Abstract
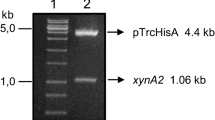
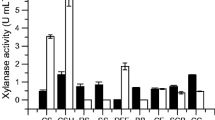
Caulobacter crescentus is able to express several enzymes involved in the utilization of lignocellulosic biomasses. Five genes, xynB1–5, that encode β-xylosidases are present in the genome of this bacterium. In this study, the xynB2 gene, which encodes β-xylosidase II (CCNA_02442), was cloned under the control of the PxylX promoter to generate the O-xynB2 strain, which overexpresses the enzyme in the presence of xylose. In addition, a null mutant strain, Δ-xynB2, was created by two homologous recombination events where the chromosomal xynB2 gene was replaced by a copy that was disrupted by the spectinomycin-resistant cassette. We demonstrated that C. crescentus cells lacking β-xylosidase II upregulates the xynB genes inducing β-xylosidase activity. Transcriptional analysis revealed that xynB1 (RT-PCR analysis) and xynB2 (lacZ transcription fusion) gene expression was induced in the Δ-xynB2 cells, and high β-xylosidase activity was observed in the presence of different agro-industrial residues in the null mutant strain, a characteristic that can be explored and applied in biotechnological processes. In contrast, overexpression of the xynB2 gene caused downregulation of the expression and activity of the β-xylosidase. For example, the β-xylosidase activity that was obtained in the presence of sugarcane bagasse was 7-fold and 16-fold higher than the activity measured in the C. crescentus parental and O-xynB2 cells, respectively. Our results suggest that β-xylosidase II may have a role in controlling the expression of the xynB1 and xynB2 genes in C. crescentus.





Similar content being viewed by others
References
Lynch, J. M. (1987). Utilization of lignocellulosic wastes. Journal of Applied Microbiology, 63, 71–83.
Jordan, D. B., & Wagschal, K. (2010). Properties and applications of microbial β-d-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Applied Microbiology and Biotechnology, 86, 1647–1658.
Ahmed, S., Saba, R., & Jamil, A. (2009). Molecular cloning of fungal xylanases: an overview. Applied Microbiology and Biotechnology, 84, 19–35.
Poindexter, J. S. (1964). Biological properties and classification of the Caulobacter group. Bacteriological Reviews, 28, 231–295.
Poindexter, J. S. (1981). The Caulobacters: ubiquitous unusual bacteria. Microbiological Reviews, 45, 123–179.
Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J., et al. (2001). Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA, 98, 4136–4141.
Marks, M. E., Castro-Rojas, C. M., Teiling, C. D. U. L., Kapatral, V., Walunas, T. L., & Crosson, S. (2010). The genetics basis of laboratory adaptation in Caulobacter crescentus. Journal of Bacteriology, 192, 3678–3688.
Santos, C. R., Polo, C. C., Corrêa, J. M., Simão, R. C. G., Seixas, F. A. V., & Murakami, M. T. (2012). Accessory domain changes accessibility and molecular topography of the catalytic interface in monomeric GH39 beta-xylosidases. Acta Cryst D, 68, 1339–1345.
Corrêa, J. M., Graciano, L., Abrahão, J., Loth, E. A., Gandra, R. F., Kadowaki, M. K., et al. (2012). Expression and characterization of a GH39 β-xylosidase II from Caulobacter crescentus. Applied Biochemistry and Biotechnology, 168, 2218–2229.
Yang, J. K., Yoon, H.-J., Ahn, H. J., Lee, B. I., Pedelacq, J.-D., Liong, E. C., et al. (2004). Crystal structure of beta-d-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. Journal of Molecular Biology, 335, 155–165.
Czjzek, M., David, A. B., Bravman, T., Shoham, G., Henrissat, B., & Shoham, Y. (2005). Enzyme-substrate complex structures of a GH39 β-xylosidase from Geobacillus stearothermophilus. Journal of Molecular Biology, 353, 838–846.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual (2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Evinger, M., & Agabian, N. (1977). Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. Journal of Bacteriology, 132, 294–301.
Simon, R., Prieffer, U., & Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology, 1, 784–790.
Gober, J. W., & Shapiro, L. (1992). A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Molecular Biology of the Cell, 3, 913–916.
Prentki, P., & Krisch, H. M. (1984). In vitro insertional mutagenesis with a selectable DNA fragments. Gene, 29, 303–313.
Ely, B. (1991). Genetics of Caulobacter crescentus. Methods in Enzymology, 204, 372–384.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature, 226, 680–685.
Towbin, H., Staehelin, T., & Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Nat Acad Sci USA, 76, 4350–4354.
Miller, J. H. (1972). Cold Spring Harbor (pp. 352–355). New York: Cold Spring Harbor Laboratory.
Graciano, L., Corrêa, J. M., Gandra, R. F., Seixas, F. A. V., Kadowaki, M. K., Sampaio, S. C., et al. (2012). The cloning, expression, purification, characterization and modeled structure of Caulobacter crescentus β - xylosidase I. World Journal of Microbiology and Biotechnology, 28, 2879–2888.
Hottes, A. K., Meewan, M., Yang, D., Arana, N., Romero, P., McAdams, H. H., et al. (2004). Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. Journal of Bacteriology, 186, 1448–1461.
Turatsinze, J. V., Thomas-Chollier, M., Defrance, M., & van Helden, J. (2008). Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nature Protocols, 3, 1578–1588.
Stephens, C., Christen, B., Fuchs, T., Sundaram, V., Watanabe, K., & Jenal, U. (2007). Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. Journal of Bacteriology, 189, 2181–2185.
Stephens, C., Christen, B., Watanabe, K., Fuchs, T., & Jenal, U. (2007). Regulation of d-xylose metabolism in Caulobacter crescentus by a LacI-type repressor. Journal of Bacteriology, 189, 8828–8834.
Schultz, J., Milpetz, F., Bork, P., & Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. PNAS, 95, 5857–5864.
Lobley, A., Sadowski, M. I., & Jones, D. T. (2009). pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics, 25, 1761–1767.
Acknowledgments
This work was supported by grants from the Araucaria Foundation and CNPq. R.C.G. Simão was partially supported by Araucaria Foundation (process 893/2012). M. R. Mingori and J. M. Corrêa are fellows of the CNPq and Coordination of Improvement of Higher Education Personnel (CAPES), respectively.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 3415 kb)
Rights and permissions
About this article
Cite this article
Corrêa, J.M., Mingori, M.R., Gandra, R.F. et al. Depletion of the xynB2 Gene Upregulates β-Xylosidase Expression in C. crescentus . Appl Biochem Biotechnol 172, 1085–1097 (2014). https://doi.org/10.1007/s12010-013-0549-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0549-8