelioration and clinical management of AD. This novel-therapeutic approach: (i) incorporates a system involving the restoration of multiple miRNA-regulated mRNA-targets via the use of selectively-stabilized AM species; and (ii) that via implementation of synthetic AMs, the abundance of only relatively small-families of miRNAs need be modulated or neutralized to re-establish neural-homeostasis in the AD-affected brain. In doing so, these strategic approaches will jointly and interactively address multiple AD-associated processes such as the disruption of synaptic communication, defects in amyloid peptide clearance and amyloidogenesis, tau pathology, deficits in neurotrophic support, alterations in the innate immune response, and the proliferation of neuroinflammatory signaling.

Similar content being viewed by others
References
Latta CH, Brothers HM, Wilcock DM (2015) Neuroinflammation in Alzheimer’s disease; a source of heterogeneity and target for personalized therapy. Neuroscience 302:103–111. https://doi.org/10.1016/j.neuroscience.2014.09.061
Lane CA, Hardy J, Schott JM (2018) Alzheimer’s disease. Eur J Neurol 25:59–70. https://doi.org/10.1111/ene.13439
Rogaev EI (2018) Different pathways to neurodegeneration. Biochemistry (Mosc) 83:1007–1008. https://doi.org/10.1134/S0006297918090018
Melis RJF, Haaksma ML, Muniz-Terrera G (2019) Understanding and predicting the longitudinal course of dementia. Curr Opin Psychiatry 32:123–129. https://doi.org/10.1097/YCO.0000000000000482
Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, Wolk D, Trojanowski JQ et al (2017) Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain 140:735–747. https://doi.org/10.1093/brain/aww319
Blennow K, Zetterberg H (2018) Biomarkers for Alzheimer’s disease: current status and prospects for the future. J Intern Med 284:643–663. https://doi.org/10.1111/joim.12816
Bhagwat N, Pipitone J, Voineskos AN, Chakravarty MM, Alzheimer’s disease neuroimaging initiative (2019) An artificial neural network model for clinical score prediction in Alzheimer disease using structural neuroimaging measures. J Psychiatry Neurosci 44:1–15
Berkowitz CL, Mosconi L, Scheyer O, Rahman A, Hristov H, Isaacson RS (2018) Precision medicine for Alzheimer’s disease prevention. Healthcare (Basel) 6(3). https://doi.org/10.3390/healthcare6030082
Hodes JF, Oakley CI, O’Keefe JH, Lu P, Galvin JE, Saif N, Bellara S, Rahman A et al (2019) Alzheimer’s “prevention” vs. “risk reduction”: transcending semantics for clinical practice. Front Neurol 9:179. https://doi.org/10.3389/fneur.2018.01179
Isaacson RS, Ganzer CA, Hristov H, Hackett K, Caesar E, Cohen R, Kachko R, Meléndez-Cabrero J et al (2018) The clinical practice of risk reduction for Alzheimer’s disease: a precision medicine approach. Alzheimers Dement 14:1663–1673. https://doi.org/10.1016/j.jalz.2018.08.004
Kauppinen S, Vester B, Wengel J (2006) Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handbook of Exp Pharmacol 173:405–422
Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T (2009) Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23:433–438. https://doi.org/10.1101/gad.1761509
Sethi P, Lukiw WJ (2009) Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett 459:100–104. https://doi.org/10.1016/j.neulet.2009.04.052
Rüegger S, Großhans H (2012) MicroRNA turnover: when, how, and why. Trends Biochem Sci 37:436–446. https://doi.org/10.1016/j.tibs.2012.07.002
Pogue AI, Hill JM, Lukiw WJ (2014) MicroRNA (miRNA): sequence and stability, viroid-like properties, and disease association in the CNS. Brain Res 1584:73–79. https://doi.org/10.1016/j.brainres.2014.03.042
Zhao Y, Alexandrov PN, Lukiw WJ (2016) Anti-microRNAs as novel therapeutic agents in the clinical management of Alzheimer’s disease. Front Neurosci 10:59. https://doi.org/10.3389/fnins.2016.00059
Exiqon-Qiagen, Germantown MD, USA; http://www.exiqon.com/lna-technology; last accessed 12 March 2019).
Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. https://doi.org/10.1038/nature09267
Towler BP, Jones CI, Newbury SF (2015) Mechanisms of regulation of mature miRNAs. Biochem Soc Trans 43:1208–1214. https://doi.org/10.1042/BST20150157
Zhao Y, Jaber VR, LeBeauf A, Sharfman NM, Lukiw WJ (2019) microRNA-34a (miRNA-34a) mediated down-regulation of the post-synaptic cytoskeletal element SHANK3 in sporadic Alzheimer’s disease (AD). Front Neurol 10:28. https://doi.org/10.3389/fneur.2019.00028
Bhattacharjee S, Zhao Y, Dua P, Rogaev EI, Lukiw WJ (2016) microRNA-34a-mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One 11(3):e0150211. https://doi.org/10.1371/journal.pone.0150211
Bartel DP (2018) Metazoan microRNAs. Cell. 173:20–51. https://doi.org/10.1016/j.cell.2018.03.006
Iwakawa HO, Tomari Y (2015) The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25:651–665. https://doi.org/10.1016/j.tcb.2015.07.011
Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16:421–433. https://doi.org/10.1038/nrg3965
Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J (2019) Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci 76:441–451. https://doi.org/10.1007/s00018-018-2940-7
Lukiw WJ, Zhao Y, Cui JG (2008) An NF-kB-sensitive miRNA-146a-mediated inflammatory circuit in AD and in stressed human brain cells. J Biol Chem 283:31315–31322. https://doi.org/10.1074/jbc.M805371200
Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, Tarr MA, Lukiw WJ (2009) Characterization of an NF-kB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem 103:1591–1595. https://doi.org/10.1016/j.jinorgbio.2009.05.012
Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ (2010) Differential regulation of IRAK-1 and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer’s disease. J Biol Chem 285:38951–38960. https://doi.org/10.1074/jbc.M110.178848
Lukiw WJ (2012) NF-κB-regulated, pro-inflammatory miRNAs in Alzheimer’s disease. Alzheimers Res Ther 4:47. https://doi.org/10.1186/alzrt150
Xiong W, Dong S, Yuan J, Li J, Liu J, Xu X (2012) miRNA-146a promotes proliferation and migration of rat vascular smooth muscle cells in vitro in a NF-κB-dependent manner. Nan Fang Yi Ke Da Xue Xue Bao 32:270–273
Czubowicz K, Jęśko H, Wencel P, Lukiw WJ, Strosznajder RP (2019) The role of ceramide and sphingosine-1-phosphate in Alzheimer’s disease and other neurodegenerative disorders. Mol Neurobiol. https://doi.org/10.1007/s12035-018-1448-3
Ng A, Tam WW, Zhang MW, Ho CS, Husain SF, McIntyre RS, Ho RC (2018) IL-1β, IL-6, TNF- α and CRP in elderly patients with depression or Alzheimer’s disease: systematic review and meta-analysis. Sci Rep 8:12050. https://doi.org/10.1038/s41598-018-30487-6
Shen XN, Niu LD, Wang YJ, Cao XP, Liu Q, Tan L, Zhang C, Yu JT (2019) Inflammatory markers in Alzheimer’s disease and mild cognitive impairment: a meta-analysis and systematic review of 170 studies. J Neurol Neurosurg Psychiatry 90:590–598. https://doi.org/10.1136/jnnp-2018-319148
LightSwitch Assay, Switchgear Genomics (an Active Motif Company), Menlo Park CA USA; https://switchgeargenomics.com/resources/science-technology; last accessed 14 March 2019.
Lukiw WJ, Alexandrov PN (2012) Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer’s disease (AD) brain. Mol Neurobiol 46:11–19
Lukiw WJ (2012) NF-кB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol 235:484–490. https://doi.org/10.1016/j.expneurol.2011.11.022
Mosconi L, Rahman A, Diaz I, Wu X, Scheyer O, Hristov HW, Vallabhajosula S, Isaacson RS et al (2018) Increased Alzheimer’s risk during the menopause transition: a 3-year longitudinal brain imaging study. PLoS One 13:e0207885. https://doi.org/10.1371/journal.pone.0207885
Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ (2002) Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res 70:462–473
Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI (2007) MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11). Biochemistry (Mosc) 72:578–582
Jaber V, Zhao Y, Lukiw WJ (2017) Alterations in micro RNA-messenger RNA (miRNA-mRNA) coupled signaling networks in sporadic Alzheimer’s disease (AD) hippocampal CA1. J Alzheimers Dis Parkinsonism 7(2):312. https://doi.org/10.4172/2161-0460.1000312
Alexandrov PN, Zhao Y, Jaber V, Cong L, Lukiw WJ (2017) Deficits in the proline-rich synapse-associated Shank3 protein in multiple neuropsychiatric disorders. Front Neurol 8:670. https://doi.org/10.3389/fneur.2017.00670
Zhao Y, Bhattacharjee S, Jones BM, Dua P, Alexandrov PN, Hill JM, Lukiw WJ (2013) Regulation of TREM2 expression by an NF-кB-sensitive miRNA-34a. Neuroreport 24:318–323. https://doi.org/10.1097/WNR.0b013e32835fb6b0
Ginsberg SD, Alldred MJ, Che S (2012) Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol Dis 45:99–107. https://doi.org/10.1016/j.nbd.2011.07.013
Zhao Y, Bhattacharjee S, Jones BM, Hill J, Dua P, Lukiw WJ (2014) Regulation of neurotropic signaling by the inducible, NF-kB-sensitive miRNA-125b in Alzheimer’s disease (AD) and in primary human neuronal-glial (HNG) cells. Mol Neurobiol 50:97–106. https://doi.org/10.1007/s12035-013-8595-3
Zhao Y, Bhattacharjee S, Dua P, Alexandrov PN, Lukiw WJ (2015) microRNA-based biomarkers and the diagnosis of Alzheimer’s disease. Front Neurol 6:162. https://doi.org/10.3389/fneur.2015.00162
Seipold L, Saftig P (2016) The emerging role of tetraspanins in the proteolytic processing of the amyloid precursor protein. Front Mol Neurosci 9:149. https://doi.org/10.3389/fnmol.2016.00149
Termini CM, Gillette JM (2017) Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol 5:34
Saint-Pol J, Eschenbrenner E, Dornier E, Boucheix C, Charrin S, Rubinstein E (2017) Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins. Biochem Soc Trans 45:937–944. https://doi.org/10.1042/BST20160296
Murru L, Moretto E, Martano G, Passafaro M (2018) Tetraspanins shape the synapse. Mol Cell Neurosci 91:76–81. https://doi.org/10.1016/j.mcn.2018.04.001
Gilmore TD, Herscovitch M (2006) Inhibitors of NF-kB signaling: 785 and counting. Oncogene. 25:6887–6899
Kaur U, Banerjee P, Bir A, Sinha M, Biswas A, Chakrabarti S (2015) Reactive oxygen species, redox signaling and neuro-inflammation in AD: the NF-κB connection. Curr Top Med Chem 15:446–457
Yu L, Li L, Medeiros LJ, Young KH (2017) NF-κB signaling pathway and its potential as a target for therapy in lymphoid neoplasms. Blood Rev 31:77–92. https://doi.org/10.1016/j.blre.2016.10.001
Khuda-Bukhsh AR, Das S, Saha SK (2014) Molecular approaches toward targeted cancer prevention with some food plants and their products: inflammatory and other signal pathways. Nutr Cancer 66:194–205. https://doi.org/10.1080/01635581.2014.864420
Wu J, Ding J, Yang J, Guo X, Zheng Y (2018) MicroRNA roles in the nuclear factor kappa B signaling pathway in cancer. Front Immunol 9:546. https://doi.org/10.3389/fimmu.2018.00546
Che J, Stark LA (2018) Crosstalk between NF-κB and nucleoli in the regulation of cellular homeostasis. Cells. 7(10):E157. https://doi.org/10.3390/cells7100157
Mendiola AS, Cardona AE (2018) The IL-1β phenomena in neuro-inflammatory diseases. J Neural Transm (Vienna) 125:781–795. https://doi.org/10.1007/s00702-017-1732-9
Hong JT (2017) NF-kB as a mediator of brain inflammation in AD. CNS Neurol Disord Drug Targets 18:3–10. https://doi.org/10.2174/1871527316666170807130011
Lukiw WJ (2007) Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 18:297–300
Lukiw WJ, Pogue AI (2007) Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem 101:1265–1269
Schipper HM, Maes OC, Chertkow HM, Wang E (2007) MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Biol 1:263–274
Lima JF, Cerqueira L, Figueiredo C, Oliveira C, Azevedo NF (2018) Anti-miRNA oligonucleotides: a comprehensive guide for design. RNA Biol 15:338–352. https://doi.org/10.1080/15476286.2018.1445959
Zhao Y, Jaber V, Lukiw WJ (2017) Secretory products of the human GI tract and their potential impact on Alzheimer’s disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front Cell Infect Microbiol 7:318. https://doi.org/10.3389/fcimb.2017.00318
Talebi A, Rahnema M, Bigdeli MR (2019) Effect of intravenous injection of antagomiR-1 on brain ischemia. Mol Biol Rep 46:1149–1155. https://doi.org/10.1007/s11033-018-04580-y
Li YY, Alexandrov PN, Pogue AI, Zhao Y, Bhattacharjee S, Lukiw WJ (2012) miRNA-155 upregulation and complement factor H deficits in Down’s syndrome. Neuroreport 23:168–173. https://doi.org/10.1097/WNR.0b013e32834f4eb4
Henry RJ, Doran SJ, Barrett JP, Meadows VE, Sabirzhanov B, Stoica BA, Loane DJ, Faden AI (2019) Inhibition of miRNA-155 limits neuro-inflammation and improves functional recovery after experimental traumatic brain injury in mice. Neurotherapeutics 16:216–230. https://doi.org/10.1007/s13311-018-0665-9
Acknowledgments
The analytical, experimental and statistical work in this research report was presented in part at the Vavilov Institute of General Genetics autumn seminar series (Серия осенних семинаров) in Moscow RUSSIA November 2018 and at the Society for Neuroscience (SFN) Annual Meeting November 2018, San Diego CA, USA. Sincere thanks are extended to the late Drs. JM Hill (JMH; Louisiana State University), TPA Kruck (TPAK; University of Toronto), C. Bergeron (CB, University of Toronto) for helpful discussions on this research area and to F Culicchia, C Eicken, C Hebel, B. Krishnan, K Navel, and L. Wong for short postmortem interval (PMI) human and other mammalian brain tissues or extracts and data interpretation and to D Guillot for expert technical assistance. Thanks are also extended to the many neuropathologists, physicians and researchers of the US, Canada, Europe and Russia who have provided high quality, short postmortem interval (PMI) human CNS or extracted brain tissue fractions for scientific study. We would like to further thank the following 18 domestic and international brain banks, and their continuing cooperation, for access to high quality postmortem tissues and valuable analytical advice: the Autism Brain Net, Los Angeles CA, USA; the Harvard University/McLean Hospital Tissue Center, Boston MA, USA; Louisiana State University, New Orleans LA, USA; the Lomonosov Institute, Moscow State University, Moscow, Russian Federation; the National Disease Research Interchange, Philadelphia PA, USA; the National Institutes of Health NIH NeuroBioBank, comprised of tissues from the National Institute of Mental Health (NIMH), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institute of Neurological Disorders and Stroke (NINDS), Bethesda MD USA; the Netherlands Brain Research Institute, Amsterdam, Netherlands; the New York State Institute for Basic Research, Staten Island NY, USA; the Oregon Health Sciences University, Portland OR, USA; the Southern Eye Bank, Metairie LA, USA; the University of California, Irvine CA, USA; the University of Kentucky Alzheimer’s disease Brain Bank, Lexington KY, USA; the University of Maryland Brain and Tissue Bank, Baltimore MD, USA; the University of Massachusetts, Worcester MA, USA; University of Pennsylvania School of Medicine, Philadelphia PA, USA, and the University of Toronto Brain Bank, Toronto ON, Canada. All authors agree on the content of this publication. Research on AM design and potential treatment strategies, metal neurotoxicity, human and murine microRNAs, small noncoding RNA (sncRNA), proinflammatory and pathogenic signaling in the Lukiw laboratory involving the innate-immune response, neuroinflammation and amyloidogenesis in AD, PrD and in other human neurological disorders was supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB); the Louisiana Biotechnology Research Network (LBRN), the Alzheimer Association and NIH grants NEI EY006311, NIA AG18031 and NIA AG038834 (WJL).
Funding
Research on AM design and potential treatment strategies, metal neurotoxicity, human and murine microRNAs, small non-coding RNA, pro-inflammatory and pathogenic signaling in the Lukiw laboratory involving the innate immune response, neuroinflammation, and amyloidogenesis in AD and PrD and in other human neurological disorders was financially supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB) and by the Louisiana Biotechnology Research Network (LBRN), the Alzheimer’s Association, and NIH grants NEI EY006311, NIA AG18031, and NIA AG038834 (WJL).
Author information
Authors and Affiliations
Contributions
VRJ, YZ, NMS, and WJL performed all microRNA and messenger RNA analysis, miRNA-mRNA verification and linking, and preliminary AM experiments, organized and tabulated all the data, and performed statistical analysis from age-, gender-, and PMI-matched control and AD brains, and collaborated interactively in the synthesis of the material presented in this Frontiers “Perspective” article. WJL wrote the paper. YZ and WJL have published in excess of 50 peer-reviewed manuscripts into this research area. All authors agree on the content of this publication.
Corresponding author
Ethics declarations
Ethics Statement
All acquisition, handling, experimental, and analytical procedures involving postmortem human brain tissues were carried out in an ethical manner in strict accordance with the ethics review board policies at brain and tissue donor institutions and at the Louisiana State University (LSU) Health Sciences Center. Informed consent from next of kin was obtained at brain and tissue donor institutions for all tissue samples prior to autopsy and donation; coded postmortem brain tissue samples (containing no personal identifying information of the donors) were obtained from the 18 brain and tissue banks listed in the “Acknowledgments” section above. The ethical use of postmortem human brain tissues and their analyses were also carried out in strict accordance with the Institutional Biosafety Committee and the Institutional Review Board Committee (IBC/IRBC) ethical guidelines IBC#18059 and IRBC#6774 at the LSU Health Sciences Center, New Orleans, LA 70112, USA.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jaber, V.R., Zhao, Y., Sharfman, N.M. et al. Addressing Alzheimer’s Disease (AD) Neuropathology Using Anti-microRNA (AM) Strategies. Mol Neurobiol 56, 8101–8108 (2019). https://doi.org/10.1007/s12035-019-1632-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1632-0