Abstract
Deep learning-based image segmentation models rely strongly on capturing sufficient spatial context without requiring complex models that are hard to train with limited labeled data. For COVID-19 infection segmentation on CT images, training data are currently scarce. Attention models, in particular the most recent self-attention methods, have shown to help gather contextual information within deep networks and benefit semantic segmentation tasks. The recent attention-augmented convolution model aims to capture long range interactions by concatenating self-attention and convolution feature maps. This work proposes a novel attention-augmented convolution U-Net (AA-U-Net) that enables a more accurate spatial aggregation of contextual information by integrating attention-augmented convolution in the bottleneck of an encoder–decoder segmentation architecture. A deep segmentation network (U-Net) with this attention mechanism significantly improves the performance of semantic segmentation tasks on challenging COVID-19 lesion segmentation. The validation experiments show that the performance gain of the attention-augmented U-Net comes from their ability to capture dynamic and precise (wider) attention context. The AA-U-Net achieves Dice scores of 72.3% and 61.4% for ground-glass opacity and consolidation lesions for COVID-19 segmentation and improves the accuracy by 4.2% points against a baseline U-Net and 3.09% points compared to a baseline U-Net with matched parameters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The novel coronavirus disease 2019 (COVID-19) emerged in early December 2019 in the Hubei province of the People’s Republic of China, being caused by the virus Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). It rapidly spread to various countries and developed into a global outbreak. The World Health Organization (WHO) declared the outbreak as a global pandemic on March 11, 2020 [1, 2]. This also led to a massive strain on the healthcare systems worldwide.
The gold standard for diagnosis of COVID-19 is reverse-transcription polymerase chain reaction (RT-PCR) test [3]. However, it suffers from various limitations, such as not being universally available in large scales, manual, time-consuming and varying accuracy [3, 4]. Alternatively, radiography techniques of chest X-rays (CXR) and CT have proven to be good diagnostic modalities for detection of COVID-19 and are being widely used for COVID-19 screening, primarily because they are faster and cheaper than the standard tests [5]. Thus literature suggests that a combination of clinical, laboratory and radiological testing should be used for prognosis and diagnosis of this disease [4, 6].
It has been reported that prognosis and diagnosis using CXR images have certain limitations in COVID-19 cases (less efficient) [1, 7]. Hence CT proves to be advantageous and an effective screening tool as it provides early stage detection of the disease and segmentation of the lungs along with their three-dimensional views [1, 4].
The important diagnostic features of COVID-19 on CT are ground-glass opacity, consolidation, nodules, [3, 4]. However, identification of these features becomes time-consuming for radiologists and clinicians, considering the rising number of cases, hence deep learning techniques and computer vision can help in automatic segmentation of these features [5, 8].
Deep learning has been implemented on COVID-19 CT data by performing automatic segmentation and classification and has supported in improving diagnosis performance significantly. Segmentation in COVID-19 cases includes lung delineation and lesion measurement and is important for diagnosis and quantitative analysis of the disease. For segmenting region of interest in CT, classic U-Net, U-Net++, VB-Net, etc., have been used widely [1]. Li et al. used U-Net to perform lung segmentation on chest CT and distinguish COVID-19 cases from community-acquired pneumonia [9]. Shan et al. used a VB-Net to perform segmentation of lung, lung lobes and infection and provided medical data such as quantitative assessment of progression of the infection in the follow-up, prediction of severity, visualization of lesion distribution, etc. [10]. Qi et al. used U-Net-based algorithm to segment lung lesions of COVID-19 patients and extracted radiomics features for predicting duration of hospital stay [11].
A number of studies have been done to classify COVID-19 patients from non-COVID-19 subjects or other types of pneumonia. A COVID-19 detection neural network, named COVNet (convolutional ResNet-50 model), was introduced in [9] to detect the disease from a mix of CT images of three types- COVID-19, community acquired pneumonia and non-pneumonia cases. Chen et al. classified COVID-19 and non-COVID-19 cases by using U-Net++-based segmentation model and making a decision based on the appearance of segmented lesions [12].
Zheng et al. [13] used a combination of two models for segmentation and classification—they employed a U-Net model to perform lung segmentation and used the result as input to 3D CNN for predicting the probability of COVID-19 and achieved an AUC of 0.959. Similarly, Jin et al. [14] proposed a U-Net++-based segmentation model for lesion localization and a ResNet50-based classification model to classify COVID-19-positive and negative cases. This model successfully achieved an accuracy of 86.7% to classify the three cases. In a similar study, three-class classification was done to classify COVID-19, bacterial pneumonia and healthy cases by integrating a pretrained ResNet-50, the Feature Pyramid Network (FPN) and the Attention module to represent more detailed aspects of the images [15]. Gozes et al. [16] used U-Net for segmentation of relevant lung regions and Resnet-50 to perform the classification. The networks obtained an AUC of 0.996 for classifying COVID-19-positive and negative cases.
2 Related work
U-Net [26] has been commonly used for segmenting lung regions and lung lesions in COVID cases [1]. Various variants of U-Net have been developed to achieve reasonable segmentation results in COVID-19 cases, and few of them have been discussed in the previous section. Along with segmentation, the attention mechanism has been reported as an efficient localization method in screening which can be adopted for application in COVID-19 cases [1]. Attention gates have also proved to be promising in 3D medical segmentation when incorporated with U-Net [23]. Hence, in this work, U-Net along with attention mechanism has been implemented to improve the performance on semantic segmentation on COVID-19 cases.
2.1 Attention mechanism
Attention models initially were first introduced through transformer models in natural language processing (NLP) domain [21]. NLP models performed better when the encoder and decoder were connected through attention blocks. Boosted NLP performance encouraged researchers for further research on attention mechanism in other domains.
Computer vision tasks subsequently benefited from attention mechanism to capture non-local interactions. Initial approaches to augment convolutional models with content-based interactions [18, 19, 23] was very promising. The seminal work by Wang et al. [20] in non-local means was one of the key milestones in progress of attention in vision applications. This was then followed by self-attention research by Bello et al. [18]. These research initiatives have hugely benefited computer vision tasks like image classification and semantic segmentation. Jo et al. [23] subsequently integrated attention-gates into U-Nets for 3D medical segmentation. Parmar et al. [17] have also demonstrated building pure self-attention vision models.
Bello et al. [18] prevail the convolution operator’s disadvantage of lack of global contexts by augmenting it with self-attention. They augmented convolution operators with a novel two-dimensional relative self-attention mechanism by concatenating self-attention feature maps with convolutional feature maps. The attention-augmented convolution attends jointly to spatial and feature subspaces. They demonstrate that attention augmentation gives significant improvement in image classification (CIFAR-100, ImageNet) and object detection tasks (COCO dataset).
Given an input activation map tensor of height H, width W and channels \(F_\mathrm{in}\), Bello at al. [18] compute multihead attention (MHA) on the flattened input matrix \({\mathbf {X}} \in {\mathbb {R}}^{HWxF_\mathrm{in}}\). The single head self-attention is calculated using the formula:
where \(W_{q}, W_{k} \in {\mathbb {R}}^{F_\mathrm{in} \times d_{k}^{h}}\) and \(W_{v} \in {\mathbb {R}}^{F_\mathrm{in} \times d_{v}^{h}}\) are the linear transformations which map the activation map features to queries Q, keys K and values V. The multi-head outputs concatenated and then reshaped to match the original dimension of (H, W, \(d_{v}\)). As the final step, the convolutional and attentional feature maps are concatenated together to get the attention-augmented convolution result [18].
The aim of this work is threefold.
-
1.
First, evaluation of whether attention-augmented convolution can be employed for semantic segmentation tasks has been performed. To our best knowledge, this work is the among the first to explore attention-augmentation for semantic segmentation specifically in medical image segmentation.
-
2.
Second, it is appraised whether attention-augmented convolution can be integrated within a U-Net [23] to improve medical image lesion segmentation for labeled data which is relatively small, a common scenario currently for COVID-19.
-
3.
Third, empirical identification of the novel network configuration (constellation) is performed where the attention augmentation convolution can be integrated to get significant improvement in segmentation performance, while still only marginally increasing the model complexity (number of learnable parameters).
The attention-augmented U-Net (AA-UNet) module is an important step to get better insight into the integration of attention-augmented convolution modules into segmentation contexts in general, and U-Nets in particular. It has been found that modifying all the convolutions blocks in U-Net into attention-augmented convolution blocks does negatively impact the complexity of semantic segmentation networks. Capturing only the necessary and essential non-local contextual information in a smart way by positioning it at the bottleneck of U-Net, yields promising segmentation results. It has been demonstrated that having the bottleneck attention block enables the network to obtain longer range dependencies on the smallest resolution of activation maps and get far improved results with minimal increase in model complexity.
3 Methods
Similar to the notations in Bello et al. [18], the following naming conventions have been used in this work: H, W and \(F_\mathrm{in}\) refer to the height, width and number of input filters of the activation maps. N h, \(d_{v}\) and \(d_{k}\) denote the number of heads, depth of values, and the depth of queries and keys in multihead-attention respectively.
3.1 Self-attention over (bottleneck) feature channel
Given an input image (tensor) of shape (H,W,\(F_\mathrm{in}\)), it is systematically processed through the proposed U-Net algorithm, to compute the feature maps using regular convolution operators. The activation maps are downsampled using maxpooling operation. After the last downsampling operator in the U-Net, when the feature maps are the smallest, the attention maps are computed on this reduced activation map. As described by Bello et al. [18], the attention-augmented convolution concatenates the convolution features and attention feature maps. The computed attention-augmented convolution feature maps are concatenated with the convolution activation maps from the last downsampling layer, and then progressively upsampled through the upsampling path of the U-Net. The details of the U-Net network architecture used in this work and the integration of attention-augmented convolution are detailed in the subsequent sections below.
3.2 Proposed AA-UNet network architecture
The architecture of the model combines the concepts of U-Net [26] and attention-augmented convolution network [18]. This novel proposed network utilizes Attention-Augmented convolutions within a medical segmentation deep learning framework so as to utilize the characteristics of attention augmentation framework to comprehend global perspective and capturing long range dependencies. A block diagram of the proposed attention-augmented U-Net (AA-UNet) is shown in Fig. 1.
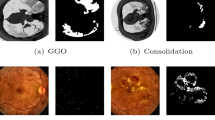
Sample slice from one of the dataset and the corresponding ground-glass opacity lesion (GGO) marking in first row and GGO and consolidation lesion marking in second row. Dataset from website [22]
A U-Net structure from Oktay et al. [23, 23] has been used, adapting it slightly by reducing one downsampling (and corresponding upsampling path), to best process the image dimension of interest (256*256). It consists of three blocks in the downsampling path and three blocks in the upsampling block. Each block consists of \(2\times \)(batch normalization—2D convolution (kernel size \(3\times 3\), stride 1, padding 1)—ReLU). The last block consists of a 2D convolution with kernel size \(1\times 1\). For downsampling, max pooling is applied in the downsampling path to halve the spatial dimension of the feature maps after each block. In the upsampling path ConvTranspose2d is used to double the size of the spatial dimension of the concatenated feature maps. The number of feature channels is increased 1–64–128–256–512 in the downsampling path and decreased again accordingly in the upsampling path. The U-Net’s last layer outputs a number of feature channels matching the number of label classes for semantic segmentation.
A block diagram of the proposed attention-augmented U-Net (AA-U-Net). Input image is progressively filtered and downsampled by factor 2 at each scale in the encoding part. The attention-augmented convolution is inserted as an extension of the U-Net’s bottleneck in order to capture contextual information from only the necessary and meaningful non-local contextual information in smart and efficient way
The local representation feature maps \({{\textbf {H}}}\) being output from the U-Net’s last block within the downsampling path serve as input of reduced dimension to the attention-augmented convolution module. The attention-augmented convolution module is inserted in the bottleneck, as the feature maps are of reduced dimension, and hence the attention maps have smaller, more manageable time and space complexity hence reducing computational cost. In the original attention-augmented convolution network [18], the attention-augmented convolution was integrated into the ResNet architecture [21]. The last three stages of the ResNet architecture were augmented with attention augmentation in each of these residual layers. In the original work, attention-augmented convolution was employed when the spatial dimension of the activation maps was \(28\times 28\), \(14\times 14\), and \(7\times 7\). Bello et al. [18] clearly documented in their paper that memory cost for attention-augmented convolution is prohibitive for large spatial dimensions—\(O((HW)^{2}N_{h}))\)
In the proposed AA-UNet, the attention-augmented convolution is computed on the activation maps after the third and the final downsampling block of the U-Net (Fig. 2). The number of activation channels is 128 at that downsampling stage, and the image dimension is \(32\times 32\). In the current realization of the integration, for the attention-augmented convolution kernel size of three has been used, four attention heads, depth of values—\(d_{v}\) is 4 and depth of queries and key—\(d_{k}\) is 40. The resulting attention activation feature maps are then concatenated with the regular convolution attention maps from the last downsampling block. The concatenated attention maps are then passed through the U-Net’s upsampling path.
The infection class in COVID-19 data is generally under represented as compared to the background class especially in early stages of the disease. This leads to a large class imbalance problem. As found in several studies, ground-glass opacities generally precede consolidations lesions. This progression of the lesion development in COVID-19 leads to the another scenario of class imbalance. In some patients, only one of the lesions is largely present and the second lesion is highly under-represented (less than 10% of the total infection labels. This also leads to a second category of class imbalance. To address all of these class-imbalance issues, especially present in COVID-19 lesion segmentation scenarios, use of inverse class-weighted cross-entropy loss has been proposed. The weights are computed to be inversely proportional to the square root of class frequency. Given a sample with class label y, this inverse class-weighted cross-entropy loss can be expressed as
with C being the total number of classes and \({\varvec{z}}\) the output from the model for all classes. The weighting factor
is determined with help of the inverse square root of the number of samples in each label class to address the problem of training from imbalanced data. The training and validation sets also have different distributions, hence we have computed the inverse weighting separately for the train and validation sets. We have also used learning rate finder [27] to find the optimal learning rate, and a 1 cycle learning rate policy scheduler, where the maximum learning rate was also determined using the learning rate finder.
4 Experimental setup and results
The publicly available COVID-19 CT segmentation dataset [22] was used for analysis. 100 axial CT images from different COVID-19 patients was considered. This first collection of data was from the Italian Society of Medical and Interventional Radiology.
The second dataset of axial volumetric CTs of nine patients was used from Radiopaedia. This second dataset with whole volumes having both positive (373 positive) and negative slices (455 negative slices).
The experiments were performed with a threefold cross-validation on this combined dataset consisting of 471 two-dimensional axial lung CT images with segmentations for ground-glass opacities (GGO) and consolidation lesions. Each fold comprised data acquired from three different patients plus one third of images from the 100 slice CT stack taken from more than 40 different patients. The CT images were cropped and rescaled to a size of \(256\times 256\). During training, for data augmentation, random affine deformations were performed.
Training was performed for 500 epochs using the Adam optimizer and an initial learning rate of 0.002. Further, a cyclic learning rate was used, with an upper boundary of 0.005 and a class-weighted cross-entropy loss to address the problem of training from imbalanced data.
For the infection region experiments and multi-class labeling, we compared the proposed model with cutting-edge U-Net models: Oktay et al. [23].
The number of trainable parameter for the U-Net [23] is 611K. For the proposed variant AA-UNet, the U-Net incorporated with the attention-augmented convolution, the parameter count was 982K. The performance with larger bandwidth U-Net was also compared with increased number of trainable parameters—1070K parameters. Four widely adopted metrics were used, i.e., Dice similarity coefficient, sensitivity (Sen.), specificity (Spec.) and mean absolute error (MAE). If the final prediction is denoted as \(\mathbf {F_p}\) and the object-level segmentation ground truth as \({\mathbf {G}}\), then the mean absolute error which measures the pixel-wise error between final prediction and ground truth is defined as
A similar approach to Fan et al. has been adopted in this study [25] and the results of the proposed AA-UNet on detecting lung infections has been presented. The network was trained on multi-class lung infection (GGO and consolidation) and during evaluation, these multiple classes were combined into one infection label. The threefold cross-validation studies’ results have been presented in Table 1, which has been averaged over multiple runs that were conducted. The results from Fan et al. [25] and DDANet by Rajamani et al. [28] were also included from our earlier work, in each of these experiments. It has to be noted that Inf-Net was only trained with the first dataset which is smaller (100 axial slices) and Semi-Inf-Net was trained with pseudolabels from unlabeled CT images. DDANet [28] is improvisation of another attention-based approach (Criss-Cross-Attention [29]) to improve the performance of segmentation, and we include the results from CCA and DDANet (in bolditalics) as baseline. AA-UNet achieves better performance than Criss-Cross-Attention (CCA) and almost similar performance like DDANet [28]. Figure. 3 captures the visual comparison of multi-class lung segmentation results, where the red and green labels indicate the GGO and Consolidation, respectively.
As captured in Table 1, the proposed AA-UNet achieves the best Dice scores in each of the folds. The best Dice score obtained is 0.809 and least mean absolute error (MAE) is 0.0189. The average infection segmentation performance of the network has been captured in the same Table 1. The proposed AA-UNet has the best infection segmentation performance in average with the average Dice score of 0.784). In terms of Dice, the proposed AA-UNet out-performs the cutting-edge U-Net model [23] by 1.05% on average infection segmentation.
The infection segmentation performance of AA-UNet on each of the patients has also been included in the supplementary materials. In each of the patients, with the exception of Patient-1, the proposed AA-UNet had the best Dice score and the minimum MAE. The average across all the patients is also captured in Table 2. The results from Fan et al. [25] as a baseline and results from DDANet by Rajamani et al. [28] has also been included. AA-UNet again achieves almost similar performance like DDANet [28]. In terms of Dice, AA-UNet method achieves the best competitive performance of 0.78 averaged across all the patients. It outperforms the baseline best U-Net model Dice by 4% on infection segmentation.
The fold-wise performance of AA-UNet on multi-class labeling has been included in the supplement section. The average multi-label segmentation performance of the network has been captured in Table 3. The obtained results have been compared with the results from Inf-Net by Fan et al. [25] and results from DDANet by Rajamani et al. [28]. The baseline U-Net [23] and proposed AA-UNet has far less trainable parameters at \((\mathbf {611K})\) and \((\mathbf {982K})\) as compared to \(\mathbf {33M}\) in Inf-Net [25]. AA-UNet again achieves almost similar performance like DDANet [28], and DDANet is a completely different attention mechanism which is more computationally intensive. The proposed AA-UNet has the best multi-label segmentation performance, also in average with the best Dice score of 0.723) for GGO lesions and best Dice score of 0.614) for consolidation lesions. The proposed AA-UNet has average best Dice score of 0.668 for detecting COVID-19 lesions. In terms of Dice, the proposed AA-UNet out-performs the cutting-edge U-Net model [23] by 4.22% on average multi-label segmentation. The trainable parameters in the proposed AA-UNet has been increased in this study in comparison to the original U-Net. Hence the results have been compared against U-Net with increased bandwidth and 1070K parameters.
The multi-label segmentation performance of AA-UNet on each of the patients has been included in the supplementary materials. In terms of Dice, AA-UNet method achieves the best competitive performance of 0.701 for GGO lesion and 0.693 for consolidation lesion averaged across all the patients. On average, the proposed AA-UNet outperforms the baseline best U-Net model Dice by 2.86% on GGO, 6.4% on consolidation and on average 4.06% on multi-label segmentation. The distribution of the GGO and consolidation lesions are not even among the different patient scans. Some patients had predominantly only GGO (Patient-8) while other patients had predominantly consolidation (Patient-3). This skew in distribution impacts the segmentation Dice scores significantly, when the lesions are minimally represented in the patients.
5 Discussion
Substantial research has been carried out on COVID-19 lesion segmentation and it still remains to be a very challenging problem. There are several challenges in obtaining accurate lesion segmentation, and one major challenge is the severe class-imbalance nature of the lesions. In the early stages of the disease, this class imbalance is very stark, and hence, it gets very difficult to get good segmentation in such contexts. Another particular challenge is when inter-class skewed representation of the lesions, as COVID-19 lesion segmentation is a multi-class segmentation problem. In some patients, severe skewed lesion classes representations were observed in this study, which is a challenge in getting good segmentation results. Even though a large corpus of research articles have been published, large public datasets is still very limited. This poses another challenge until recently, to develop compelling algorithms with superior performances. Steadily the number of publicly available COVID-19 datasets is increasing and this scenario could change quite dramatically in the future. This would then enable further research into more compelling algorithms to address this challenging problem.
The proposed attention-augmented U-Net (AA-UNet) is only one of the potential ways to integrate attention augmentation that are smarter, elegant and have better performance than earlier proposed U-Net. There are many research possibilities to make this even better. There is no requirement or limitation to integrate attention only at the bottleneck location as is being computed in this work, primarily to make it computationally tractable. The attention augmentation could be integrated smartly into more locations. One of the next research problems could be to explore what could be the optimal or minimal number of non-local attention that needs to be gathered to get the best results. It would also be interesting to establish theoretical upper and lower bounds for number of locations to get non-local attention and its impact on performance. This work opens up all these and more possible research directions and can be the trigger for more fundamental work on attention augmentation in semantic segmentation tasks.
6 Conclusion
In this paper, a novel adaptation to the U-Net module with attention augmentation has been proposed. This modified U-Net framework (AA-UNet) improves the segmentation of lesion regions in COVID-19 CT scans. The novel solution and smart combination of attention augmentation in the bottleneck of U-Net has shown to be a working combination yielding superior and promising results. It has immense potential in better aiding clinicians with state-of-art infection segmentation models. AA-UNet achieves better performance than Criss-Cross-Attention (CCA) and almost similar performance like DDANet [28]. For future studies, incorporating attention augmentation in other stages of the U-Net will be explored. Deforming attention augmentation is another potential direction of research that can be explored. are also parameters in attention augmentation like the number of attention heads, number of attention channels, which can dramatically impact the performance of the segmentation while also steeply increasing the number of trainable parameters and hence the GPU requirements. We will conduct further research to find better constellations of utilizing attention augmentation more effectively. The source-code and trained models will be made publicly available.
The infection segmentation performance of AA-UNet on each of the patients, multi-label segmentation performance of AA-UNet on each of the patients and average of multi-label segmentation performance across the patients have been captured in the supplementary materials. Our proposed AA-UNet has excellent Dice score with competitive performance of 0.78 and MAE of 0.014 for infection segmentation have been averaged across all patients.
The performance of AA-UNet on multi-class labeling has also been captured. The threefold cross-validation studies’ results has been presented in Table 5, which was averaged over multiple runs. The results from Fan et al. [25] has also been included in each of the experiments. As captured in Table 5, the proposed AA-UNet achieves the best Dice scores in each of the folds. The best Dice score achieved for GGO is 0.7793 and best Dice score for consolidation is 0.7348. The proposed model outperforms the cutting-edge U-Net model, in terms of Dice, by 0.86% in GGO lesion and 8.48% in consolidation lesion segmentation in average. The proposed attention-augmented U-Net is able to segment GGO and consolidation lesions far better than the state-of-art models or baseline U-Net models.
References
Shi, F., Wang, J., Shi, J., Wu, Z., Wang, Q., Tang, Z., He, K., Shi, Y., Shen, D.: Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 14, 4–15 (2020)
Dong, D., Tang, Z., Wang, S., et al.: The role of imaging in the detection and management of COVID-19: a review. IEEE Rev. Biomed. Eng. (2020). https://doi.org/10.1109/RBME.2020.2990959
Ulhaq, A., Khan, A., Gomes, D., Paul, M.: Computer vision For COVID-19 control: a survey. arXiv preprint (2020)
Li, K., Wu, X., Zhong, Y., Qin, W., Zhang, Z.: Diagnostic performance of CT and its key signs for COVID-19: a systematic review and meta-analysis. medRxiv (2020)
Naudé, W.: Artificial Intelligence against COVID-19: an early review. IZA Discussion Paper, No. 13110 (2020)
Nair, A., Rodrigues, J.C.L., et al.: A British Society of Thoracic Imaging statement: considerations in designing local imaging diagnostic algorithms for the COVID-19 pandemic. Clin. Radiol. 75(5), 329–334 (2020)
Burlacu, A., Crisan-Dabija, R., et al.: Curbing the AI-induced enthusiasm in diagnosing COVID-19 on chest X-Rays: the present and the near-future. medRxiv 2020.04.28.20082776
Nguyen, T.T.: Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions. arXiv (2020)
Li, L., Qin, L., Xu, Z., Yin, Y., Wang, X., Kong, B., Bai, J., Lu, Y., Fang, Z., Song, Q., Cao, K., Liu, D., Wang, G., Xu, Q., Fang, X., Zhang, S., Xia, J., Xia, J.: Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiol. 296(2), E65–E71 (2020). https://doi.org/10.1148/radiol.2020200905
Shan, F., Gao, Y., et al.: Lung infection quantification of COVID-19 in CT images with deep learning. arXiv (2020)
Qi, X., et al.: Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study, medRxiv (2020)
Chen, J., et al.: Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv (2020)
Zheng, C., et al.: Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv (2020)
Jin, S., et al.: AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv (2020)
Song, Y., et al.: Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv (2020)
Gozes, O., et al.: Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection and patient monitoring using deep learning CT image analysis. arXiv preprint arXiv:2003.05037 (2020)
Parmar, N., Ramachandran, P., Vaswani, A., Bello, I.: Stand-alone self-attention in vision models. In: Advances in Neural Information Processing Systems (2019)
Bello, I., Zoph, B., et al.: Attention augmented convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV) (2019)
Chen, Y., Kalantidis, Y., et al.: A\(^2\)-Nets: double attention networks, In: Advances in Neural Information Processing Systems, vol. 31 (2018)
Wang, X., Girshick, R.B., Gupta, A., He, K.: Non-local neural networks. In: IEEE Conference on Computer Vision and Pattern Recognition (2018)
Vaswani, A., Shazeer, N., Parmar, N.U., Jakob, J., Llion, G., Aidan, Kaiser, N., Polosukhin, I.: Attention is all you need. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (2017)
MedicalSegmentation.com, COVID-19 CT segmentation dataset. http://medicalsegmentation.com/covid19/
Schlemper, J., Oktay, O., Schaap, M., Heinrich, M., Kainz, B., Glocker, B., Rueckert, D.: Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019)
Oktay, O., Schlemper, J., et al.: Attention U-Net: learning where to look for the pancreas. CoRR abs arXiv:1804.03999 (2018)
Fan, D., et al.: Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging 39(8), 2626–2637 (2020)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing MICCAI (2015)
Smith, L.N.: Cyclical learning rates for training neural networks. In: IEEE winter conference on applications of computer vision (WACV) (2017)
Rajamani, K.T., Siebert, H., Heinrich, M.P.: Dynamic deformable attention (DDANet) for semantic segmentation. medRxiv 2020.08.25.20181834 (2020)
Huang, Z., Wang, X., Huang, L., Huang, C., Wei, Y., Liu, W. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV), CCNet: Criss-Cross Attention for Semantic Segmentation (2019)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rajamani, K.T., Rani, P., Siebert, H. et al. Attention-augmented U-Net (AA-U-Net) for semantic segmentation. SIViP 17, 981–989 (2023). https://doi.org/10.1007/s11760-022-02302-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02302-3