Abstract
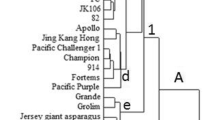
Field pea (Pisum sativum L.) is an important cool season legume crop widely grown around the world. This research provides a basis for selection of pea germplasm across geographical regions in current and future breeding and genetic mapping efforts for pea improvement. Eleven novel genic markers were developed from pea expressed sequence tag (EST) sequences having significant similarity with gene calls from Medicago truncatula spanning at least one intron. In this study, 96 cultivars widely grown or used in breeding programs in the USA and Canada were analyzed for genetic diversity using 31 microsatellite or simple sequence repeat (SSR) and 11 novel EST-derived genic markers. The polymorphic information content varied from 0.01–0.56 among SSR markers and 0.04–0.43 among genic markers. The results showed that SSR and EST-derived genic markers displayed one or more highly reproducible, multi-allelic, and easy to score loci ranging from 200 to 700 bp in size. Genetic diversity was assessed through unweighted neighbor-joining method, and 96 varieties were grouped into three main clusters based on the dissimilarity matrix. Four subpopulations were determined through STRUCTURE analysis with no significant geographic separation of the subpopulations. The findings of the present study can be used to select diverse genotypes to be used as parents of crosses aimed for breeding improved pea cultivars.




Similar content being viewed by others
Abbreviations
- PIC:
-
Polymorphic information content
- UNJ:
-
Unweighted neighbor-joining
- EST:
-
Expressed sequence tags
- PCR:
-
Polymerase chain reaction
- CAPS:
-
Cleaved amplified polymorphic sequences
- SSR:
-
Simple sequence repeats
- PAGE:
-
Polyacrylamide gel electrophoresis
References
Bastianelli, D., Grosjean, F., Peyronnet, C., Duparque, M., & Regnier, J. M. (1998). Feeding value of pea (Pisum sativum, L.) 1. Chemical composition of different categories of pea. Animal Science, 67, 609–619.
Zohary, D. (1996). The mode of domestication of the founder crops of near east agriculture. In D. R. Harris (Ed.), The origin and spread of agriculture and pastoralism in Eurasia (pp. 142–158). London: University College London Press.
Smýkal, P., Aubert, G., Burstin, J., Coyne, C. J., Ellis, N. T. H., Flavell, A. J., et al. (2012). Pea (Pisum sativum L.) in the genomic era. Agronomy, 2, 74–115.
Tar’an, B., Zhang, C., Warkentin, T., Tullu, A., & Vandenberg, A. (2005). Genetic diversity among varieties and wild species accessions of pea (Pisum sativum L.) based on molecular markers, morphological and physiological characters. Genome, 48, 257–272.
Bahrman, N., Le Gouis, J., Hariri, D., Guilbaud, L., & Jestin, L. (1999). Genetic diversity of old French six-rowed winter barley varieties assessed with molecular, biochemical and morphological markers and its relation to BaMMV resistance. Heredity, 83, 568–574.
O’Neill, R., Snowdon, R. J., & Kohler, W. (2003). Population genetics aspects of biodiversity. Progress in Botany, 64, 115–137.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, A. J. A., & Tingey, S. V. (1990). DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 22, 6531–6535.
Jacobson, A., & Hedr´en, M. (2007). Phylogenetic relationships in Alisma (Alismataceae) based on RAPDs, and sequence data from ITS and trnL. Plant Systematics and Evolution, 265, 27–44.
Soller, M., & Beckmann, J. S. (1983). Genetic polymorphism in varietal identification and genetic improvement. Theoretical and Applied Genetics, 67, 25–33.
Becker, J., & Heun, M. (1994). Barley microsatellites: Allele variation and mapping. Plant Molecular Biology, 274, 835–845.
Vos, P. R., Hogers, R., Bleeker, M., Reijans, M., Lee, T., Hornes, M., et al. (1995). AFLP: A new technique for fingerprinting. Nucleic Acids Research, 21, 4407–4414.
Zietkiewicz, E., Rafalski, A., & Labuda, D. (1994). Genome fingerprinting by simple sequence repeat SSR-anchored polymerase chain reaction amplification. Genomics, 202, 176–183.
Smykal, P., Hybl, M., Corander, J., Jarkovsky, J., Flavell, A. J., & Griga, M. (2008). Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theoretical and Applied Genetics, 117, 413–424.
Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Smykal, P., Marshall, D., et al. (2010). The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evolutionary Biology, 10, 44.
Martin-Sanz, A., Caminero, C., Jing, R., Flavell, A. J., & Perez de la Vega, M. (2011). Genetic diversity among Spanish pea (Pisum sativum L.) landraces, pea cultivars and the world Pisum sp. core collection assessed by retrotransposon based insertion polymorphisms (RBIPs). Spanish Journal of Agricultural Research, 9, 166–178.
Zhuang, X., McPhee, K. E., Coram, T. E., Peever, T. L., & Chilvers, M. I. (2012). Rapid transcriptome characterization and parsing of sequences in a non-model host–pathogen interaction; pea-Sclerotinia sclerotiorum. BMC Genomics, 13, 668.
Tessier, C., David, J., Boursiquot, P. J. M., & Charrier, A. (1999). Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics, 98, 171–177.
Garcia, A. A. F., Benchimol, L. L., Barbosa, A. M. M., Geraldi, I. O., Souza, C. L. J., & Souza, A. P. (2004). Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genetics and Molecular Biology, 27, 579–588.
Agrama, H. A., & Tuinstra, M. R. (2003). Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. African Journal of Biotechnology, 2, 334–340.
Davierwala, A. P., Chowdari, K. V., Kumar, S., Reddy, A. P. K., Ranjekar, P. K., & Gupta, V. S. (2000). Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica, 108, 269–284.
Chao, S., Zhang, W., Dubcovsky, J., & Sorrells, M. (2007). Evaluation of genetic diversity and genome wide linkage disequilibrium among US wheat Triticum aestivum (L.) germplasm representing different market classes. Crop Science, 47, 1018–1030.
Sarwat, M., Das, S., & Srivastava, P. S. (2008). Analysis of genetic diversity through AFLP, SAMPL, ISSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Reports, 27, 519–528.
Belaj, A., Satovic, Z., Cipriani, G., Baldoni, L., Testolin, R., Rallo, L., et al. (2003). Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theoretical and Applied Genetics, 107, 736–744.
Pejic, I., Ajmone-Marsan, P., Morgante, M., Kozumplick, V., Castiglioni, P., Taramino, G., et al. (1998). Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theoretical and Applied Genetics, 97, 1248–1255.
Powell, W., Morgante, M., Andre, C., Hanafey, M., Vogel, J., Tingey, S., et al. (1996). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding, 2, 225–238.
Russell, J. R., Fuller, J. D., Macaulay, M., Hatz, B. G., Jahoor, A., Powell, W., et al. (1997). Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theoretical and Applied Genetics, 95, 714–722.
Cuevas, H. E., & Prom, L. K. (2013). Assessment of molecular diversity and population structure of the Ethiopian sorghum [Sorghum bicolor (L.) Moench] germplasm collection maintained by the USDA–ARS National Plant Germplasm System using SSR markers. Genetic Resources and Crop Evolution, 60, 1817–1830.
Izzah, N. K., Lee, J., Perumal, S., Park, J. Y., Ahn, K., Fu, D., et al. (2013). Microsatellite-based analysis of genetic diversity in 91 commercial Brassica oleracea L. cultivars belonging to six varietal groups. Genetic Resources and Crop Evolution, 60, 1967–1986.
Mishra, R. K., Gangadhar, B. H., Nookaraju, A., Kumar, S., & Park, S. W. (2012). Development of EST-derived SSR markers in pea (Pisum sativum) and their potential utility for genetic mapping and transferability. Plant Breeding, 131, 118–124.
Varshney, R. K., Graner, A., & Sorrells, M. E. (2005). Genic microsatellite markers in plants: Features and applications. Trends in Biotechnology, 23, 48–55.
Karakas, O., Gurel, F., & Uncuoglu, A. A. (2011). Assessment of genetic diversity of wheat genotypes by resistance gene analog-EST markers. Genetics and Molecular Research, 10, 1098–1110.
Marconi, T. G., Costa, E. A., Miranda, H. R., Mancini, M. C., Cardoso-Silva, C. B., Oliveira, K. M., et al. (2011). Functional markers for gene mapping and genetic diversity studies in sugarcane. BMC Research Notes, 4, 264.
Park, Y. H., Alabady, M. S., Ulloa, M., Sickler, B., Wilkins, T. A., Yu, J., et al. (2005). Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Molecular Genetics and Genomics, 274, 428–441.
Phan, H. T. T., Ellwood, S. R., Ford, R., Thomas, S., & Oliver, R. (2006). Differences in syntenic complexity between Medicago truncatula with Lens culinaris and Lupinus albus. Functional Plant Biology, 33, 775–782.
Choi, H. K., Mun, J. H., Kim, D. J., Zhu, H., Baek, J. M., Mudge, J., et al. (2004). Estimating genome conservation between crop and model legume species. Proceedings of the National Academy of Sciences of the United States of America, 101, 15289–15294.
Brunel, D., Froger, N., & Pelletier, G. (1999). Development of amplified consensus genetic markers (ACGM) in Brassica napus from Arabidopsis thaliana sequences of known biological function. Genome, 42, 387–402.
Bertioli, D. J., Moretzsohn, M. C., Madsen, L. H., Sandal, N., Leal-Bertioli, S. C. M., Guimarães, P. M., et al. (2009). An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics, 10, 45.
Brauner, S., Murphy, R. L., Walling, J. G., Przyborowski, J., & Weeden, N. F. (2002). STS markers for comparative mapping in legumes. Journal of American Society of Horticultural Sciences, 127, 616–622.
Alo, F., Furman, B. J., Akhunov, E., Dvorak, J., & Gepts, P. (2011). Leveraging genomic resources of model species for the assessment of diversity and phylogeny in wild and domesticated lentil. Journal of Heredity, 102, 315–329.
Rogers, S. O., & Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium, and mummified plant tissues. Plant Molecular Biology, 5, 69–76.
Jain, S., & McPhee, K. E. (2013). Isolation and characterization of novel EST-derived genic markers in Pisum sativum (Fabaceae). Application in Plant Sciences, 1(11), 1300026. doi:10.3732/apps.1300026.
Jain, S., Weeden, N. F., Porter, L. D., Eigenbrode, S. D., & McPhee, K. (2013). Finding linked markers to En for efficient selection of pea enation mosaic virus resistance in pea. Crop Science, 53, 1–8.
Gascuel, O. (1997). Concerning the NJ algorithm and its unweighted version, UNJ. In B. Mirkin, F. R. McMorris, F. Roberts, & A. Rzhetsky (Eds.), Mathematical hierarchies and biology. DIMACS workshop, series in discrete mathematics and theoretical computer science, vol. 37 (pp. 149–170). Providence, RI: American Mathematical Society.
Liu, K., & Muse, S. (2005). PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics, 21, 2128–2129.
Perrier, X., Flori, A., & Bonnot, F. (2003). Data analysis methods. In P. Harnon, M. Seguin, X. Perrier, & J. C. Glaszmann (Eds.), Genetic diversity of cultivated plants (pp. 43–76). Enfield: Science Publishers.
Pritchard, J. K., Stephens, M., & Donelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620.
Earl, D. A., & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics resources, 4, 359–361.
Gilpin, B. J., McCallum, J. A., Frew, T. J., & Timmerman-Vaughan, G. M. (1997). A linkage map of the pea (Pisum sativum L.) genome containing cloned sequences of known function and expressed sequence tags (ESTs). Theoretical and Applied Genetics, 95, 1289–1299.
Loridon, K., McPhee, K., Morin, J., Dubreuil, P., Pilet-Nayel, M. L., Aubert, G., et al. (2005). Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theoretical and Applied Genetics, 111, 1022–1031.
Aubert, G., Morin, J., Jacquin, F., Loridon, K., Quillet, M. C., Petit, A., et al. (2006). Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theoretical and Applied Genetics, 112, 1024–1041.
Deulvot, C., Charrel, H., Marty, A., Jacquin, F., Donnadieu, C., Lejeune-Henaut, I., et al. (2010). Highly multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics, 11, 468.
Eujayl, I., Sledge, M. K., Wang, L., May, G. D., Chekhovskiy, K., Zwonitzer, J. C., et al. (2004). Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theoretical and Applied Genetics, 108, 414–422.
Hougaard, B. K., Madsen, L. H., Sandal, N., Moretzsohn, M. C., Fredslund, J., Schauser, L., et al. (2008). Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics, 179, 2299–2312.
Young, N. D., & Udvardi, M. (2009). Translating Medicago truncatula genomics to crop legumes. Current Opinion in Plant Biology, 12, 193–201.
Gao, L., Tang, J., Li, H., & Jia, J. (2003). Analysis of microsatellites in major crops assessed by computational and experimental approaches. Molecular Breeding, 12, 1–17.
Konovalov, F. A., Toshchakova, E. V., & Gostimsky, S. A. (2009). CAPS markers for the identification of garden pea (Pisum sativum L.) cultivars. Genetika, 45, 251–254.
Burstin, J., Deniot, G., Potier, J., Weinachter, C., Aubert, G., & Baranger, A. (2001). Microsatellite polymorphism in Pisum sativum. Plant Breeding, 120, 311–317.
Ford, R., Roux, K. L., Itman, C., Brouwer, J. B., & Taylor, P. W. J. (2002). Diversity analysis and genotyping in Pisum with sequence tagged microsatellite site (STMS) primers. Euphytica, 124, 397–405.
Baranger, A., Aubert, G., Arnau, G., Laine, A. L., Deniot, G., Potier, J., et al. (2004). Genetic diversity within Pisum sativum using protein and PCR-based markers. Theoretical and Applied Genetics, 108, 1309–1321.
Haghnazari, A., Samimifard, R., Najafi, J., & Mardi, M. (2005). Genetic diversity in pea (Pisum sativum L.) accessions detected by sequence tagged microsatellite markers. Journal of Genetics and Breeding, 59, 145–152.
Choudhury, R. P., Tanveer, H., & Dixit, G. P. (2006). Identification and detection of genetic relatedness among important varieties of pea (Pisum sativum L.), grown in India. Genetica, 130, 183–191.
Nasiri, J., Haghnazari, A., & Saba, J. (2009). Genetic diversity among varieties and wild species accessions of pea (Pisum sativum L.) based on SSR markers. African Journal of Biotechnology, 8, 3405–3417.
Ahmad, S., Singh, M., Lamb-Palmer, N. D., Lefsrud, M., & Singh, J. (2012). Assessment of genetic diversity in 35 Pisum sativum accessions using microsatellite markers. Canadian Journal of Plant Science, 92, 1075–1081.
Datta, S., Tiwari, S., Kaashyap, M., Gupta, P. P., Choudhury, P. R., Kumari, J., et al. (2011). Genetic similarity analysis in lentil using cross-genera legume sequence tagged microsatellite site markers. Crop Science, 51, 2412–2422.
He, C., Poysa, V., & Yu, K. (2003). Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theoretical and Applied Genetics, 106, 363–373.
Panwar, P., Nath, M., Yadav, V. K., & Kumar, A. (2010). Comparative evaluation of genetic diversity using RAPD, SSR and cytochrome P450 gene based markers with respect to calcium content in finger millet (Eleusine coracana L. Gaertn.). Journal of Genetics, 89, 121–133.
Sarikamis, G., Yanmaz, R., Ermis, S., Bakir, M., & Yuksel, C. (2010). Genetic characterization of pea (Pisum sativum) germplasm from Turkey using morphological and SSR markers. Genetics and Molecular Research, 9, 591–600.
Hoey, B. K., Crowe, K. R., Jones, V. M., & Polans, N. O. (1996). A phylogenetic analysis of Pisum based on morphological characters, and allozyme and RAPD markers. Theoretical and Applied Genetics, 92, 92–100.
Lu, J., Knox, M. R., Ambrose, M. J., Brown, J. K. M., & Ellis, T. H. N. (1996). Comparative analysis of genetic diversity in pea assessed by RFLP- and PCR-based methods. Theoretical and Applied Genetics, 93, 1103–1111.
Jing, R., Johnson, R., Seres, A., Kiss, G., Ambrose, M. J., Knox, M. R., et al. (2007). Gene-based sequence diversity analysis of field pea (Pisum). Genetics, 177, 2263–2275.
Heath, M., & Hebblethwaite, P. (1985). Agronomic problems associated with the pea crop. In P. D. Hebblehwaite, M. C. Heath, & T. C. K. Dawkins (Eds.), The pea crop: A basis for improvement (pp. 19–29). London: Butterworths.
Jha, A. B., Arganosa, G., Tar’an, B., Diederichsen, A., & Warkentin, T. D. (2013). Characterization of 169 diverse pea germplasm accessions for agronomic performance, Mycosphaerella blight resistance and nutritional profile. Genetic Resources and Crop Evolution, 60, 747–761.
Tanksley, S. D., & McCouch, S. R. (1997). Seed bank and molecular maps: Unlocking genetic potential from the wild. Science, 277, 1063–1066.
Acknowledgments
The authors are grateful to funding (Grant No. 2008-511010-4522) by the RAMP (Risk Assessment and Mitigation Program) of NIFA (National Institute for Food and Agriculture). The authors would like to thank Drs. Ted Helms and Javed Iqbal, North Dakota State University, Fargo, ND for critical reading of manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jain, S., Kumar, A., Mamidi, S. et al. Genetic Diversity and Population Structure Among Pea (Pisum sativum L.) Cultivars as Revealed by Simple Sequence Repeat and Novel Genic Markers. Mol Biotechnol 56, 925–938 (2014). https://doi.org/10.1007/s12033-014-9772-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-014-9772-y