Abstract
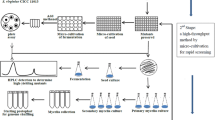
Genome shuffling is a powerful approach for efficiently engineering industrial microbial strains with interested phenotypes. Here we reported a high producer of nuclease P1, Penicillium citrinum G-16, that was bred by the classical physics-mutagenesis and genome shuffling process. The starting populations were generated by 60Co γ-irradiation mutagenesis. The derived two protoplast fractions were inactivated by heat-treatment and ultraviolet radiation respectively, then mixed together and subjected to recursive protoplast fusion. Three recombinants, E-16, F-71, and G-16, were roughly obtained from six cycles of genome shuffling. The activity of nuclease P1 by recombinant G-16 was improved up to 1,980.22 U4/ml in a 5-l fermentor, which was 4.7-fold higher than that of the starting strain. The sporulation of recombinant G-16 was distinguished from the starting strain. Random amplified polymorphic DNA assay revealed genotypic differences between the shuffled strains and the wild type strain. The close similarity among the high producers suggested that the genetic basis of high-yield strains was achieved by genome shuffling.







Similar content being viewed by others
References
Munetaka, S., Jun, I., Shigenmi, A., & Hiroo, F. (2000). Endonucleases. Plant Molecular Biology, 44, 387–397.
Gangadhara, B. N., Kumar, P. R., & Prakash, V. (2008). Enhancement of nuclease P1 activity in low concentration of denaturants. Enzyme and Microbial Technology, 43, 336–342.
Burdock, G. A., Flamm, W. G., & Carabin, I. G. (2000). Toxicity and mutagenicity studies of DN-50000® and RP-1® enzymes. Food and Chemical Toxicology, 38, 429–442.
Warnecke, J. M., Sontheimer, E. J., Piccirilli, J. A., & Hartmann, R. K. (2000). Active site constraints in the hydrolysis reaction catalyzed by bacterial RNase P: analysis of precursor tRNAs with a single 3′-S-phosphorothiolate internucleotide linkage. Nucleic Acids Research, 28, 720–727.
Li, H. H., He, Y. H., Yuan, Y., & Guan, Z. (2011). Nuclease p1: a new biocatalyst for direct asymmetric aldol reaction under solvent-free conditions. Green Chemistry, 13, 185–189.
Shi, L. E., Yi, Y., Tang, Z. X., Xiong, W. Y., Mei, J. F., & Ying, G. Q. (2010). Nuclease p1 immobilized on deae cellulose. Brazilian Journal of Chemical Engineering, 27, 31–39.
Li, B., Chen, Y., Chen, X., Liu, D., Niu, H., Xiong, J., et al. (2012). A novel immobilization method for nuclease P1 on macroporous absorbent resin with glutaraldehyde cross-linking and determination of its properties. Process Biochemistry, 47, 655–670.
He, Q. T., Li, N., Chen, X. C., Ye, Q., Bai, J. X., Xiong, J., et al. (2011). Mutation breeding of nuclease p1 production in Penicillium citrinum by low-energy ion beam implantation. Korean Journal of Chemical Engineering, 28, 544–549.
Li, K., Zeng, Q., & Xiang, Z. (2007). Selection of Biochemical Mutants that Overproduce Nuclease P1 and Optimization of the Fermentation Conditions. Journal of Yunnan Agricultural University, 6, 898–904.
Matthew, T. W., & Laurence, D. H. (2012). Direct and indirect consequences of meiotic recombination: implications for genome evolution. Trends in Genetics, 28, 101–109.
Zhang, Y. X., Perry, K., Vinci, V. A., Powell, K., Stemmer, W. P., & Del Cardayré, S. B. (2002). Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature, 415, 644–646.
Jin, Z. H., Xu, B., Lin, S. Z., Jin, Q. C., & Cen, P. L. (2009). Enhanced Production of Spinosad in Saccharopolyspora spinosa by Genome Shuffling. Applied Biochemistry and Biotechnology, 159, 655–663.
Wang, H. K., Zhang, J., & Wang, X. J. (2012). Genome shuffling improves production of the low-temperature alkalophilic lipase by Acinetobacter johnsonii. Biotechnology Letters, 34, 145–151.
El-Bondkly, A. M. A. (2012). Molecular identification using ITS sequences and genome shuffling to improve 2-deoxyglucose tolerance and xylanase activity of marine-derived fungus, Aspergillus sp. NRCF5. Applied Biochemistry and Biotechnology, 167, 2160–2173.
Li, S., Li, F., Chen, X. S., Wang, L., Xu, J., Tang, L., et al. (2012). Genome shuffling enhanced ε-poly-l-lysine production by improving glucose tolerance of Streptomyces graminearus. Applied Biochemistry and Biotechnology, 166, 414–423.
Burkhard, O., Eike, G., Osama, M., & Stefan, J. (2009). Genome shuffling in Clostridium diolis DSM 15410 for improved 1,3-propanediol production. Applied and Environmental Microbiology, 75, 7610–7616.
Gregory, S. (2002). Metabolic engineering by genome shuffling. Nature Biotechnology, 20, 666–668.
Lv, X. A., Jin, Y. Y., Li, Y. D., Zhang, H., & Liang, X. L. (2013). Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Applied Microbiology and Biotechnology, 97, 641–648.
Futoshi, N., Ichiro, W., & Nobufusa, S. (1993). Development of transformation system for the filamentous, ML-236B (compactin) - producing fungus Penicilliurn citrinum. Current Genetics, 23, 28–32.
Tahoun, M. K. (1993). Gene manipulation by protoplast fusion and penicillin production by Penicillium chrysogenum. Applied Biochemistry and Biotechnology, 39(40), 445–453.
Savitha, S., Sadhasivam, S., & Swaminathan, K. (2010). Regeneration and molecular characterization of an intergeneric hybrid between Graphium putredinis and Trichoderma harzianum by protoplasmic fusion. Biotechnology Advances, 28, 282–292.
Abe, Y., Baba, S., Suzuke, T., Ono, C., Iwamoto, K., & Hosobuchi, M. (2004). Molecular basis of ML-236B production in the high-producing mutant No. 41520 of Penicillium citrinum. Journal of General Applied Microbiology, 50, 169–176.
Ying, G. Q., Shi, L. E., Yua, Y., Tang, Z. X., & Chen, J. S. (2006). Production, purification and characterization of nuclease p1 from Penicillium citrinum. Process Biochemistry, 41, 1276–1281.
Patnaik, R., Louie, S., Gavrilovic, V., Perry, K., Stemmer, W. P. C., Ryan, C. M., et al. (2002). Genome shuffling of Lactobacillus for improved acid tolerance. Nature, 20, 707–712.
Yu, L., Pei, X., Lei, T., Wang, Y., & Feng, Y. (2008). Genome shuffling enhanced l-lactic acid production by improving glucose tolerance of Lactobacillus rhamnosus. Journal of Biotechnology, 134, 154–159.
Cheng, Y., Song, X., Qin, Y., & Qu, Y. (2009). Genome shuffling improves production of cellulase by Penicillium decumbens JU-A10. Journal of Applied Microbiology, 107, 1738–1846.
Hide, H., Yamada, T., & Yamada, Y. (2007). Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitic acid. Applied Microbiology and Biotechnology, 73, 1387–1393.
Wucherpfennig, T., Kiep, K. A., Driouch, H., Wittmann, C., & Krull, R. (2010). Chapter 4 - morphology and rheology in filamentous cultivations. Advances in Applied Microbiology, 73, 89–136.
Wucherpfennig, T., Hestler, T., & Krull, R. (2001). Morphology engineering - osmolality and its effect on Aspergillus niger morphology and productivity. Microbial Cell Factories, 10, 58.
Angel, M., & Ruben, A. (2008). Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Applied and Environmental Microbiology, 12, 3877–3886.
Xu, B., Jin, Z. H., Wang, H. Z., Jin, Q. C., Jin, X., & Cen, P. L. (2008). Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Applied Microbiology and Biotechnology, 80, 261–267.
El-Gendy, M. M., & EL-Bondkly, A. M. (2011). Genome shuffling of marine derived bacterium Nocardia sp. ALAA 2000 for improved ayamycin production. Antonie Van Leeuwenhoek, 99, 773–780.
Acknowledgments
The work was financially supported by the National Nature and Science Foundation of China (3117175) and Nature and Science Foundation of Zhejiang Province (Y3100609).
Conflict of interests
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, C., Wu, G., Li, Y. et al. Genome Shuffling of Penicillium citrinum for Enhanced Production of Nuclease P1. Appl Biochem Biotechnol 170, 1533–1545 (2013). https://doi.org/10.1007/s12010-013-0297-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0297-9