Abstract
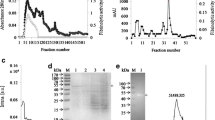
Bacterial fibrinolytic proteases achieved more attention in the prevention and treatment of cardiovascular diseases, so purification, characterization, and activity enhancement are of prime importance. In this study, a fibrinolytic serine metalloprotease was purified from the culture supernatant from Bacillus sp. BC1. It was purified to homogeneity by a two-step procedure with a 24-fold increase in specific activity and a 33.1% yield. It showed 28 kDa molecular weight, while its optimal pH and temperature were obtained 8 and 50–60 °C. The cross-link enzyme aggregates of this fibrinolytic BC1 successfully immobilized on magnetic chitosan nanoparticles. A 52% activity enhancement was obtained by immobilized enzyme at pH 6.0, compared to free protease. Km values of the free and immobilized proteases were obtained about 0.638 and 0.61 mg/ml, respectively. The free and immobilized enzymes did not show any activity concerning transferrin, γ-globulins, and hemoglobin, as blood plasma proteins. The in vitro blood clot lysis test of the free and immobilized proteases showed a maximum of 42 and 50% clot lysis, which was comparatively higher than that revealed by streptokinase and heparin at the same condition. These results indicated that the free and immobilized proteases have the potential to be effective fibrinolytic agents.








Similar content being viewed by others
References
Bajaj, B. K., Singh, S., Khullar, M., Singh, K., & Bhardwaj, S. (2014). Optimization of fibrinolytic protease production from Bacillus subtilis I-2 using agro-residues. Brazilian Archives of Biology and Technology, 57(5), 653–662.
Vijayaraghavan, P., Arasu, M. V., Rajan, R. A., & Al-Dhabi, N. A. (2019). Enhanced production of fibrinolytic enzyme by a new Xanthomonas oryzae IND3 using low-cost culture medium by response surface methodology. Saudi Journal of Biological Sciences, 26(2), 217–224.
World Health Organization (2017). (http://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)) Accessed on 13 June 2018.
Yong, K. J., Jeong, U. P., Hyun, B., Sung, H. P., In Soo, K., Dong, W. K., & Joo, W. H. (2001). Purification and biochemical characterization of a fibrinolytic enzyme from Bacillus subtilis BK-17. World Journal of Microbiology and Biotechnology, 17, 89–92.
Raafat, A. I., Araby, E., & Lotfy, S. (2012). Enhancement of fibrinolytic enzyme production from Bacillus subtilis via immobilization process onto radiation synthesized starch/dimethylaminoethyl methacrylate hydrogel. Carbohydrate Polymer, 87(2), 1369–1374.
Kotb, E. (2013). Activity assessment of microbial fibrinolytic enzymes. Applied Microbiology and Biotechnology, 97(15), 6647–6665.
Krishnamurthy, A., & Belur, P. D. (2018). A novel fibrinolytic serine metalloprotease from the marine Serratia marcescens subsp. sakuensis: Purification and characterization. International Journal of Biological Macromolecules, 112, 110–118.
Krishnamurthy, A., Mundra, S., & Belur, P. D. (2018). Improving the catalytic efficiency of fibrinolytic enzyme from Serratia marcescens subsp. sakuensis by chemical modification. Process Biochemistry, 72, 79–85.
Balaraman, K., & Prabakaran, G. (2007). Production and purification of a fibrinolytic enzyme (thrombinase) from Bacillus sphaericus. Indian Journal of Medical Research, 126, 459–464.
Liu, X. L., Du, L. X., Lu, F. P., Zheng, X. Q., & Xiao, J. (2005). Purification and characterization of a novel fibrinolytic enzyme from Rhizopus chinensis 12. Applied Microbiology and Biotechnology, 67, 209–214.
Simkhada, J. R., Maner, P., Cho, S. S., & Ydoo, J. C. (2010). A novel fibrinolytic protease from Streptomyces sp. CS684. Process Biochemistry, 45(1), 88–93.
Ju, X., Cao, X., Sun, Y., Wang, Z., Cao, C., Liu, J., & Jiang, J. (2012). Purification and characterization of a fibrinolytic enzyme from Streptomyces sp. XZNUM 00004. World Journal of Microbiology and Biotechnology, 28(7), 2479–2486.
Pan, S., Chen, G., Zeng, J., Cao, X., Zheng, X., Zeng, W., & Liang, Z. (2019). Fibrinolytic enzyme production from low-cost substrates by marine Bacillus subtilis: Process optimization and kinetic modeling. Biochemical Engineering Journal, 141, 268–277.
Peng, Y., Huang, Q., Zhang, R. H., & Zhang, Y. Z. (2003). Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douchi, a traditional Chinese soybean food. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 134(1), 45–52.
Chandramohan, M., Chang, Y. Y., Beatrice, P. H. K., Ponnaiah, P., Narendrakumar, G., & Samrot, A. V. (2019). Production, characterization and optimization of fibrinolytic protease from Bacillus pseudomycoides strain MA02 isolated from poultry slaughter house soils. Biocatalysis and Agricultural Biotechnology, 22, 101371–101378.
Khursade, P. S., Galande, S. H., Krishna, P., Prakasham, S., & Prakashama, R. S. (2019). Stenotrophomonas maltophilia Gd2: A potential and novel isolate for fibrinolytic enzyme production. Saudi Journal of Biological Sciences, 26(7), 1567–1575.
Wang, C. T., Ji, B. P., Li, B., Nout, R., Li, P. L., Ji, H., & Chen, L. F. (2006). Purification and characterization of a fibrinolytic enzyme of Bacillus subtilis DC33, isolated from Chinese traditional Douchi. Journal of Industrial Microbiology & Biotechnology, 33(9), 750–758.
Kumar, S. S., Haridas, M., & Abdulhameed, S. (2020). A novel fibrinolytic enzyme from marine Pseudomonas aeruginosa KU1 and its rapid in vivo thrombolysis with little haemolysis. International Journal of Biological Macromolecules, 162, 470–479.
Kim, H. C., Choi, B. S., Sapkota, K., Kim, S., Lee, H. J., Yoo, J. C., & Kim, S. J. (2011). Purification and characterization of a novel, highly potent fibrinolytic enzyme from Paecilomyces tenuipes. Process Biochemistry, 46(8), 1545–1553.
Avhad, D. N., Vanjari, S. S., & Rathod, V. K. (2013). A novel fibrinolytic enzyme from Bacillus sphaericus MTCC 3672: Optimization and purification studies. International Journal of Current Microbiology and Applied Sciences, 1, 1–13.
Taneja, K., Bajaj, B. K., Kumar, S., & Dilbaghi, N. (2017). Production, purification and characterization of fibrinolytic enzyme from Serratia sp. KG-2-1 using optimized media. 3 Biotechnology, 7, 184–199.
Yang, H., Liu, Y., Ning, Y., Wang, C., Zhang, X., Weng, P., & Wu, Z. (2020). Characterization of an intracellular alkaline serine protease from Bacillus velezensis SW5 with fibrinolytic activity. Current Microbiology, 77(8), 1610–1621.
Sahoo, A., Mahanty, B., Daverey, A., & Dutt, K. (2020). Nattokinase production from Bacillus subtilis using cheese whey: Effect of nitrogen supplementation and dynamic modelling. Journal of Water Process Engineering, 38, 101533.
Wang, C., Du, M., Zheng, D., Kong, F., Zu, G., & Feng, Y. (2009). Purification and characterization of Nattokinase from Bacillus subtilis natto B-12. Journal of Agricultural and Food Chemistry, 57(20), 9722–9729.
Pan, C., Hu, B., Li, W., Sun, Y., Ye, H., & Zeng, X. (2009). Novel and efficient method for immobilization and stabilization of β-d-galactosidase by covalent attachment onto magnetic Fe3O4–chitosan nanoparticles. Journal of Molecular Catalysis B: Enzymatic, 61(3-4), 208–215.
Zhou, Z., & Hartmann, M. (2012). Recent progress in biocatalysis with enzymes immobilized on mesoporous hosts. Topics in Catalysis, 55(16-18), 1081–1100.
Rodrigues, R. C., Ortiz, C., Berenguer-Murcia, A., Torres, R., & Fernandez-Lafuente, R. (2013). Modifying enzyme activity and selectivity by immobilization. Chemical Society Reviews, 42(15), 6290–6307.
Homaei, A. A., Sariri, R., Vianello, F., & Stevanato, R. (2013). Enzyme immobilization: An update. Journal of Chemical Biology, 6(4), 185–205.
Zhou, Z., & Hartmann, M. (2013). Progress in enzyme immobilization in ordered mesoporous materials and related applications. Chemical Society Reviews, 42(9), 3894–3912.
Liang, S., Wu, X. L., Xiong, J., Zong, M. H., & Lou, W. Y. (2020). Metal-organic frameworks as novel matrices for efficient enzyme immobilization: An update review. Coordination Chemistry Reviews, 406, 213149.
Nadar, S. S., Vaidya, L., & Rathod, V. K. (2020). Enzyme embedded metal organic framework (enzyme–MOF): De novo approaches for immobilization. International Journal of Biological Macromolecules, 149(15), 861–876.
Ye, N., Kou, X., Shen, J., Huang, S., Chen, G., & Ouyang, G. (2020). Metal-organic frameworks: A new platform for enzyme immobilization. ChemBioChem, 21(18), 2585–2590.
Matsuno, R., Yamamoto, K., Otsuka, H., & Takahara, A. (2004). Polystyrene- and poly(3-vinylpyridine)-grafted magnetite nanoparticles prepared through surface-initiated nitroxide-mediated radical polymerization. Macromolecules, 37(6), 2203–2209.
Liu, Y., Jia, S., Wu, Q., Ran, J., Zhang, W., & Wu, S. (2011). Studies of Fe3O4-chitosan nanoparticles prepared by co-precipitation under the magnetic field for lipase immobilization. Catalysis Communications, 12(8), 717–720.
Susanto, H., Samsudin, A. M., Rokhati, N., & Widiasa, I. N. (2013). Immobilization of glucose oxidase on chitosan-based porous composite membranes and their potential use in biosensors. Enzyme and Microbial Technology, 52(6-7), 386–392.
Liu, Q., Hua, Y., Kong, X., Zhang, C., & Chen, Y. (2013). Covalent immobilization of hydroperoxide lyase on chitosan hybrid hydrogels and production of C6 aldehydes by immobilized enzyme. Journal of Molecular Catalysis B: Enzymatic, 95, 89–98.
Yewale, T., Singhal, R. S., & Vaidya, A. A. (2013). Immobilization of inulinase from Aspergillus niger NCIM 945 on chitosan and its application in continuous inulin hydrolysis. Biocatalysis and Agricultural Biotechnology, 2(2), 96–101.
Srivastava, P. K., & Anand, A. (2014). Immobilization of acid phosphatase from Vigna aconitifolia seeds on chitosan beads and its characterization. International Journal of Biological Macromolecules, 64, 150–154.
Wang, X. Y., Jiang, X.-P., Li, Y., Zeng, S., & Zhang, Y. W. (2015). Preparation Fe3O4@chitosan magnetic particles for covalent immobilization of lipase from Thermomyces lanuginosus. International Journal of Biological Macromolecules, 75, 44–50.
Wu, Y., Wang, Y., Luo, G., & Dai, Y. (2009). In situ preparation of magnetic Fe3O4-chitosan nanoparticles for lipase immobilization by cross-linking and oxidation in aqueous solution. Bioresource Technology, 100(14), 3459–3464.
Kuo, C.-H., Liu, Y.-C., Chang, C.-M. J., Chen, J.-H., Chang, C., & Shieh, C.-J. (2012). Optimum conditions for lipase immobilization on chitosan-coated Fe3O4 nanoparticles. Carbohydrate Polymer, 87(4), 2538–2545.
Assa, F., Jafarizadeh-Malmiri, H., Ajamein, H., Vaghari, H., Anarjan, N., Ahmadi, O., & Berenjian, A. (2016). A chitosan magnetic nanoparticles for drug delivery systems. Critical Reviews in Biotechnology, 37(4), 492–509.
Wulandari, I. O., Sulistyarti, H., Safitri, A., Santjojo, D. J. D. H., & Sabarudin, A. K. (2019). Development of synthesis method of magnetic nanoparticles modified by oleic acid and chitosan as a candidate for drug delivery agent. Journal of Applied Pharmaceutical Science, 9(07), 1–11.
Jafarizadeh-Malmiri, H., & Ghaz.Jahanian, M.A., Berenjian, A. (2012). Potential applications of chitosan nanoparticles as novel support in enzyme immobilization. American Journal of Biochemistry and Biotechnology, 8(4), 203–219.
Li, X., Zeng, D., Ke, P., Wang, G., & Zhang, D. (2020). Synthesis and characterization of magnetic chitosan microspheres for drug delivery. RSC Advances, 10(12), 7163–7169.
Talekar, S., Ghodake, V., Ghotage, T., Rathod, P., Deshmukh, P., Nadar, S., Mulla, M., & Ladole, M. (2012). Novel magnetic cross-linked enzyme aggregates (magnetic-CLEAs) of alpha amylase. Bioresource Technology, 123, 542–547.
Sheldon, R. (2011). Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Applied Microbiology and Biotechnology, 92(3), 467–477.
Torabizadeh, H., & Mikani, M. (2018). Nano-magnetic cross-linked enzyme aggregates of naringinase an efficient nanobiocatalyst for naringin hydrolysis. International Journal of Biological Macromolecules, 117, 134–143.
Badoei-dalfard, A., Karami, Z., & Malekabadi, S. (2019). Construction of CLEAs-lipase on magnetic graphene oxide nanocomposite: An efficient nanobiocatalyst for biodiesel production. Bioresource Technology, 278, 473–476.
Badoei-dalfard, A., Malekabadi, S., Karami, Z., & Sargazi, G. (2019). Magnetic cross-linked enzyme aggregates of Km12 lipase: A stable nanobiocatalyst for biodiesel synthesis from waste cooking oil. Renewable Energy, 141, 874–882.
Holt, J. G., Krieg, N. R., Sneath, P. H., Staley, J. T., & Williams, S. T. (1994). Bergey’s manual of determinative bacteriology, ninth ed. Baltimore: Lippincott Williams & Wilkins.
Laemmli, U. K. (1970). Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature, 227(5259), 680–685.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1-2), 248–254.
Badoei-dalfard, A., & Karami, Z. (2013). Screening and isolation of an organic solvent tolerant-protease from Bacillus sp. JER02: Activity optimization by response surface methodology. Journal of Molecular Catalysis B: Enzymatic, 89, 15–23.
Badoei-dalfard, A., Karami, Z., Ravan, H. (2015). Purification and characterization of a thermo- and organic solvent-tolerant alkaline protease from Bacillus sp. JER02. Preparative biochemistry and biotechnology, (45), 128-143.
Kim, W., Choi, K., Kim, Y., Park, H., Choi, J., Lee, Y., Oh, H., Kwon, I., & Lee, S. (1996). Purification and characterization of a fibrinolytic enzyme produced from Bacillus sp. strain CK 11-4 screened from Chungkook-Jang. Applied and Environmental Microbiology, 62(7), 2482–2488.
Kotb, E. (2015). Purification and partial characterization of serine fibrinolytic enzyme from Bacillus megaterium KSK 07 isolated from Kishk, a traditional Egyptian fermented food. Applied Biochemistry and Microbiology, 51(1), 34–43.
Choi, J. H., Kim, J. E., Kim, S., Yoon, J., Park, D. H., Shin, H. J., Leeb, H. J., & Cho, S. S. (2017). Purification and partial characterization of a low molecular fibrinolytic serine metalloprotease C142 from the culture supernatant of Bacillus subtilis C142. International Journal of Biological Macromolecules, 104(Pt A), 724–731.
Ko, J. H., Yan, J. P., Zhu, L., & Qi, Y. P. (2004). Identification of two novel fibrinolytic enzymes from Bacillus subtilis QK02: Comp. Comparative Biochemistry and Physiology-Part C: Toxicology, 137, 65–74.
Kim, S. H., & Choi, N. S. (2000). Purification and characterization of subtilisin DJ-4 secreted by Bacillus sp. strain DJ-4 screened from Doen-Jang. Bioscience, Biotechnology, and Biochemistry, 64(8), 1722–1725.
Wang, S. L., Wu, Y. Y., & Liang, T. W. (2011). Purification and biochemical characterization of a nattokinase by conversion of shrimp shell with Bacillus subtilis TKU007. New Biotechnology, 28(2), 196–202.
Hashemabadi, M., & Badoei-Dalfard, A. (2019). Fabrication of magnetic CLEA-protease nanocomposite: High progression in biotechnology and protein waste management. Catalysis Letter, 149(7), 1753–1764.
Peng, Y., Yang, X., & Zhang, Y. (2005). Microbial fibrinolytic enzymes: An overview of source, production, properties, and thrombolytic activity in vivo. Applied Microbiology and Biotechnology, 69(2), 126–132.
Mahajan, M. P., Nayak, S., & Lele, S. S. (2012). Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. Journal of Bioscience and Bioengineering, 113(3), 307–314.
Lei, L., Bai, Y., Li, Y., Yi, L., Yang, Y., & Xia, C. (2009). Study on immobilization of lipase onto magnetic microspheres with epoxy groups. Journal of Magnetism and Magnetic Materials, 321(4), 252–258.
Bedade, D. K., Muley, A. B., & Singhal, R. S. (2019). Magnetic cross-linked enzyme aggregates of acrylamidase from Cupriavidus oxalaticus ICTDB921 for biodegradation of acrylamide from industrial waste water. Bioresource Technology, 272, 137–145.
Xu, M., Ji, D., Deng, Y., & Agyei, D. (2020). Preparation and assessment of cross-linked enzyme aggregates (CLEAs) of β-galactosidase from Lactobacillus leichmannii 313. Food and Bioproducts Processing, 124, 82–96.
Singh, R. S., Saini, G. K., & Kennedy, J. F. (2010). Covalent immobilization and thermodynamic characterization of pullulanase for the hydrolysis of pullulan in batch system. Carbohydrate Polymers, 81(2), 252–259.
Sangeetha, K., & Emilia Abraham, T. (2008). Preparation and characterization of cross-linked enzyme aggregates (CLEA) of subtilisin for controlled release applications. International Journal of Biological Macromolecules, 43(3), 314–319.
Nguyen, L. T., & Yang, K. L. (2017). Combined cross-linked enzyme aggregates of horseradish peroxidase and glucose oxidase for catalyzing cascade chemical reactions. Enzyme and Microbial Technology, 100, 52–59.
Kumar, A., Wu, G., & Liu, Z. (2018). Synthesis and characterization of cross linked enzyme aggregates of serine hydroxyl methyltransferase from Idiomerina leihiensis. International Journal of Biological Macromolecules, 117, 683–690.
Kumar, S., Jana, A. K., Dhamija, I., Singla, Y., & Maiti, M. (2013). Preparation, characterization and targeted delivery of serratiopeptidase immobilized on amino-functionalized magnetic nanoparticles. European Journal of Pharmaceutics and Biopharmaceutics, 85(3), 413–426.
Tutar, H., Yilmaz, E., Pehlivan, E., & Yilmaz, M. (2009). Immobilization of Candida rugosa lipase on sporopollenin from Lycopodium clavatum. International Journal of Biological Macromolecules, 45(3), 315–320.
Mander, P., Cho, S. S., Simkhada, J. R., Choi, Y. H., & Yoo, J. C. (2011). A low molecular weight chymotrypsin-like novel fibrinolytic enzyme from Streptomyces sp. CS624. Process Biochemistry, 46(7), 1449–1455.
Moon, S., Kim, J., Kim, H., Choi, M. S., Park, B. R., Kim, S., Ahn, H., Chun, H. S., Shin, Y. K., Kim, J., Kim, D. K., Lee, S., Seo, Y., Kim, Y. H., & Kim, C. S. (2014). Purification and characterization of a novel fibrinolytic a chymotrypsin like serine metalloprotease from the edible mushroom, Lyophyllum shimeji. Journal of Bioscience and Bioengineering, 117(5), 544–550.
Yogesh, D., & Halami, P. M. (2015). A fibrin degrading serine metallo protease of Bacillus circulans with α-chain specificity. Food Bioscience, 11, 72–78.
Choi, D., Cha, W.-S., Park, N., Kim, H.-W., Lee, J. H., Park, J. S., & Park, S.-S. (2011). Purification and characterization of a novel fibrinolytic enzyme from fruiting bodies of Korean Cordyceps militaris. Bioresource Technology, 102(3), 3279–3285.
Palmer, T., Bonner, P. L. (2007). Enzymes: Biochemistry, biotechnology, clinical chemistry: Elsevier, 2nd Edition, Paperback
Li, Q., Yi, L., Marek, P., & Iverson, B. L. (2013). Commercial proteases: Present and future. FEBS letter, 587(8), 1155–1163.
Rahman, R. A., Zaliha, R. N., Geok, L. P., Basri, M., & Salleh, A. B. (2006). An organic solvent-stable alkaline protease from Pseudomonas aeruginosa strain K: Enzyme purification and characterization. Enzyme and Microbial Technology, 39(7), 1484–1491.
Klibanov, A. M. (2001). Improving enzymes by using them in organic solvents. Nature, 409(6817), 241–246.
Liu, X., Kopparapu, N. K., Li, Y., Deng, Y., & Zheng, X. (2017). Biochemical characterization of a novel fibrinolytic enzyme from Cordyceps militaris. International Journal of Biological Macromolecules, 94, 793–801.
Prasad, S., Kashyap, R. S., Deopujari, J. Y., Purohit, H. J., Taori, G. M., & Daginawal, H. F. (2006). Development of an in vitro model to study clot lysis activity of thrombolytic drugs. Thrombosis Journal, 4, 1–4.
Funding
The authors express their gratitude to the Research Council of the Shahid Bahonar University of Kerman (Iran) and Iran National Science Foundation (INSF) for their financial support during the course of this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Ethics Approval
This article does not contain any studies done with human or animal participants performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khankari, S., Badoei-dalfard, A. & Karami, Z. Cross-linked Enzyme Aggregates of Fibrinolytic Protease BC1 Immobilized on Magnetic Chitosan Nanoparticles (CLEAs-Fib-mChi): Synthesis, Purification, and Characterization. Appl Biochem Biotechnol 193, 2004–2027 (2021). https://doi.org/10.1007/s12010-021-03494-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03494-z