Abstract
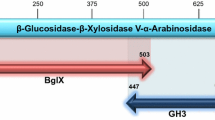
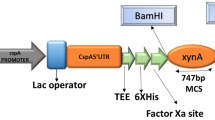
The xynB1 gene (CCNA 01040) of Caulobacter crescentus that encodes a bifunctional enzyme containing the conserved β-Xylosidase and α-l-Arabinofuranosidase (β-Xyl I-α-l-Ara) domains was amplified by PCR and cloned into the vector pJet1.2Blunt. The xynB1 gene was subcloned into the vector pPROEX-hta that produces a histidine-fused translation product. The overexpression of recombinant β-Xyl I-α-l-Ara was induced with IPTG in BL21 (DE3) and the resulting intracellular protein was purified with pre-packaged nickel-Sepharose columns. The recombinant β-Xyl I-α-l-Ara exhibited a specific β-Xylosidase I activity of 1.25 U mg−1 to oNPX and a specific α-l-Arabinofuranosidase activity of 0.47 U mg−1 to pNPA. The predominant activity of the recombinant enzyme was its β-Xylosidase I activity, and the enzymatic characterization was focused on it. The β-Xylosidase I activity was high over the pH range 3–10, with maximal activity at pH 6. The enzyme activity was optimal at 45 °C, and a high degree of stability was verified over 240 min at this temperature. Moreover, β-Xylosidase activity was inhibited in the presence of the metals Zn2+ and Cu2+, and the enzyme exhibited KM and VMax values of 2.89 ± 0.13 mM and 1.4 ± 0.04 μM min−1 to oNPX, respectively. The modeled structure of β-xylosidase I showed that its active site is highly conserved compared with other structures of the GH43 family. The increase in the number of contact residues responsible for maintaining the dimeric structure indicates that this dimer is more stable than the tetramer form.




Similar content being viewed by others
References
Ahmed S, Riaz S, Jamil A (2009) Molecular cloning of fungal xylanases: an overview. Appl Microbiol Biotechnol 84:19–35. doi:10.1007/s00253-009-2079-4
Basaran P, Ozcan M (2008) Characterization of β-Xylosidase enzyme from a Pichia stipitis mutant. Bioresour Technol 99:38–43. doi:10.1016/j.biortech.2006.11.056
Bravman T, Mechaly A, Shulami S, Belakhov V, Baasov T, Shoham G, Shoham Y (2001) Glutamic acid 160 is the acid-base catalyst of beta-xylosidase from Bacillus stearothermophilus T-6: a family 39 glycoside hydrolase. FEBS Lett 495:115–119. doi:10.1016/S0014-5793(01)02371-7
Brunzelle JS, Jordan DB, McCaslin DR, Olczak A, Wawrzak Z (2008) Structure of the two-subsite beta-d-xylosidase from Selenomonas ruminantium in complex with 1,3-bis [tris (hydroxymethyl) methylamino] propane. Arch Biochem Biophys 474:157–166. doi:10.1016/j.abb.2008.03.007
Brüx C, Ben-David A, Shallom-Shezifi D, Leon M, Niefind K, Shoham G, Shoham Y, Schomburg D (2006) The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J Mol Biol 359:97–109. doi:10.1016/j.jmb.2006.03.005
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi:10.1093/nar/gkn663
CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi:10.1107/S0907444994003112
Chen WP, Kuo TT (1993) A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res 21:2260
Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36(suppl. 2):W197–W201. doi:10.1093/nar/gkn238
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinform Chapter 5: Unit 5.6. doi: 10.1002/0471140864.ps0209s50
Evinger M, Agabian N (1977) Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132:294–301
Fan Z, Wagschal K, Lee CC, Kong Q, Shen KA, Maiti IB, Yuan L (2008) The construction and characterization of two xylan degrading chimeric enzymes. Biotechnol Bioeng 102:684–692. doi:10.1002/bit.22112
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Jordan DB, Wagschal K (2010) Properties and applications of microbial β-d-Xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl Microbiol Biotechnol 86(6):1647–1658. doi:10.1007/s00253-010-2538-y
Jordan DB, Li XL, Dunlap CA, Whitehead TR, Cotta M (2007) A structure function relationships of a catalytically efficient β-D-Xylosidase. Appl Biochem Biotechnol 141:51–76
Knob A, Terrasan CRF, Carmona EC (2010) β-Xylosidases from filamentous fungi: an overview. World J Microbiol Biotechnol 26:389–407
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi:10.1016/j.jmb.2007.05.022
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 226:680–685
Lama L, Calandrelli V, Gambacorta A, Nicolaus B (2004) Purification and characterization of thermostable Xylanase and β-Xylosidase by the thermophilic bacterium Bacillus thermantarcticus. Res Microbiol 155:283–289. doi:10.1016/j.resmic.2004.02.001
Li YK, Yao HJ, Cho Y (2000) Effective induction, purification and characterization of Trichoderma koningii G-39 β-Xylosidase with high transferase activity. Biotechnol Appl Biochem 31:119–125. doi:10.1042/BA19990072
MacKerell AD Jr, Feig M, Brooks CL III (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25:1400–1415. doi:10.1002/jcc.20065
Marks ME, Castro-Rojas CM, Teiling CDUL, Kapatral V, Walunas TL, Crosson S (2010) The genetics basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192:3678–3688. doi:10.1128/JB.00255-10
Mc Ilvaine TC (1921) A buffer solution for colorimetric comparison. J Biol Chem 49:183–186
Mc Lachlan AD (1982) Rapid comparison of protein structres. Acta Crystallogr 38:871–873
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem 31:426–428
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. doi:10.1002/jcc.20289
Poindexter JS (1964) Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295
Polizeli MLTM, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS (2005) Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol 67:577–591. doi:10.1007/s00253-005-1904-7
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. doi:10.1093/nar/gki442
Quintero D, Velasco Z, Hurtado E-G, Neira JL, Contreras LM (2007) Isolation and characterization of a thermostable β-Xylosidase in the thermophilic bacterium Geobacillus pallidus. Biochem Biophys Acta 1774:510–518. doi:10.1016/j.bbapap.2007.02.002
Rodionova NA, Tavobilov IM, Bezborodov AM (1983) β-Xylosidase from Aspergillus niger 15 Purification and properties. J Appl Biochem 5:300–312
Sanz-Aparicio J, Hermoso J, Grangeiro TB, Calvete JJ, Cavada BS (1997) The crystal structure of Canavalia brasiliensis lectin suggests a correlation between its quaternary conformation and its distinct biological properties from Concanavalin A. FEBS Lett 405:114–118. doi:10.1016/S0014-5793(97)00137-3
Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D (2011) BRENDA, the enzyme information system in 2011. Nucleic Acids Res 39:D670–D676. doi:10.1093/nar/gkq1089
Shao W, Xue Y, Wu A, Kataeva I, Pei J, Wu H, Wiegel J (2011) Characterization of a novel beta-xylosidase XylC from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl Environ Microbiol 77:719–726. doi:10.1128/AEM.01511-10
Suryani K, Sakka K, Ohmiya K (2004) Sequencing and expression of the gene encoding the Clostridium stercorarium beta-xylosidase Xyl43B in Escherichia coli. Biosci Biotechnol Biochem 68:609–614. doi:10.1271/bbb.68.609
Ximenes FA, Silveira FQP, Filho EX (1996) Production of beta-xylosidase activity by Trichoderma harzianum strains. Curr Microbiol 33:71–77
Xue Y, Shao W (2004) Expression and characterization of a thermostable β-Xylosidase from the hyperthermophile Thermotoga maritima. Biotechnol Lett 26:1511–1515. doi:10.1023/B:BILE.0000044454.70768.81
Acknowledgments
L. Graciano and J. M. Corrêa are fellows of Fundação Araucária and Fundação Parque Tecnológico Itaipu (PTI C&T, FTPI-BR), respectively. This work was supported by “Fundação Araucária, Fundo Paraná/Secretaria da Ciência Tecnologia e Ensino Superior (SETI)” and “Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).”
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Graciano, L., Corrêa, J.M., Gandra, R.F. et al. The cloning, expression, purification, characterization and modeled structure of Caulobacter crescentus β-Xylosidase I. World J Microbiol Biotechnol 28, 2879–2888 (2012). https://doi.org/10.1007/s11274-012-1099-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-012-1099-x