Abstract
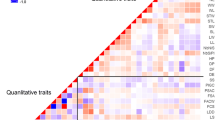
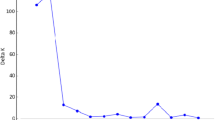
A collection of 148 Pisum accessions, mostly from Western Europe, and including both primitive germplasm and cultivated types, was structured using 121 protein- and PCR-based markers. This molecular marker-based classification allowed us to trace back major lineages of pea breeding in Western Europe over the last decades, and to follow the main breeding objectives: increase of seed weight, introduction of the afila foliage type and white flowers, and improvement of frost tolerance for winter-sown peas. The classification was largely consistent with the available pedigree data, and clearly resolved the different main varietal types according to their end-uses (fodder, food and feed peas) from exotic types and wild forms. Fodder types were further separated into two sub-groups. Feed peas, corresponding to either spring-sown or winter-sown types, were also separated, with two apparently different gene pools for winter-sown peas. The garden pea group was the most difficult to structure, probably due to a continuum in breeding of feed peas from garden types. The classification also stressed the paradox between the narrowness of the genetic basis of recent cultivars and the very large diversity available within P. sativum. A sub-collection of 43 accessions representing 96% of the whole allelic variability is proposed as a starting point for the construction of a core collection.


Similar content being viewed by others
References
Arnau G, Lallemand J, Bourgoin M (2002) Fast and reliable strawberry cultivar identification using inter simple sequence repeat (ISSR) amplification. Euphytica 129:69–79
Auld DL, Murray GA, O’Keeffe LE, Campbell AR, Markarian D (1978) Registration of Melrose field pea. Crop Sci 18:913
Bourgoin-Grenèche M, Lallemand J (1993) Electrophoresis and its application to the description of varieties: a presentation of the techniques used by GEVES. GEVES Editions, La Miniere
Blixt S (1972) Mutations genetics in Pisum. Agric Hortic Genet 30:1–293
Burstin J, Charcosset A (1997) Relationship between phenotypic and marker distances: theoretical and experimental investigations. Heredity 79:477–483
Burstin J, Deniot G, Potier J, Weinachter C, Aubert G, Baranger A (2001) Microsatellite polymorphism in Pisum sativum. Plant Breed 120:311–317
Christiansen MJ, Andersen SB, Ortiz R (2002) Diversity changes in an intensively bred wheat germplasm during the 20th century. Mol Breed 9:1–11
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA micropreparation: version II. Plant Mol Biol Rep 1:19–21
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dubreuil P, Charcosset A (1999) Relationships among maize inbred lines and populations from European and North American origins as estimated using RFLP markers. Theor Appl Genet 99:473–480
Dubreuil P, Dillmann C, Warburton M, Crossa J, Franco J, Baril C (2003) User’s manual for the LCDMV software (calculation of molecular distances between varieties) for fingerprinting and genetic diversity studies http://www.cimmyt.org/abc/manual/controls.htm
Ellis THN, Poyser SJ, Knox MR, Vershinin AV, Ambrose MJ (1998) Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol Gen Genet 260:9–19
Gilpin BJ, McCallum JA, Frew TJ, Timmerman-Vaughan GM (1997) A linkage map of the pea (Pisum sativum L.) genome containing cloned sequences of known function and expressed sequence tags (ESTs). Theor Appl Genet 95:1289–1299
Gouesnard B, Bataillon TM, Decoux G, Rozale C, Schoen DJ, David JL (2001) MSTRAT: an algorithm for building germplasm core collections by maximizing allelic or phenotypic richness. J Hered 9:93–94
Hodgkin T (1995) Some current issues in the conservation and use of plant genetic resources. In: Ayad WG, Hodgkin T, Jaradat A, Rao VR (eds) Molecular genetic techniques for plant genetic resources. Report of an IPGRI workshop, Rome, 9–11 October 1995
Hoey BK, Crowe KR, Jones VM, Polans MO (1996) A phylogenetic analysis of Pisum based on morphological characters, allozyme and RAPD markers. Theor Appl Genet 92:92–100
Innan I, Terauchi R, Miyashita NT (1997) Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaudoise Sci Nat 44:223–270
Koebner RMD, Donini P, Reeves JC, Cooke RJ, Law JR (2003) Temporal flux in the morphological and molecular diversity of UK barley. Theor Appl Genet 106:550–558
Lallemand J, Arnau G (2000) Marqueurs moléculaires (ISSR) pour l’identification des variétés de pois et la caractérisation des ressources génétiques: comparaison avec l’approche biochimique. In: Deuxièmes journées méthodologiques du GEVES, 24 November 2000, GEVES Editions, La Miniere, pp 21–22
Laucou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea. I. RAPD-based genetic linkage map of Pisum sativum. Theor Appl Genet 97:905–915
Lejeune-Hénaut I, Bourion V, Etévé G, Cunot E, Delhaye K, Desmyter C (1999) Floral initiation in field-grown forage peas is delayed to a greater extent by short photoperiods than in other types of European varieties. Euphytica 109:201–211
Lewis BG, Matthews P (1984) The world germplasm of Pisum sativum: could it be used more effectively to produce healthy crops ? In: Hebblethwaite PD, Heath MC, Dawkins TCK (eds) The pea crop, a basis for crop improvement. Butterworths, London, pp 215–229
Liu F, Sun GL, Salomon B, von Bothmer R (2001) Distribution of allozymic alleles and genetic diversity in the American Barley Core Collection. Theor Appl Genet 102:606–615
Lu H, Bernardo R (2001) Molecular marker diversity among current and historical maize inbreds. Theor Appl Genet 103:613–617
Lu J, Knox MR, Ambrose MJ, Brown JKM, Ellis THN (1996) Comparative analysis of genetic diversity in pea assessed by RFLP and PCR based methods. Theor Appl Genet 93:1103–1111
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:311–335
Métais I, Hamon B, Jalouzot R, Peltier D (2002) Structure and level of genetic diversity in various bean types evidenced with microsatellite markers isolated from a genomic enriched library. Theor Appl Genet 104:1346–1352
Narvel JM, Walter RF, Chu WC, Grant D, Shoemaker RC (2000) Simple sequence repeat diversity among soybean plant introductions and elite genotypes. Crop Sci 40:1452–1458
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, Coyne CJ (2002) Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor Appl Genet 106:28–39
Posvec Z, Griga M (2000) Utilisation of isozyme polymorphism for cultivar identification of 45 commercial peas (Pisum sativum L.). Euphytica 113:251–258
Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Prioul S, Onfroy C, Tivoli B, Baranger A (2002) Controlled environment assessment of partial resistance to Mycosphaerella pinodes in pea (Pisum sativum L.) seedlings. Euphytica 131:121–130
Rogers JS (1972) Measures of similarities and genetic distances. In: Studies in genetics VII. University of Texas Publication 7213:145–153
Rongwen J, Akkaya MS, Bhagwat AA, Lavi U, Cregan PB (1995) The use of microsatellite DNA markers for soybean genotype identification. Theor Appl Genet 90:43–48
Russell JR, Fuller JD, Young G, Thomas B, Taramino G, Macaulay M, Waugh R, Powell W (1997) Discriminating between barley genotypes using microsatellite markers. Genome 40:442–450
Russell JR, Ellis RP, Thomas WTB, Waugh R, Provan J, Booth A, Fuller J, Lawrence P, Young G, Powell W (2000) A retrospective analysis of spring barley germplasm development from ‘foundation genotypes’ to currently successful cultivars. Mol Breed 6:553–568
Samec P, Nasinec V (1995) Detection of DNA polymorphism among pea cultivars using RAPD technique. Biol Planta 37:321–327
Santalla M, Power JB, Davey MR (1998) Genetic diversity in mung bean revealed by RAPD markers. Plant Breed 117:473-478
SAS Institute (1988) SAS/STAT User’s Guide: release 603. SAS Institute, Cary, N.C.
Simioniuc D, Uptmoor R, Friedt W, Ordon F (2002) Genetic diversity and relationships among pea cultivars revealed by RAPDs and AFLPs. Plant Breed 121:429–435
Slinkard AE, Murray GA (1972) Registration of Fenn field pea. Crop Sci 12:127
Smartt J (1984) Evolution of grain legumes. I. Mediterranean pulses. Exp Agric 20:275–296
Smartt J (1990) Pulses of the classical world. In: Grain legumes: evolution and genetic resources. Cambridge University Press, Cambridge, p 176
Soleimani VD, Baum BR, Johnson DA (2002) AFLP and pedigree-based genetic diversity estimates in modern cultivars of durum wheat [Triticum turgidum L. subsp. durum (Desf.) Husn.]. Theor Appl Genet 104:350–357
Steiger DL, Nagai C, Moore PH, Morden CW, Osgood RV, Ming R (2002) AFLP analysis of genetic diversity within and among Coffea arabica cultivars. Theor Appl Genet 105:209–215
Ward JH (1963) Hierarchical grouping to optimize an objective function. Am Stat Assoc J 56:236–244
Weeden NF, Ellis THN, Timmerman-Vaughan GM, Swiecicki WK, Rozov SM, Berdnikov VA (1998) A consensus linkage map for Pisum sativum. Pisum Genet 30:1–4
White S, Doebley J (1998) Of genes and genomes and the origin of maize. Trends Genet 14:327–332
Acknowledgements
The authors thank M. Ambrose (John Innes Centre, Norwich), S. Anguelova (International Programme for Genetic Resources, Plovdiv), A. Burghoffer (Institut National pour la Recherche Agronomique, Versailles), C. Coyne (United States Department of Agriculture, Pullman), A. Ramos Monreal (University of Valladolid), M. Vishnyakova (NI Vavilov Research Institute of Plant Industry, St Petersburg), and X. Zong (Chinese Academy of Agricultural Sciences, Beijing) for providing accessions with related information. They also thank M. Duparque, P. Marget and G. Morin for the description of the accessions, T. Bataillon for his help with the MSTRAT analysis, P. Dubreuil, M. Manzanares, M.L. Pilet-Nayel, R. Thompson and M. Trottet for helpful comments on the manuscript. Finally, we are very grateful to R. Cousin for spending time in tracing back the history of pea breeding with us, and to M. Chauvet for sharing his knowledge on the history of pea and its uses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Baranger, A., Aubert, G., Arnau, G. et al. Genetic diversity within Pisum sativum using protein- and PCR-based markers. Theor Appl Genet 108, 1309–1321 (2004). https://doi.org/10.1007/s00122-003-1540-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1540-5