Abstract
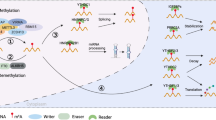
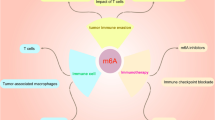
RNA modification has garnered increasing attention in recent years due to its pivotal role in tumorigenesis and immune surveillance. N6-methyladenosine (m6A) modification is the most prevalent RNA modification, which can affect the expression of RNA by methylating adenylate at the sixth N position to regulate the occurrence and development of tumors. Dysregulation of m6A affects the activation of cancer-promoting pathways, destroys immune cell function, maintains immunosuppressive microenvironment, and promotes tumor cell growth. In this review, we delve into the latest insights into how abnormalities in m6A modification in both tumor and immune cells orchestrate immune evasion through the activation of signaling pathways. Furthermore, we explore how dysregulated m6A modification in tumor cells influences immune cells, thereby regulating tumor immune evasion via interactions within the tumor microenvironment (TME). Lastly, we highlight recent discoveries regarding specific inhibitors of m6A modulators and the encapsulation of m6A-targeting nanomaterials for cancer therapy, discussing their potential applications in immunotherapy.




Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- m6A:
-
N6-methyladenosine
- TME:
-
Tumor microenvironment
- m5C:
-
5-Methylcytosine
- m7G:
-
N7-methylguanosine
- m1A:
-
N1-methyladenosine
- ncRNA:
-
Non-coding RNA
- METTL3:
-
Methyltransferase-like 3
- METTL14:
-
Methyltransferase-like 14
- WTAP:
-
Wilms tumor-associated protein
- RBM15:
-
RNA-binding motif 15
- METTL16:
-
Methyltransferase-like 16
- FTO:
-
Fat mass and obesity-associated protein
- ALKBH5:
-
Alkb homolog 5
- ALKBH3:
-
Alkb homolog 5
- YTH:
-
YTH-domain family protein
- IGF2BP:
-
Insulin growth factor-2 mRNA-binding proteins
- HNRNP:
-
Heterogeneous nuclear ribo nucleo protein
- SAM:
-
S-adenosylmethionine
- IFN:
-
Interferon
- TBK1:
-
TANK-binding kinase 1
- IRF3:
-
Interferon regulatory factor 3
- PD-L1:
-
Programmed death ligand-1
- PD-1:
-
Programmed death-1
- JAK:
-
Janus kinase
- STAT:
-
Signal transducer and activator of transcription
- JNK:
-
C-Jun N-terminal kinase
- HBV:
-
Hepatitis B virus
- PTEN:
-
Phosphatase and tensin homolog
- RIG-I:
-
Retinoic acid-inducible gene-I
- IFN-α:
-
Interferon α
- YTHDF3:
-
YTH-domain-containing family protein-3
- PD-L1:
-
Programmed death-ligand-1
- circRNA:
-
Circular RNA
- YTHDC2:
-
YTH-domain-containing-2
- IFN-γ:
-
Interferon γ
- ETV5:
-
ETS Variant Transcription Factor 5
- MAPK:
-
Mitogen-activated protein kinase
- NK cell:
-
Natural killer cell
- IL-15:
-
Interleukin-15
- ERK:
-
Extracellular regulated protein kinase
- mTOR:
-
Mechanistic target of rapamycin kinase
- NF-κB:
-
Nuclear factor-κB
- SOCS:
-
Suppressor Of Cytokine Signaling
- IL-7:
-
Interleukin 7
- Treg:
-
Regulatory T-cell
- IL-2:
-
Interleukin 7
- TAM:
-
Tumor-associated macrophages
- IL-27:
-
Interleukin 27
- IL-35:
-
Interleukin 35
- lncRNA:
-
Long non-coding RNA
- DC cell:
-
Dendritic cell
- TLR4:
-
Toll like receptor 4
- TIM cell:
-
Tumor-infiltrating myeloid cell
- MDSCs cell:
-
Myeloid-derived suppressor cells
- IL-8:
-
Interleukin 8
- TGF-β:
-
Transforming growth factor β
- p65:
-
Rela, RELA Proto-Oncogene, NF-KB Subunit
- CXCL1:
-
C-X-C Motif Chemokine Ligand 1
- EMT:
-
Epithelial-mesenchymal transformation
- IFNGR1:
-
Interferon γ receptor 1
- TNF-α:
-
Tumor necrosis factor α
References
An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21(1):14.
Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22(2):119–31.
Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9(2):124.
Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M, et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21(5):651–61.
Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–12.
Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49(13):7239–55.
Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. 2022;21(1):52.
Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–50.
Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A transcripts by the 3’→5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017;68(2):374-87.e12.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507–19.
Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y, et al. The evolving landscape of N(6)-methyladenosine modification in the tumor microenvironment. Mol Ther. 2021;29(5):1703–15.
van den Homberg DAL, van der Kwast R, Quax PHA, Nossent AY. N-6-Methyladenosine in Vasoactive microRNAs during Hypoxia; A Novel Role for METTL4. Int J Mol Sci. 2022;23(3).
Gasparri ML, Besharat ZM, Farooqi AA, Khalid S, Taghavi K, Besharat RA, et al. MiRNAs and their interplay with PI3K/AKT/mTOR pathway in ovarian cancer cells: a potential role in platinum resistance. J Cancer Res Clin Oncol. 2018;144(12):2313–8.
Kim GW, Imam H, Khan M, Mir SA, Kim SJ, Yoon SK, et al. HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology. 2021;73(2):533–47.
Jin S, Li M, Chang H, Wang R, Zhang Z, Zhang J, et al. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKε/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol Cancer. 2022;21(1):97.
Esteva FJ, Hubbard-Lucey VM, Tang J, Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20(3):e175–86.
Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–82.
Kuang Z, Liu X, Zhang N, Dong J, Sun C, Yin M, et al. USP2 promotes tumor immune evasion via deubiquitination and stabilization of PD-L1. Cell Death Differ. 2023;30(10):2249–64.
Zhao Y, Huang S, Tan X, Long L, He Q, Liang X, et al. N(6) -methyladenosine-modified CBX1 regulates nasopharyngeal carcinoma progression through heterochromatin formation and STAT1 activation. Adv Sci (Weinh). 2022;9(36):e2205091.
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):105.
Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20(1):105.
Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M, et al. JNK signaling promotes bladder cancer immune escape by regulating METTL3-mediated m6A modification of PD-L1 mRNA. Cancer Res. 2022;82(9):1789–802.
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21(1):60.
Wen J, Xue L, Wei Y, Liang J, Jia W, Yong T, et al. YTHDF2 is a therapeutic target for HCC by suppressing immune evasion and angiogenesis through ETV5/PD-L1/VEGFA axis. Adv Sci (Weinh). 2024:e2307242.
Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021;33(6):1221-33.e11.
Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19(3):200–18.
Gong Y, Klein Wolterink RGJ, Wang J, Bos GMJ, Germeraad WTV. Chimeric antigen receptor natural killer (CAR-NK) cell design and engineering for cancer therapy. J Hematol Oncol. 2021;14(1):73.
Yuan F, Cai X, Cong Z, Wang Y, Geng Y, Aili Y, et al. Roles of the m(6)A modification of RNA in the glioblastoma microenvironment as revealed by single-cell analyses. Front Immunol. 2022;13:798583.
Li X, Ma S, Deng Y, Yi P, Yu J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol Cancer. 2022;21(1):76.
Becknell B, Caligiuri MA. Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv Immunol. 2005;86:209–39.
Carson WE, Fehniger TA, Haldar S, Eckhert K, Lindemann MJ, Lai CF, et al. A potential role for interleukin-15 in the regulation of human natural killer cell survival. J Clin Invest. 1997;99(5):937–43.
Carson WE, Giri JG, Lindemann MJ, Linett ML, Ahdieh M, Paxton R, et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 1994;180(4):1395–403.
Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. 2021;12(1):5522.
Ma S, Yan J, Barr T, Zhang J, Chen Z, Wang LS, et al. The RNA m6A reader YTHDF2 controls NK cell antitumor and antiviral immunity. J Exp Med. 2021;218(8).
Liu XS, Zhou LM, Yuan LL, Gao Y, Kui XY, Liu XY, et al. NPM1 is a prognostic biomarker involved in immune infiltration of lung adenocarcinoma and associated with m6A modification and glycolysis. Front Immunol. 2021;12:724741.
Gardner A, de Mingo PÁ, Ruffell B. Dendritic cells and their role in immunotherapy. Front Immunol. 2020;11:924.
Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–4.
Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun. 2019;10(1):1898.
Zhang F, Liu H, Duan M, Wang G, Zhang Z, Wang Y, et al. Crosstalk among m(6)A RNA methylation, hypoxia and metabolic reprogramming in TME: from immunosuppressive microenvironment to clinical application. J Hematol Oncol. 2022;15(1):84.
Park SL, Gebhardt T, Mackay LK. Tissue-resident memory T cells in cancer immunosurveillance. Trends Immunol. 2019;40(8):735–47.
Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2(5):309–22.
Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–42.
Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28(2):253–6.
Li Y, Zheng JN, Wang EH, Gong CJ, Lan KF, Ding X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ. 2020;8:e10385.
Lu L, Zheng D, Qu J, Zhuang Y, Peng J, Lan S, et al. METTL16 predicts a favorable outcome and primes antitumor immunity in pancreatic ductal adenocarcinoma. Front Cell Dev Biol. 2022;10:759020.
Ding C, Xu H, Yu Z, Roulis M, Qu R, Zhou J, et al. RNA m(6)A demethylase ALKBH5 regulates the development of γδ T cells. Proc Natl Acad Sci USA. 2022;119(33):e2203318119.
Qin S, Liu G, Jin H, Chen X, He J, Xiao J, et al. The comprehensive expression and functional analysis of m6A modification “readers” in hepatocellular carcinoma. Aging (Albany NY). 2022;14(15):6269–98.
Na YR, Je S, Seok SH. Metabolic features of macrophages in inflammatory diseases and cancer. Cancer Lett. 2018;413:46–58.
Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, et al. Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci Adv. 2021;7(18).
Cai L, Li D, Feng Z, Gu X, Xu Q, Li Q. YTHDF2 regulates macrophage polarization through NF-κB and MAPK signaling pathway inhibition or p53 degradation. Dis Markers. 2022;2022:3153362.
Wang X, Ji Y, Feng P, Liu R, Li G, Zheng J, et al. The m6A reader IGF2BP2 regulates macrophage phenotypic activation and inflammatory diseases by stabilizing TSC1 and PPARγ. Adv Sci (Weinh). 2021;8(13):2100209.
Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell. 2021;39(7):945-57.e10.
Ma S, Sun B, Duan S, Han J, Barr T, Zhang J, et al. YTHDF2 orchestrates tumor-associated macrophage reprogramming and controls antitumor immunity through CD8(+) T cells. Nat Immunol. 2023.
Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021;12(1):1394.
Liu Y, Shi M, He X, Cao Y, Liu P, Li F, et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2022;15(1):52.
Gong PJ, Shao YC, Yang Y, Song WJ, He X, Zeng YF, et al. Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer. Front Oncol. 2020;10:578963.
Siemińska I, Poljańska E, Baran J. Granulocytes and cells of granulocyte origin-the relevant players in colorectal cancer. Int J Mol Sci. 2021;22(7).
Su G, Liu T, Han X, Sun H, Che W, Hu K, et al. YTHDF2 is a potential biomarker and associated with immune infiltration in kidney renal clear cell carcinoma. Front Pharmacol. 2021;12:709548.
Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive analysis of the PD-L1 and immune infiltrates of m(6)A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids. 2020;21:299–314.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82(9):1660-77.e10.
Wang L, Dou X, Chen S, Yu X, Huang X, Zhang L, et al. YTHDF2 inhibition potentiates radiotherapy antitumor efficacy. Cancer Cell. 2023.
Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933–59.
Zhang F, Guo J, Zhang Z, Qian Y, Wang G, Duan M, et al. Mesenchymal stem cell-derived exosome: a tumor regulator and carrier for targeted tumor therapy. Cancer Lett. 2022;526:29–40.
Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. 2022;163(4):891–907.
He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29(5):1821–37.
Zeng X, Chen K, Li L, Tian J, Ruan W, Hu Z, et al. Epigenetic activation of RBM15 promotes clear cell renal cell carcinoma growth, metastasis and macrophage infiltration by regulating the m6A modification of CXCL11. Free Radic Biol Med. 2022;184:135–47.
Dong F, Qin X, Wang B, Li Q, Hu J, Cheng X, et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. 2021;81(23):5876–88.
You Y, Wen D, Zeng L, Lu J, Xiao X, Chen Y, et al. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int J Biol Sci. 2022;18(13):5001–18.
Sun Z, Su Z, Zhou Z, Wang S, Wang Z, Tong X, et al. RNA demethylase ALKBH5 inhibits TGF-β-induced EMT by regulating TGF-β/SMAD signaling in non-small cell lung cancer. Faseb j. 2022;36(5):e22283.
Tan B, Zhou K, Liu W, Prince E, Qing Y, Li Y, et al. RNA N(6) -methyladenosine reader YTHDC1 is essential for TGF-beta-mediated metastasis of triple negative breast cancer. Theranostics. 2022;12(13):5727–43.
Hua W, Ten Dijke P, Kostidis S, Giera M, Hornsveld M. TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol Life Sci. 2020;77(11):2103–23.
Liu Z, Zheng N, Li J, Li C, Zheng D, Jiang X, et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat. 2022;65:100886.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95.
Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. 2023;72(8):1497–509.
Fiore PF, Di Matteo S, Tumino N, Mariotti FR, Pietra G, Ottonello S, et al. Interleukin-15 and cancer: some solved and many unsolved questions. J Immunother Cancer. 2020;8(2).
Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK, et al. Allosteric regulation of IGF2BP1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent Sci. 2022;8(8):1102–15.
Bai X, Wong CC, Pan Y, Chen H, Liu W, Zhai J, et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J Immunother Cancer. 2022;10(2).
Zhou B, Bie F, Zang R, Zhang M, Song P, Liu L, et al. RNA modification writer expression profiles predict clinical outcomes and guide neoadjuvant immunotherapy in non-small cell lung cancer. EBioMedicine. 2022;84:104268.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28.
Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy B, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. 2016;44(6):1255–69.
Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–1.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103.
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601.
Yu R, Wei Y, He C, Zhou P, Yang H, Deng C, et al. Integrative analyses of m6A regulators identify that METTL3 is associated with HPV status and immunosuppressive microenvironment in HPV-related cancers. Int J Biol Sci. 2022;18(9):3874–87.
Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38(1):79-96.e11.
Liu Z, Zhong J, Zeng J, Duan X, Lu J, Sun X, et al. Characterization of the m6A-associated tumor immune microenvironment in prostate cancer to aid immunotherapy. Front Immunol. 2021;12:735170.
Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782.
Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134(43):17963–71.
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373–84.
Zheng G, Cox T, Tribbey L, Wang GZ, Iacoban P, Booher ME, et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem Neurosci. 2014;5(8):658–65.
Wang T, Hong T, Huang Y, Su H, Wu F, Chen Y, et al. Fluorescein derivatives as bifunctional molecules for the simultaneous inhibiting and labeling of FTO protein. J Am Chem Soc. 2015;137(43):13736–9.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172(1–2):90-105.e23.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677-91.e10.
Xie G, Wu XN, Ling Y, Rui Y, Wu D, Zhou J, et al. A novel inhibitor of N (6)-methyladenosine demethylase FTO induces mRNA methylation and shows anti-cancer activities. Acta Pharm Sin B. 2022;12(2):853–66.
Chen J, Xu C, Yang K, Gao R, Cao Y, Liang L, et al. Inhibition of ALKBH5 attenuates I/R-induced renal injury in male mice by promoting Ccl28 m6A modification and increasing Treg recruitment. Nat Commun. 2023;14(1):1161.
Fang Z, Mu B, Liu Y, Guo N, Xiong L, Guo Y, et al. Discovery of a potent, selective and cell active inhibitor of m(6)A demethylase ALKBH5. Eur J Med Chem. 2022;238:114446.
Wang YZ, Li HY, Zhang Y, Jiang RX, Xu J, Gu J, et al. Discovery of pyrazolo[1,5-a]pyrimidine derivative as a novel and selective ALKBH5 inhibitor for the treatment of AML. J Med Chem. 2023;66(23):15944–59.
Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci USA. 2020;117(33):20159–70.
Ouyang Z, Gao Y, Yang R, Shen M, Shi X. Genetic engineering of dendritic cells using partially zwitterionic dendrimer-entrapped gold nanoparticles boosts efficient tumor immunotherapy. Biomacromol. 2022;23(3):1326–36.
Zhang X, Su T, Wu Y, Cai Y, Wang L, Liang C, et al. N6-methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. 2024.
Acknowledgements
The figures are created with Biorender.com.
Funding
This work was supported partially by grants from the National Natural Science Foundation of China (81903138) and the Natural Science Foundation of Hunan Province (2019JJ50778).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The first draft of the manuscript was written by C.Y.W, L.Y.L, and Q.L.T and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, C., Li, L., Tang, Q. et al. Role of m6A modifications in immune evasion and immunotherapy. Med Oncol 41, 159 (2024). https://doi.org/10.1007/s12032-024-02402-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-024-02402-9