Abstract
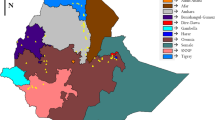
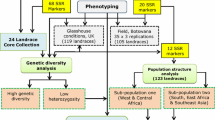
The molecular diversity and population structure present in the Ethiopian sorghum collection maintained at the USDA–ARS National Plant Germplasm System (NPGS) has not been studied. In addition, 83 % of the accessions in the Ethiopian collection lack passport information which has constrained their evaluation and utility. Therefore, 137 Ethiopian accessions from NPGS were randomly selected and characterized with 20 strategically selected simple sequence repeat markers. These markers indentified 289 alleles with average polymorphic information content of 0.78. The allele frequency distribution reflects that 62 % of the alleles were rare (<0.05), 17 % range from 0.05 to 0.10, and 22 % were higher than 0.10. Expected and observed heterozygosity were estimated at 0.78 and 0.23, respectively, demonstrating Ethiopia has high sorghum genetic diversity germplasm. Population structure analysis indentified two subpopulations of 77 and 41 accessions, respectively, while a third group was constituted by 19 accessions whose classifications were not defined (i.e. hybrids). Analysis of molecular variances determined variation within subpopulations as the major source of variation. Likewise, genetic differentiation between subpopulations was moderate (Fst = 0.10). These results indicated that a continuous exchange of genes between subpopulations of sorghum exists in Ethiopia. The absence of a well defined population structure positioned this germplasm as an important resource for the study and dissection of agricultural traits by association mapping. However, genetic redundancy analysis indicated the presence of highly related accessions, therefore, strategic selected accessions must be consider prior to phenotype evaluation of larger number of accessions. The Ethiopian collection is composed of highly genetically diverse germplasm, and the genetic information presented herein is valuable to ex situ and in situ conservation programs to promote the use of this germplasm for breeding programs.


Similar content being viewed by others
References
Abdi A, Bekele E, Asfaw Z, Teshome A (2002) Patterns of morphological variation of sorghum (Sorghum bicolor (L.) Moench) landraces in qualitative characters in North Shewa and South Welo, Ethiopia. Hereditas 137:161–172
Ali ML, Rajewski JF, Baenziger PS, Gill KS, Eskridge KM, Dweikat I (2008) Assessment of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Mol Breeding 21:497–509
Ayana A, Bekele E (1998) Geographical patterns of morphological variation in Sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea: qualitative characters. Hereditas 129:195–205
Ayana A, Bekele E (1999) Multivariate analysis of morphological variation in sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea. Genet Resour Crop Evol 46:273–284
Ayana A, Bekele E, Bryngelsson T (2000) Genetic variation in wild sorghum (Sorghum bicolor ssp. verticilliflorum (L.) Moench) germplasm from Ethiopia assessed by random amplified polymorphic DNA (RAPD). Hereditas 132:249–254
Bhosale SU, Stich B, Rattunde HF, Weltzien E, Haussmann BIG, Hash CT, Melchinger AE, Parzies HK (2011) Population structure in sorghum accessions from West Africa differing in race and maturity class. Genetica 139: 453–463
Bohasale SU, Stich B, Rattunde HF, Weltzien E, Haussmann BIG, Hash CT, Melchinger AE, Parzies HK (2011) Population structure in sorghum accessions from West Africa differing in race and maturity class. Genetica 139:453–463
Casa AM, Pressoir G, Brown P, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48:30–40
Dahlberg J (2000) Classification and characterization of sorghum. In: Smith CW, Frederisken RA (eds) Sorghum. Wiley, New York, pp 99–130
Degu E (1996) Sorghum breeding and achievements in Ethiopia. In: Deressa A, Seboka B (eds) Research achievements and technology transfer attempts: vignettes from Shoa. In: Proceedings from the first technology generation, transfer, and gap analysis workshop, 25–27 Dec 1995, Nazret, Ethiopia
Deu M, Sagnard F, Chantereau J, Calatayud C, Hérault D, Mariac C, Pham JL, Vigouroux Y, Traore PS, Mamadou A, Gerard B, Ndjeunga J, Bezancon G (2008) Niger-wide assessment of in situ sorghum diversity with microsatellite markers. Theor Appl Genet 116:903–913
Djè Y, Heuertz M, Ater M, Lefèbvre C, Vekemans X (2004) In situ estimation of out crossing rate in sorghum landraces using microsatellite markers. Euphytica 138:205–212
Doggett H (1988) Sorghum, 2nd edn. Longman Scientific and Technical, London
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ejeta J, Grenier C (2005) Sorghum and its weedy hybrids. In: Gressel J (ed) Crop ferality and volunteerism. CRC Press, Boca Raton, pp 123–135
Elshire RJ, Glaubitz JC, Qi S, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping by sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379. doi:10.1371/journal.pone.0019379
England PR, Cornuet JM, Berthier P, Tallmon DA, Luikart G (2006) Estimating effective population size from linkage disequilibrium: severe bias in small samples. Conserv Genet 7:303–308
Erpelding J (2009) Anthracnose diseases response for photoperiod-insensitive Ethiopian germplasm from the U.S. sorghum collection. World J Agric Sci 5:707–713
Erpelding J, Wang ML (2007) Response to anthracnose infection for a random selection of sorghum germplasm. Plant Pathol J 6:127–133
Evano G, Regnaut S, Goudet J (2005) Detecting the number of cluster of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetic analysis. Evol Bioinform Online 1:47–50
Food and Agriculture Organization of the United Nation (FAO) (2010) FAOSTAT. http://faostat.fao.org/site/339/default.aspx. Accessed April 2012
Gebrekidan B (1973) The importance of the Ethiopian sorghum germplasm in the world sorghum collection. Econ Bot 27:442–445
Ghebru B, Schmidt RJ, Bebbetzen JL (2002) Genetic diversity sorghum landraces assessed with simple sequence repeat (SSR) markers. Theor Appl Genet 105:229–236
Grenier C, Bramel-Cox PJ, Hamon P (2001a) Core collection of sorghum: I. Stratification based on eco-geographical data. Crop Sci 41:234–239
Grenier C, Hamon P, Bramel-Cox PJ (2001b) Core collection of sorghum: II. Comparison of three random sampling strategies. Crop Sci 41:241–246
GRIN (2012) USDA–ARS National Genetic Resources Program, Germplasm Resource Information Network (GRIN). Online database. National Germplasm Resources Laboratory, Beltsville, MD, USA. http://www.ars-grin.gov/. Accessed May 2012
Han Z, Jian-Cheng W, Dong-Jian W, Feng-Xia Y, Jin-Fang XU, Guo-An S, Yan-An G, Ru-Yu L (2011) Assessment of genetic diversity in Chinese sorghum landraces using SSR markers as compared with foreign accessions. Acta Agron Sin 37:224–234
Harlan JR (1992) Indigenous African agriculture. In: Cowman CW, Watson PJ (eds) The origins of agriculture: an international perspective. Smithsonian Institution Press, Washington, DC, pp 59–70
Harlan JR, de Wet JMJ (1972) Simplified classification of cultivated sorghum. Crop Sci 12:172–176
Harlan JR, Stemler A (1976) The races of sorghum in Africa. In: Harlan JR, de Wet JMJ, Stemler A (eds) Origins of African plant domestication. Mouton, The Hague
Hash CT, Ramu P, Upadhyaya HD, Folkertsma RT, Billot C, Rami J-F, Rivallan R, Deu M, Chantereau J, Gardes L, Li, Y, Wang T, Lu P (2008) Diversity analysis of sorghum global composite collection and reference set. Poster presented in generation challenge program (GCP) annual review meeting (ARM), 16–20 Sept 2008, Bangkok, Thailand. http://www.generationcp.org/UserFiles/File/2008_Poster-abstracts_final.pdf
Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res (Camb) 38:209–216
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:475–478
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Mace ES, Buhariwalla KK, Crouch JH (2003) A high-throughput DNA extraction protocol for tropical molecular breeding programs. Plant Mol Biol Rep 21:459–460
Mann JA, Kimber CT, Miller FR (1983) The origin and early cultivation of sorghum in Africa. Texas Agricultural Experimental Station Bulletin, vol 1454. Texas A&M University, College Station
Manzelli M, Pileri L, Lacerenza N, Benedettelli S, Vecchio V (2007) Genetic diversity assessment in Somalia sorghum (Sorghum bicolor (L.) Moench) accessions using microsatellite markers. Biodivers Conserv 16:1715–1730
McGuire S (2000) Farmers’ management of sorghum diversity in Eastern Ethiopia. In: Almekinder CJM, de Boef W (eds) Encouraging diversity: the conservation and development of plant genetic resources. Intermediate Technology, London, pp 43–48
McGuire S (2002) Farmers’ view and management of sorghum diversity in Western Harerghe, Ethiopia: implications for collaboration with formal breeding. In: Cleveland DA, Soleri D (eds) Farmers, scientist and plant breeding. CAB International, pp 107
Mekbib F (2008) Genetic erosion of sorghum (Sorghum bicolor (L.) Moench) in the centre of diversity, Ethiopia. Genet Resour Crop Evol 55:351–364
Menz MA, Klein RR, Unruh NC, Rooney WL, Klein PE, Mullet JE (2004) Genetic diversity of public inbreeds of sorghum determined by mapped AFLP and SSR markers. Crop Sci 44:1236–1244
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data II. Gene frequency data. J Mol Evol 19:153–170
Ng’Uni D, Geleta M, Bryngelsson T (2011) Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) accessions of Zambia as revealed by simple sequence repeats (SSR). Hereditas 148:52–62
Pederson JF, Toy JJ, Johnson B (1998) Natural outcrossing of sorghum and sudangrass in the Central Great Plains. Crop Sci 38:937–939
Peel D, Ovenden JR, Peel SL (2004) NeEstimator: software for estimating effective population size, Version 1.3. Queensland Government, Department of Primary Industries and Fisheries
Petit RJ, El Mousadick A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prom LK, Erpelding J, Perumal R, Isakeit T, Cuevas HE (2012) Response of sorghum accessions from four African countries against Colletotrichum sublineolum, causal agent of sorghum anthracnose. Am J Plant Sci 3:125–129
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Snowden JD (1936) The cultivated races of Sorghum. Adlard and Son, London
Stemler ABL, Harlan JR, de Wet JMJ (1977) The sorghums of Ethiopia. Econ Bot 31:446–460
Taffesse AS, Dorosh P, Asrat S (2011) Crop production in Ethiopia: regional patterns and trends. Ethiopia strategy support program II (ESSP II): Working paper no. 0016
Teshome A, Baum BR, Fahrig L, Torrance JK, Arnason TJ, Lambert JD (1997) Sorghum (Sorghum bicolor (L.) Moench) landraces variation and classification in north Shewa and south Welo, Ethiopia. Euphytica 97:255–263
Tesso T, Kapran I, Grenier C, Snow A, Sweeney P, Pedersen J, Marx D, Bothma G, Ejeta G (2008) The potential for crop-to-wild gene flow in sorghum in Ethiopia and Niger: a geographic survey. Crop Sci 48:1425–1431
Upadhyaya HD, Pundir RPS, Dwivedi SL, Godwa CLL, Gopal Reddy V, Singh S (2009) Developing mini core collection of sorghum (Sorghum bicolor (L.) Moench) for diversified utilization of germplasm. Crop Sci 49:1769–1780
Wang ML, Dean R, Erpelding JE, Pederson G (2006) Molecular genetic evaluation of sorghum germplasm differing in response to fungal diseases: rust (Puccinia purpurea) and anthracnose (Collectotrichum graminicola). Euphytica 148:319–330
Wang ML, Zhu C, Barkley NA, Chen Z, Erpelding JE, Murray SC, Tuinstra MR, Tesso T, Pederson GA, Yu J (2009) Genetic diversity and population structure analysis of accessions in the US historic sweet sorghum collection. Theor Appl Genet 120:13–23
Yao D, Myriam H, Mohammed T, Claude L, Xavier V (2004) In situ estimation of out crossing rate in sorghum landraces using micro-satellite markers. Euphytica 138:205–212
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospect of association mapping in plants. Plant Genome 1:5–20
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cuevas, H.E., Prom, L.K. Assessment of molecular diversity and population structure of the Ethiopian sorghum [Sorghum bicolor (L.) Moench] germplasm collection maintained by the USDA–ARS National Plant Germplasm System using SSR markers. Genet Resour Crop Evol 60, 1817–1830 (2013). https://doi.org/10.1007/s10722-013-9956-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-013-9956-5