Abstract
Curcumin, a potent phytochemical derived from the spice element turmeric, has been identified as a herbal remedy decades ago and has displayed promise in the field of medicinal chemistry. However, multiple traits associated with curcumin, such as poor bioavailability and instability, limit its effectiveness to be accepted as a lead drug-like entity. Different reactive sites in its chemical structure have been identified to incorporate modifications as attempts to improving its efficacy. The diketo group present in the center of the structural scaffold has been touted as the group responsible for the instability of curcumin, and substituting it with a heterocyclic ring contributes to improved stability. In this study, four heterocyclic curcumin analogues, representing some broad groups of heterocyclic curcuminoids (isoxazole-, pyrazole-, N-phenyl pyrazole- and N-amido-pyrazole-based), have been synthesized by a simple one-pot synthesis and have been characterized by FTIR, 1H-NMR, 13C-NMR, DSC and LC–MS. To predict its potential anticancer efficacy, the compounds have been analyzed by computational studies via molecular docking for their regulatory role against three key proteins, namely GSK-3β—of which abnormal regulation and expression is associated with cancer; Bcl-2—an apoptosis regulator; and PR which is a key nuclear receptor involved in breast cancer development. One of the compounds, isoxazole-curcumin, has consistently indicated a better docking score than the other tested compounds as well as curcumin. Apart from docking, the compounds have also been profiled for their ADME properties as well as free energy binding calculations. Further, the in vitro cytotoxic evaluation of the analogues was carried out by SRB assay in breast cancer cell line (MCF7), out of which isoxazole-curcumin (IC50–3.97 µM) has displayed a sevenfold superior activity than curcumin (IC50–21.89 µM). In the collation of results, it can be suggested that isoxazole-curcumin behaves as a potential lead owing to its ability to be involved in a regulatory role with multiple significant cancer proteins and hence deserves further investigations in the development of small molecule-based anti-breast cancer agents.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The quest for antitumor compounds is an ongoing process that necessitates combinatorial chemistry and high-throughput screening for the identification of potent leads. Cancer, being a major health concern, has intrigued researchers to intensively investigate drugs targeting the various receptors in cell proliferation pathways. In the recent past, the pharmaceutical industry has shifted its focus to the repurposing of existing drug moieties and recognizing natural products as bioactive molecules (Demain and Vaishnav 2011). Natural product drug discovery constitutes the identification of bioactivity studies, structure–activity relationship studies for derivatives/analogues, along with the synchronized mechanism of action directed synthesis, isolation and characterization for rational drug design-based modifications (Dholwani et al. 2008). Leading the research of bioactive natural products at the forefront of this quest is curcumin, which is isolated from turmeric and made from the rhizomes of Curcuma longa L. turmeric, apart from being a primary spice in the South East Asian cuisine, has also had the repute of being a herbal remedy and has been mentioned in Ayurveda, Siddha, Unani and Traditional Chinese Medicine (Prasad S. 2011).
Curcumin was first isolated in 1815 and was identified as 1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadien-3,5-dione and is the major constituent of turmeric (Vogel and Pelletier 1815)(Goel et al. 2008). It is attributed to be the entity that contributes to the multiple activities displayed by turmeric such as antibacterial (Moghadamtousi 2014), anti-inflammatory (Menon and Sudheer 2007) (Chainani-Wu 2003), antithrombotic (Srivastava et al. 1985), antitumor (Ravindran et al. 2009), neuroprotective (Cole et al. 2007), etc. Apart from playing a role in multiple activities, curcumin indicates minimal toxicity even on the administration of doses as high as 10 g/day in humans (Aggarwal et al. 2003). With respect to anticancer activity, curcumin has an edge over other compounds because of its effectiveness in various types of cancers owing to its chemotherapeutic potential. Furthermore, it is also known to have a pleiotropic modulatory behavior for several molecular targets in turn regulating gene expressions that play key roles in cell proliferation pathways (Shishodia et al. 2007). These molecular targets range from inflammatory cytokines, growth factors, apoptosis-related proteins, cell cycle proteins, transcription factors, etc. (Shishodia 2013). Irrespective of the numerous positive traits exhibited by curcumin, a larger number of limitations hinder its ability to be exploited to its full potential. These limitations are usually enlisted as poor aqueous solubility, poor bioavailability and rapid elimination from the systemic circulation (Priyadarsini 2014). One of the approaches adopted to improve its pharmacological profile is an exhaustive structure activity relationship of the molecule so as to understand the contributing aspect of each reactive site in the skeletal structure of curcumin. This allows for the identification of the sites that are responsible for the instability of the molecule and retention of the sites which attribute to the superlative activity of the molecule to facilitate lead optimization. The privileged scaffold of curcumin can be categorically divided as four major reactive sites, namely the aryl side ring, the carbonyl chain/olefin bonds, the diketo moiety and the active methylene site (Fig. 1). Numerous analogues and derivatives have been designed targeting each of these sites to analyze the corresponding change in activity and stability of the molecule (Rodrigues et al. 2019) (Vyas et al. 2013). Multiple papers have indicated that the diketo moiety has contributed to the instability of curcumin and its rapid metabolism in vivo and hence must be targeted by incorporating scaffolds which can add to the potency of the compound. In a structure–activity relationship analysis of the β-diketone chain of curcumin, Reddy et al. postulated that the introduction of hydrazine to curcumin leads to derivatives wherein the central 1,3-diketo-enol is masked or made rigid, which in turn improves the antitumor activity (Reddy et al. 2013). Furthermore, Jankun et al. have also postulated by SAR studies that cyclizing the central part of the compound could lead to improved antiangiogenic and antitumor activity of curcumin (Jankun 2016). Heterocyclic moieties such as isoxazole and pyrazoles have had a great impact in the improvement of the therapeutic potential of natural products and act as ideal scaffolds for natural products in medicinal chemistry (Zhang et al. 2018) (Kumar et al. 2013) (Naim 2016) (Jain, et al. 2006). It is postulated that the β-diketone site is an ideal scaffold for the integration of the heterocyclic moiety and based on previously ordered studies isoxazole, pyrazole and N-phenyl pyrazole and N-amido-pyrazoles are among the most commonly synthesized heterocyclic curcuminoids that have been evaluated for a plethora of activities (Mishra et al. 2008) (Sahu 2012) (Ahsan et al. 2015) (Sahu et al. 2016) (Ahmed 2017) (Rodrigues et al. 2021). Such modifications allow for improved stability and bioavailability as compared to curcumin adding to the chance of retention of curcumin-like efficacy.
In this study, four representatives of heterocyclic curcumin analogues, i.e., isoxazole-, pyrazole-, N-phenyl pyrazole- and N-amido-pyrazole-based curcuminoids, have been identified and the four analogues have been synthesized via a simple and cost-effective one-pot synthesis protocol and were further characterized. As a predictive auxiliary study to understand the regulatory behavior of these compounds, the compounds were screened for their ADME profile and were docked in silico against major key proteins, i.e. GSK-3β, Bcl-2 and PR, which are actively involved in the cell proliferation pathway. To confirm the predictive model study, the compounds were further evaluated for their cytotoxic capabilities in vitro against human breast cancer cell line MCF 7 and were compared with curcumin to identify their potency.
Experimental section
Materials and methods
To facilitate the present work, analytical-grade chemicals were obtained from Aldrich and Merck and were used without any purification. The purity of the substrates and monitoring of the reaction was carried out by using thin-layer chromatography (TLC) on TLC silica gel 60 F254 aluminum sheets, Merck. The FTIR (Fourier-transform infrared) spectra were determined on FTIR Spectrometer 6300, JASCO-UK. For nuclear magnetic resonance (NMR) evaluation, all 1H NMR and 13C NMR data were recorded using a Bruker Ascend 400 MHz. To record the melting point of the synthesized compounds, differential scanning colorimetry (DSC) was recorded on DSC-60 plus (Shimadzu) for a temperature range of 20°–250 °C with 10 °C/min heating rate, and LC–MS spectra were recorded on Waters, Synapt G2 High Detection Mass spectrometer.
General procedure for the synthesis of the heterocyclic curcumin analogues
As depicted in Scheme 1, to a solution of curcumin (1 mmol, 1 eq.) and glacial acetic acid (10 ml), appropriate amounts of hydrazine derivatives (1 mmol, 1 eq.) were added and stirred at 90 °C for 12–14 h to enable the synthesis of products in good yields. The progress of the reaction was monitored by TLC. Once the reaction was complete, the product was extracted by a two solvent system—hexane and ethyl acetate. The extracted solution was passed through a bed of anhydrous sodium acetate, and the filtrate was stripped of the solvent in the rotor vapor. The extract so obtained after the evaporation was then subjected to ethanol recrystallization. The filtrate was then left to dry and then scraped through after a petroleum ether wash to give a resultant powder. All the synthesized derivatives were further characterized by FTIR, 1H NMR, 13C NMR, DSC and LC–MS for purity.
C1, Curcumin-pyrazole
4,4'-[(1E)-1H-pyrazole-3,5-diyldi-2,1-ethenediyl]bis[2-methoxy-phenol] Dark yellow-orange colored powder. To curcumin (1.2 mmol, 0. 441 g), hydrazine hydrate (1.5 mmol, 0.7 ml) was taken in a round-bottom flask in the solvent glacial acetic acid (7 ml). The mixture was continuously stirred under reflux conditions for 14 h at 85 °C in an oil bath. IR (Vmax, cm−1): 3560 (C–OH), 3020, 1660 (C=N), 1516 (C–C), 1010 (C–O–C). 1H-NMR (400 MHz, DMSO) δ ppm: d 3.88 (s, 6H, 2 OCH3), 6.67 (s, 1H, C4-H), 6.85–6.83 (d, 2H, J = 8 Hz), 6.99–6.97 (d, 2H, J = 7.6 Hz), 7.03–7.19 (m, 6H, Ar–H) 7.43–7.41 (d, 2H, J = 8 Hz, Ar–H). 13C NMR (400 MHz, DMSO)—173.3, 147.8, 146.9, 129.5, 128.3, 127.6, 120.0, 115.8, 115.6, 115.2, 109.5, 99.1, 55.5. M.p.—216 °C; LC–MS for C21H20O4N2—365.08.
C2, Curcumin-isoxazole
4-[2-[3-[2-(4-hydroxy-3-methoxyphenyl)ethenyl]-5-isoxazolyl]ethenyl]-2-methoxy-phenol]. Orange-brown colored powder. To curcumin (1 mmol, 0.368 g), hydroxylamine hydrochloride (1 mmol, 0.69 ml) was taken in a round-bottom flask in the solvent glacial acetic acid (10 ml). The mixture was continuously stirred under reflux conditions for 14 h at 85 °C in an oil bath. IR (Vmax, cm−1): 3550 (C–OH), 3035, 1597 (C=N), 1518 (C–C), 1090 (C–O–C), 1270 (C–O, of five-membered ring). 1H-NMR (400 MHz, DMSO) δ ppm: d 3.84 (s, 6H, OCH3), 6.38 (s, 1H, C4-H), 6.61–6.96 (m, 4H), 7.00–7.28 (m, 4H, Ar–H), 7.51–7.30 (d, 1H), 9.42–9.36 (2H). 13C NMR (400 MHz, DMSO) 168.8, 162.7, 148.6, 148.4, 148.3, 136.9, 135.2, 127.8, 127.4, 122.1, 121.7, 116.08, 116.02, 115.7, 113.1, 110.86, 110.81, 110.6, 98.3, 56.1. M.p.—182 °C; LC–MS for C21H19O5N—366.09.
C3, Curcumin-2,4DNPH
4,4′-((1E,1′E)-(1-(2,4-Dinitrophenyl)-1H-pyrazole-3,5-diyl)bis(ethene-2,1-diyl))bis(2-methoxy-phenol). Dark orange powder. A mixture of the curcumin compound (1 mmol, 0.368 g) and 2, 4-dintrophenylhydrazine (2 mmol, 0.396 ml) was taken in the presence of solvent glacial acetic acid (10 ml) in a round-bottom flask. The mixture was continuously stirred at reflux conditions for 12 h at 90 °C. IR (Vmax, cm−1): 3510 (C–OH), 3013, 1680 (C=N), 1521 (N–O for nitro group), 1260 (aromatic C=C), 1028 (C–O–C). 1H-NMR (400 MHz, DMSO) δ ppm: 3.79–3.84 (s, 6H, OCH3), 6.77–7.28 (m, 12H, Ar–H), 8.02–8.04 (d, 1 H, J = 8.8 Hz, Ar–H containing NO2), 8.688–8.665 (d, 1 H, J = 9.2 Hz, Ar–H containing NO2), 9.25–9.37 (s, 2H, OH). 13C NMR (400 MHz, DMSO)—56.24, 56.07, 102.07, 110.05, 110.91, 111.32, 115.98, 116.12, 121.35, 121.81, 123.66, 127.87, 128.41, 128.88, 130.22, 130.50, 133.03, 136.68, 137.05, 144.51, 145.46, 146.62, 147.66, 148.24, 148.28, 148.39, 153.99. M.p.—120 °C; LC–MS for C27H22O8 N4—531.05.
C4, Curcumin-semicarbazide
3,5-Bis[(E)-2-(4-hydroxy-3-methoxyphenyl)vinyl]-1H-pyrazole-1-carboxamide. Yellow powder. A mixture of the curcumin compound (1 mmol, 0.368 g) and semicarbazide hydrochloride (2 mmol, 0.396 ml) was taken in the presence of solvent glacial acetic acid (10 ml) in a round-bottom flask. The mixture was continuously stirred at reflux conditions for 12 h at 90 °C. IR (Vmax, cm−1): 3362 (C–OH), 3012, 1641 (C=N), 1520 (C–C), 1290 (aromatic C=C), 1100 (C–O–C). 1H-NMR (400 MHz, DMSO) δ ppm: 3.83 (s, 6H, OCH3), 6.62–7.38 (m, 11H, Ar–H), 9.112–9.235 (s, 2 H, OH), 12.814 (s, 1H, NH). 13C NMR (400 MHz, DMSO)—23.605, 38.883, 39.092, 39.301, 39.509, 39.718, 39.927, 40.135, 55.567, 99.186, 109.530, 115.257, 115.608, 115.861, 120.030, 127.602, 128.145, 129.524, 146.991, 147.894, 173.383. M.p.—210 °C; LC–MS for C22H21O5N3—409.02.
In silico analysis
ADME profiling
To predict and analyze the ADME (absorption, distribution, metabolism and excretion) characteristics of the synthesized compounds, the QikProp module by Schrӧdinger 2019-1 was used. Selected descriptors were analyzed to obtain an understanding of the solubility and absorption levels of the compounds. Descriptors like QPlogPo/w, QPlogS, QPPCaco, rule of 5 and % human oral absorption were calculated using this module.
Ligand preparation
The structures of the synthesized curcumin analogues were sketched via the 2DSketcher tool. For the ligand optimization, LigPrep tool was employed using OPLS 2005 force field. Default parameters of retaining specific chiralities and desalting were selected.
Protein optimization
Protein structure
X-ray crystal structure of the proteins GSK-3β (PDB ID: 4ACC), with resolution 2.21 Å; R-value: 0.191 (obs.)(Arfeen et al. 2015), Bcl-2 (PDB ID: 2W3L) with resolution 2.10 Å; R-value: 0.218 (obs.) (Ponnulakshmi 2020)(Murray 2019) and PR (PDB ID: 4OAR) with resolution 2.41 Å; R-value: 0.211 (obs.) (Acharya 2019) were retrieved from the Protein Data Bank (Research Collaboratory for Structural Bioinformatics (RCSB) (http://www.rcsb.org/pdb) (Berman 2000).
Protein preparation
In the Maestro interface of Schrödinger 2019-1, the ligand–protein complex was subjected to docking, wherein the proteins had to undergo prior pre-processing using the Protein Preparation Wizard Tool. At this step, optimization using ProtAssign was allotted, water that made fewer hydrogen bonds than the specified number was deleted, hydrogens were added, and heavy atoms were removed.
Receptor Grid Generation
Following protein preparation, a receptor grid was generated with the GLIDE module of Schrödinger keeping the van der Waals scaling factor as 1.00 and charge cutoff of 0.25 subjected to OPLS 2005 force field with default parameters and without any constraints. A cubic box with defined dimensions fixed around the centroid of the active site was generated for each protein.
Molecular docking protocol
The analogues were screened using extra precision (XP) mode flexible ligand docking which is implemented in GLIDE. Van der Waals scaling factor and partial charge cutoff were selected to be 0.80 and 0.15, respectively, for ligand atoms. The optimized ligands were utilized for this purpose. The best-docked pose with Glide score values was recorded for each ligand.
Free energy calculations
To calculate the free energy of the protein–ligand complex of the titular compounds, MM-GBSA (molecular mechanics energies combined with generalized Born and surface area continuum solvation) method was implemented via the prime module of Schrödinger’s molecular modeling package. Prime employs the VSGB 2.0 solvation model and the OPLS2005 force field to simulate these interactions (Li et al. 2011)(Shivakumar 2010).
Cytotoxic evaluation
Cell culture
The human breast cancer cell line (MCF 7) was obtained from National Centre of Cell Science, Pune, India. The cells were maintained at 37 °C in a humidified atmosphere (90%) containing 5% CO2 and then cultured in DMEM (Dulbecco's modified Eagle's medium) with 10% (v/v) FBS (fetal bovine serum), with 2% antibiotic–antimycotic solution. The cells were seeded in 96-well plates for 24 h and then incubated with different concentrations of the synthesized analogues.
Cell viability
The synthesized analogues were evaluated for their anticancer potential by the SRB (sulforhodamine B) assay method using the cancer cell line MCF7 to obtain their IC50 values. SRB assay is a highly sensitive in vitro cytotoxicity screening assay with an ability to detect densities as low as 1000–2000 cells/well (Skehan 1990). All the compounds were tested at concentrations ranging from 500 to 7.81 µM. The compounds were tested against curcumin and doxorubicin as control and model drug, respectively. The assay was executed in accordance with previously reported methods along with minor modifications (Houghton 2007)(Vichai and Kirtikara 2006). The cells were harvested with trypsin and seeded in 96-well plates in a concentration of 5000 cells/well. Simultaneously, the synthesized analogues were dissolved in DMSO, and different concentrations were added into the designated wells. On completion of 48 h, ice-cold solution of 100 µl of 10% TCA (trichloroacetic acid) was added per well and incubated for 1 h at 4 °C. Following this, the wells were washed three times with distilled water and air-dried. A 100 µl aliquot of the sulforhodamine B solution (0.057% w/v in 1% v/v acetic acid) was added to each well, and the plates were left for incubation in the dark for 30 min. On completion of the incubation period, the plates were rinsed thrice with 1% v/v acetic acid to remove any unbound dye. After sufficiently drying the plates at room temperature, the protein-bound dye was solubilized by adding 200 µl of 10 mM Tris base to each well. The absorbance was determined at 540 nm wavelength on a scanning multiplate reader (ELx800, BioTek Instruments Inc., Winooski, VT, USA), and the percentage cell viability was calculated using excel sheet and IC50 values were determined using GraphPad Prism and Origin.
Statistical analysis
All experiments were carried out in triplicates to assess reproducibility and robustness. Mean and standard deviation were computed. T test was performed to determine p-value and results, indicating p < 0.05 were deemed statistically significant.
Results and discussion
Chemistry
The preparation of the heterocyclic representatives of the four major groups of analogues, i.e., isoxazole-, pyrazole-, substituted phenyl pyrazole- and amido-pyrazole-based analogues of curcumin, is illustrated in accordance with Scheme 1. The diketone moiety of curcumin was converted into its isoxazole with hydroxylamine hydrochloride by refluxing in glacial acetic acid for 14 h. Pyrazole and substituted phenyl pyrazole derivatives were synthesized by refluxing curcumin with hydrazine hydrate and 2,4 dinitrophenyl hydrazine, respectively, in glacial acetic acid for 12–14 h. The amido-pyrazole derivative was achieved by refluxing curcumin with semicarbazide hydrochloride for 12 h. The completion of the reaction was monitored by TLC using mobile phase hexane/ethyl acetate (3:2). The impurities were isolated by passing the product through a bed of anhydrous sodium acetate to remove any insoluble salts, and further purification was carried out by ethanol recrystallization. The yield of the titular compounds was good, ranging from 67 to 78%, and the melting points as recorded by DSC are mentioned in Table 1. The structures of the synthesized curcumin analogues were confirmed by analytical and spectral studies (IR, 1H-NMR, 13C-NMR, DSC and LC–MS), and the synthesized compounds were in full accordance with the proposed structures. The spectral peaks and empirical data have been explicitly mentioned in the Supplementary Information.
All of the synthesized compounds have indicated the characteristic peak of the –OH group in the range of 3500–3650 cm−1 with respect to the FTIR data. The absorption bands in the 3010–3100 cm−1 and 1590–1680 cm−1 regions correspond to N–H stretching and bending peaks, respectively. The –NH group of curcumin-pyrazole (C1) was observed at 3020 cm−1. For curcumin-isoxazole (C2), a shift in the peak was observed to 1597 cm−1 from curcumin’s peak at 1627 cm−1, which is indicative of the change of ketone group to an imine. Compound C3 indicates the additional peaks of 1521 cm−1 correspond to the nitro group. And curcumin-semicarbazide (C4) indicated the presence of its C=N groups at 3012 and 1641 cm−1. On analysis of the 1H NMR spectra, all compounds indicated the OCH3 peak between the ranges of δ 3.79–3.89 ppm. The aromatic protons could be attributed to δ 6.62–8.02 ppm. Sharp bands at δ 6.5–9.5 ppm attributed to the N–H proton. The OH proton could be accounted for at δ 9.11–9.37 ppm. At some places, the presence of the NH proton could be observed at δ 12.00–12.81 ppm. 13C NMR also accurately indicated the presence of the accounted carbons (such as C=O and CH3), and this could be reflected in the mass spectra recorded. The spectral data and melting points have been comparatively analyzed along with previous reports and seem to be in congruence with the same (Mishra et al. 2008)(Sahu et al. 2012)(Changtam et al. 2010).
In silico analysis
ADME studies
ADME profiling for the synthesized compounds was facilitated via the QikPrep module of Schrödinger. The selected descriptors, such as molecular weight, QPlogPo/w, QPlogS, QPPCaco, rule of 5 and % human oral absorption, were calculated and analyzed. The majority of the synthesized compounds have indicated ADME characteristics that lie within the recommended values and have obeyed the Lipinski rule of five. C3 or N-Phenyl pyrazole-curcumin has indicated poor predicative permeability and poor aqueous solubility. Also, isoxazole-curcumin (C2) has indicated the highest human oral absorption percentage of 93.862% (QikProp 2012) (Table 2).
Molecular docking
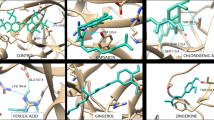
The binding sites of the proteins were located after the grid generation, and the ligands were docked in the pockets by extra precision ligand docking. For this study, each protein was individually evaluated by the three parameters, i.e. Dock score, XP score and Glide GScore, and the scores were compared to that of curcumin to identify which ligand posed an ideal fit. The selected proteins have been known to play key roles in the cancer proliferation pathway. GSK3β (glycogen synthase kinase 3-beta) has been linked to tumorigenesis and is a regulator of apoptosis in tumor cells and modulating centrosome function (Yoshino and Ishioka 2015). Studies have also indicated that inhibition of GSK3β could lead to the induction of apoptosis and restraining cell motility (Zeng et al. 2014). GSK is known to be phosphorylated by AKT, modulate NF-КB and interact with other crucial signaling pathways which are vital to cancer cells (Duda 2020). On comparing the scores generated, isoxazole-curcumin (C2) has indicated an overall better scoring than other compounds and curcumin. The synthesized compounds have also displayed interactions with key residues of GSK 3β and have interacted with 6 amino acids which take part in enzyme inhibitory activities (Fig. 2) (Table 3). Other studies have carried out a residue wise energy decomposition analysis and have enlisted interactions with Lys85 as an ideal selective inhibitor which was seen in the ligand–protein interaction in 3 (pyrazole-curcumin (C1), isoxazole-curcumin (C2) and N-phenyl pyrazole-curcumin (C3)) of the synthesized compounds (Arfeen et al. 2015)(Lescot 2005).
a GSK-3β protein (PDB ID—4ACC), b molecular surface structure of protein, c representation of docking of the ligand within suitably identified pocket, d ligand interaction of curcumin, e ligand interaction of pyrazole-curcumin (C1), f ligand interaction of isoxazole-curcumin (C2), g ligand interaction of N-phenyl pyrazole-curcumin (C3), h ligand interaction of N-amido pyrazole-curcumin (C4)
Bcl-2 (B cell lymphoma 2) family of proteins plays a central role in the regulation of cell death mechanisms such as apoptosis, necrosis and autophagy and work closely along the intrinsic mitochondrial pathway (Frenzel et al. 2009)(Pentimalli 2019). Bcl-2 protein expression plays a significant role in the prognosis of cancers, and since it has the ability to suppress cell death, it is identified as a noteworthy target for cancer drug discovery (Yip and Reed 2008). The ligands were docked in the site generated by the grid and the scores were recorded, based on which it was observed that all the synthesized analogues have indicated a better docking profile than curcumin. Interactions have been observed with 7 amino acids and 8 residues and some crucial residue interactions are Phe63 and Ala108 owing to Bcl-2 highly conservative amino acid sites (Xu et al. 2019). PR or progesterone receptor, belonging to the nuclear receptor family, is a receptor of paramount importance in breast cancer and is an important marker in breast cancer prognosis and subsequent hormone therapy. The overexpression of PR is indicative of the expression of estrogen and in turn plays a vital role in the understanding of the initiation and progression of breast cancer (Lim et al. 2016). The four synthesized compounds have indicated docking equivalent, if not better than curcumin and C2 or isoxazole-curcumin, has indicated the most overall score performance than the other compounds screened. The compounds have also indicated interactions with key residues such as Met759, Phe778 and Gln725 which has been enlisted in previous reports (Acharya et al. 2019) (Kiani 2006) (Table 2). In an overall analysis, it has been noted that most of the bonds formed between the ligands and the residues are H-bonds or pi-pi stacking and pi-cation bonds and a trend has been observed in which isoxazole-curcumin has indicated promising binding scores to the selected proteins which can be a predictive indication of its potential role as a promising inhibitor. Furthermore, the free energy of the binding of small ligands to the biological macromolecules was calculated by the MM-GBSA (molecular mechanics/generalized born and surface area) method via the Prime module. The study resulted in indicating that the binding energies of the tested compounds are comparable to that of curcumin and sometimes the identified ligands have more negative dG bind values which is indicative of a tendency for stronger binding and formation of a more stable ligand–protein complex (Basu Mallik et al. 2017).
Cytotoxicity evaluation
To investigate its cytotoxic potential, curcumin and its synthesized representative heterocyclic analogues were screened in vitro. Since the proteins selected for the molecular docking study play a vital role in the signaling pathways of breast cancer, a breast cancer cell line was chosen for this study. Breast cancer is one among the leading causes of cancer deaths in females, and constant efforts have been made by researchers in the quest for suitable drugs and therapies to treat breast cancer. MCF7 cell line is the most ubiquitously used xenograft model of breast cancer for in vitro studies (Comşa, Cîmpean 2015). MCF7 cells are ideal for research as they retain several characteristics to the mammary epithelium and are considered to be the first hormone responding breast cancer cell line (Camarillo 2015). The SRB assay was the chosen assay for cytotoxicity screening because of its high sensitivity, efficiency, reproducibility and better signal-to-noise ratio (Houghton et al. 2007). The average IC50 (µM) values are presented for three independent experiments in Table 4.
IC50 value of 0.0652 µM and are in correspondence with previously reported studies (Zaidi et al. 2011) (Poma 2007). Among the four synthesized compounds, two compounds (i.e., isoxazole-curcumin, C2, and N-phenyl pyrazole-curcumin, C3) have indicated a better activity than curcumin. However, even though the compound N-phenyl pyrazole-curcumin (C3) has specified a potent activity of 1.48 µM, it has indicated the formation of black precipitate on visual representation. This could be attributed to the non-polar nature of the compound, the presence of nitro groups and DMSO solubility (Popa-Burke and Russell 2014) (Ismail 2017). Additionally, on correlation with the ADME studies, N-phenyl pyrazole-curcumin (C3) has indicated a molecular weight of 530 which is higher than 500 and is usually considered to be a disadvantage for pharmaceutical development purposes (Bos and Meinardi 2000). Hence, N-phenyl pyrazole-curcumin (C3) has not been considered as a potent anticancer lead. Further, pyrazole-curcumin (C1) and N-amido-pyrazole-curcumin (C4) have indicated very high IC50 values (< 50 µM) and can be considered to be less potent than curcumin. This study is suggestive of the poor cytotoxic potency of pyrazole-curcumin and its derivatives when paralleled to curcumin.
The compound isoxazole-curcumin (C2) has indicated potential inhibitory activity and has displayed a sevenfold better IC50 value (isoxazole-curcumin (C2), IC50–3.97 µM) when compared to curcumin (curcumin, IC50–21.89 µM). A dose-dependent analytical study was carried out for curcumin, isoxazole-curcumin and doxorubicin ranging from 7.81 to 500 µM. Isoxazole-curcumin (C2) has indicated better activity when compared to curcumin at smaller doses and comparable activity in higher doses. The highest cell death of isoxazole-curcumin (C2) could be seen at 125 µM at 85.93%. The study has also revealed that isoxazole-curcumin displays cell death similar to that of doxorubicin over the different concentrations tested between 31.25 and 250 µM (Fig. 3).
This study hypothesized that incorporating a heterocyclic scaffold in the central part of curcumin’s structural skeleton could stabilize the otherwise unsteady nature of curcumin which is usually credited to the β-diketone moiety (Liang 2009)(Liao 2019). Previous studies have also postulated that apart from contributing to the improved stability and bioavailability of the compound it could also lead to better antitumor and antiangiogenic activity (Jankun et al. 2016). The selected synthesized compounds had incorporations of the heterocyclic moiety in the center of the compound and were screened for their ADME characteristics in silico, and most of the compounds indicated comparable activity with curcumin. In the collation of the ADME, molecular docking and in vitro analysis, isoxazole-curcumin has indicated consistent potency as an anticancer agent. The five-membered cyclic N–O analogue has previously indicated superior activities multiple times. A study was conducted to analyze the antioxidant potential of curcumin-isoxazole and curcumin-pyrazole, and it resulted in the findings that the two compounds indicated a better antioxidant potential than the model drug Trolox and the compounds showed better COX-1/COX-2 selectivity (Selvam et al. 2005). The superior antioxidant potential of these was further reiterated by other groups (Lozada-García 2017)(Shaikh 2019). A study by Narlawar et al. was carried out in which the improved neuroprotective activity by isoxazole-curcumin was attributed to the decreased rotational freedom and absence of stereoisomers, along with minimization of metal-chelation properties as compared to curcumin (Narlawar et al. 2008). In an antimycobacterial activity investigation, curcumin indicated a minimum inhibitory concentration of 100 μg/ml against M. tuberculosis H37Ra strain, whereas curcumin-isoxazole indicated a value of 1.56 μg/ml. the study further studied various other isoxazole analogues of curcumin and concluded that the presence of an isoxazole group and its nitrogen functionality improves the antimycobacterial activity by several fold (Changtam et al. 2010). In investigating the relevance of heterocyclic analogues in anticancer activity, the relevance of curcumin’s Michael acceptor properties and its ability to act as an anticancer agent in MCF7 and SKBR3 cell line and the study revealed that isoxazole-curcumin indicated a lower IC50 value than curcumin in MCF7 cancer cell line (Amolins and Peterson 2009). The mechanistic approach of the interactions between the thiol groups of cells and curcumin and its derivatives was done, and the study resulted in the finding that in HA22T/VGH cells, the IC50 of curcumin and curcumin-isoxazole was 17.4 ± 1.2 and 12.8 ± 1.5 μM, respectively, indicating isoxazole-curcumin’s better potency than curcumin (Labbozzetta 2009). In a comparative study carried out by Chakraborti et al., curcumin-isoxazole and curcumin-pyrazole not only indicated better anticancer activity in A549 cells but also indicated a stable IC50 value over the passage of time as compared to that of curcumin which increased gradually. This study is indicative of the stability of these analogues as compared to curcumin in the absence of serum (Chakraborti et al. 2013). A more relevant study to elucidate the effects of curcumin and curcumin-isoxazole on breast cancer cell line MCF7 and its multi-drug-resistant variant MCF7R was also carried out. The IC50 of curcumin was 29.3 ± 1.7 μM in MCF7 and 26.2 ± 1.6 μM in MCF7R, whereas the IC50 of curcumin-isoxazole was 13.1 ± 1.6 μM in MCF7 and 12.0 ± 2.0 μM in MCF7R. In this study, curcumin-isoxazole analogue has shown more potent antitumor and molecular activities both in parental and in MDR tumor cells (Poma et al. 2007).
Hence, the results of this study are in congruence with previously reported studies in which isoxazole-curcumin has shown better stability and growth inhibitory potential when screened for cytotoxicity. In summary, this study speculates the retention of curcumin's key residues of its skeletal structure contributing to curcumin's multiple potencies and modifying at the site associated with the instability of curcumin. On incorporation of such modifications, this study also displays the improvement of the efficacy of one such analogue of curcumin, i.e., isoxazole-curcumin, when tested for its potential as breast cancer agent owing to its cytotoxic potential in MCF 7 cell line. To the best of our knowledge, this is the first study establishing the role of isoxazole-curcumin as a potential modulator of key proteins such as GSK-3β, Bcl-2 and PR which are crucial for the breast cancer pathway cascade. Hence, to further establish this compound as a lead molecule, it must be evaluated by gene expression analysis which can then translate to in vivo studies.
Conclusion
In summary, a series of heterocyclic curcumin analogues mainly isoxazole-curcumin and pyrazole-curcumin and its variants were synthesized by a simple one-pot synthesis method. The structures of these compounds were verified by spectrophotometric routine evaluations. The titular compounds were analyzed for their pharmacokinetic profiles and their ability to bind to key proteins such as GSK-3β, Bcl-2 and PR which are actively involved in the various cascades of cancer proliferation pathways. The synthesized compounds have indicated ADME characteristics that lie within the recommended values. The in silico investigation consistently identified the potency of isoxazole-curcumin across the three proteins when analyzed for its docking scores and compared with curcumin. The study also revealed the ligand binding with major amino acids which could lead to potential regulatory and inhibitory activities when tested further. The compounds also indicated comparable free binding energy to curcumin indicative of strong and stable ligand–protein complex. Cytotoxic evaluation of the compounds has reflected the in vitro study analysis as isoxazole-curcumin (IC50–3.97 µM) indicated a promising sevenfold better activity than curcumin (IC50–21.89 µM) in breast cancer cell line MCF7. A dose-dependent evaluation has also established the efficacy of isoxazole-curcumin at lower doses and comparable activity with standard drug doxorubicin at higher concentrations. Thus, this study is indicative of the potency of isoxazole-curcumin, which can further act as a lead for the development of newer breast cancer agents or adjuvants with improved potency and selectivity.
References
Acharya R et al (2019) Structure Based Multitargeted Molecular Docking Analysis of Selected Furanocoumarins against Breast Cancer. Sci Rep 9(1):15743
Aggarwal BB, Kumar A, Bharti AC (2003) Anticancer Potential of Curcumin: Preclinical and Clinical Studies. Anticancer Res 23(1A):363–398
Ahmed M et al (2017) Azomethines, isoxazole, N-substituted pyrazoles and pyrimidine containing curcumin derivatives: urease inhibition and molecular modeling studies. Biochem Biophys Res Commun 490(2):434–440
Ahsan Nuzhat et al (2015) Curcumin Pyrazole and its derivative (N-(3-Nitrophenylpyrazole) curcumin inhibit aggregation, disrupt fibrils and modulate toxicity of wild type and Mutant a-Synuclein. Scientific Reports. https://doi.org/10.1038/srep09862
Amolins MW, Peterson LB, Blagg BSJ (2009) Synthesis and evaluation of electron-rich curcumin analogues. Bioorg Med Chem 17(1):360–367
Arfeen M, Patel R, Khan T, Bharatam PV (2015) Molecular Dynamics Simulation Studies of GSK-3β ATP Competitive Inhibitors: understanding the factors contributing to selectivity. J Biomol Struct Dyn 33(12):2578–2593
Berman HM (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242
Bos JD, Meinardi MMHM (2000) The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp Dermatol 9(3):165–169
Camarillo Ignacio G et al (2015) Electroporation-based therapies for cancer: from basics to clinical applications Low and High Voltage Electrochemotherapy for Breast Cancer: an in Vitro Model Study. Woodhead Publishing Limited. https://doi.org/10.1533/9781908818294.55
Chainani-Wu N (2003) Safety and Anti-Inflammatory Activity of Curcumin: A Component of Tumeric ( Curcuma Longa ). J Altern Complement Med 9(1):161–168
Chakraborti S et al (2013) Stable and potent analogs derived from the modification of the dicarbonyl moiety of curcumin. Biochemistry 52(42):7449–7460
Changtam C, Hongmanee P, Suksamrarn A (2010) Isoxazole analogs of curcuminoids with highly potent multidrug-resistant antimycobacterial activity. Eur J Med Chem 45(10):4446–4457
Cole GM, Teter B, Frautschy SA (2007) Neuroprotective effects of Curcumin. In: The molecular targets and therapeutic uses of curcumin in health and disease. Springer US, Boston, MA, pp 197–212
Comşa Ş, Cîmpean AM, Raica M (2015) The Story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res 35(6):3147–3154
Demain AL, Vaishnav P (2011) Natural products for cancer chemotherapy. Microb Biotechnol 4(6):687–699
Dholwani KK, Saluja AK, Gupta AR, Shah DR (2008) A review on plant-derived natural products and their analogs with anti-tumor activity. Indian J Pharmacol 40(2):49
Duda P et al (2020) Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 9(5):1110
Frenzel A, Grespi F, Chmelewskij W, Villunger A (2009) Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 14(4):584–596
Goel A, Kunnumakkara AB, Aggarwal BB (2008) Curcumin as ‘Curecumin’: from kitchen to clinic. Biochem Pharmacol 75(4):787–809
Houghton P et al (2007) The sulphorhodamine (SRB) assay and other approaches to testing plant extracts and derived compounds for activities related to reputed anticancer activity. Methods 42(4):377–387
Ismail Z, Ahmad A, Muhammad TST (2017) Phytochemical screening of in vitro Aglaonema simplex plantlet extracts as inducers of SR-B1 ligand expression. J Sustainability Sci Manag 12(2):34–44
Jain KS, Chitre TS, Miniyar PB, Kathiravan MK, Bendre VS, Veer VS, Shahane SR, Shishoo CJ (2006) Biological and medicinal significance of pyrimidines. Curr Sci 90(6):793–803
Jankun J et al (2016) Determining whether curcumin degradation/condensation is actually bioactivation (Review). Int J Mol Med 37(5):1151–1158
Jianing Li et al (2011) The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinform 79(10):2794–2812
Kiani J et al (2006) Estrogen receptor α-negative and progesterone receptor-positive breast cancer: lab error or real entity? Pathol Oncol Res 12(4):223–227
Kumar V, Kaur K, Gupta GK, Sharma AK (2013) Pyrazole containing natural products: synthetic preview and biological significance. Eur J Med Chem 69:735–753
Labbozzetta M et al (2009) Lack of nucleophilic addition in the isoxazole and pyrazole diketone modified analogs of curcumin; implications for their antitumor and chemosensitizing activities. Chem Biol Interact 181(1):29–36
Lescot E et al (2005) 3D-QSAR and docking studies of selective GSK-3β inhibitors. Comparison with a Thieno[2,3-b]pyrrolizinone Derivative, a New Potential Lead for GSK-3β Ligands. J Chem Inf Model 45(3):708–715
Liang G et al (2009) Synthesis, crystal structure and anti-inflammatory properties of curcumin analogues. Eur J Med Chem 44(2):915–919
Liao L et al (2019) Activation of anti-oxidant of curcumin pyrazole derivatives through preservation of mitochondria function and Nrf2 signaling pathway. Neurochem Int 125:82–90
Lim E, Palmieri C, Tilley WD (2016) Renewed interest in the progesterone receptor in breast cancer. Br J Cancer 115(8):909–911
Lozada-García M et al (2017) Synthesis of curcuminoids and evaluation of their cytotoxic and antioxidant properties. Molecules 22(4):633
Mallik B, Sanchari AP, Shenoy RR, Jayashree BS (2017) Novel flavonol analogues as potential inhibitors of JMJD3 histone demethylase: a study based on molecular modelling. J Mol Graph Model 72:81–87
Menon VP, Sudheer AR (2007) Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol 595:105–125
Mishra, Satyendra, Krishanpal Karmodiya, Avadhesha Surolia (2008) “Synthesis and Exploration of Novel Curcumin Analogues as Anti-Malarial Agents. 16: 2894–2902.
Murray JB et al (2019) Establishing drug discovery and identification of hit series for the anti-apoptotic proteins, Bcl-2 and Mcl-1. ACS Omega 4(5):8892–8906
Naim MohdJaved et al (2016) Current status of pyrazole and its biological activities. J Pharm Bioallied Sci 8(1):2
Narlawar R et al (2008) Curcumin-derived pyrazoles and isoxazoles: Swiss Army Knives or blunt tools for alzheimer’s disease? 3(1):1172
Pentimalli F et al (2019) Cell death pathologies: targeting death pathways and the immune system for cancer therapy. Genes Immun 20(7):539–554
Poma P et al (2007) The antitumor activities of curcumin and of its isoxazole analogue are not affected by multiple gene expression changes in an MDR model of the MCF-7 breast cancer cell line: analysis of the possible molecular basis. Int J Mol Med 20(3):329–335
Ponnulakshmi R et al (2020) Molecular docking analysis of Bcl-2 with phytocompounds. Bioinformation 16(6):468–473
Popa-Burke I, Russell J (2014) Compound precipitation in high-concentration DMSO solutions. J Biomol Screen 19(9):1302–1308
Prasad S, Aggarwal BB (2011) Turmeric, the Golden spice: from traditional medicine to modern medicine. In: Herbal medicine: biomolecular and clinical aspects, chap 13. CRC Press/Taylor & Francis
Priyadarsini KI (2014) The chemistry of curcumin: from extraction to therapeutic agent. Molecules 19(12):20091–20112
QikProp V (2012) Schrödinger, LLC, New York, NY
Reddy AR et al (2013) A comprehensive review on SAR of curcumin. Mini-Rev Med Chem. https://doi.org/10.2174/1389557511313120007
Ravindran J, Prasad S, Aggarwal BB (2009) Curcumin and Cancer Cells: How Many Ways Can Curry Kill Tumor Cells Selectively? AAPS J 11(3):495–510
Rodrigues FC, Anil Kumar NV, Thakur G (2019) Developments in the anticancer activity of structurally modified curcumin: an up-to-date review. Eur J Med Chem 177:76–104
Rodrigues FC, Anilkumar NV, Thakur G (2021) The potency of heterocyclic curcumin analogues: an evidence-based review. Pharmacol Res 166:105489
Sahu PK et al (2012) Synthesis and evaluation of antimicrobial activity of 4 H-pyrimido [2,1-b] benzothiazole, pyrazole and benzylidene derivatives of curcumin. Eur J Med Chem 54:366–378. https://doi.org/10.1016/j.ejmech.2012.05.020
Sahu PK, Sahu PK, Sahu PL, Agarwal DD (2016) Structure activity relationship, cytotoxicity and evaluation of antioxidant activity of curcumin derivatives. Bioorg Med Chem Lett 26(4):1342–1347. https://doi.org/10.1016/j.bmcl.2015.12.013
Selvam, C., Sanjay M. Jachak, Ramasamy Thilagavathi, and Asit. K. Chakraborti. 2005. “Design, Synthesis, Biological Evaluation and Molecular Docking of Curcumin Analogues as Antioxidant, Cyclooxygenase Inhibitory and Anti-Inflammatory Agents.” Bioorganic & Medicinal Chemistry Letters 15(7): 1793–97. https://www.sciencedirect.com/science/article/pii/S0960894X0500212X (April 18, 2018).
Shaikh SAM et al (2019) Unravelling the effect of β-diketo group modification on the antioxidant mechanism of curcumin derivatives: a combined experimental and DFT approach. J Mol Struct 1193:166–176
Shishodia S (2013) Molecular mechanisms of curcumin action: gene expression. BioFactors 39(1):37–55
Shishodia S, Singh T, Chaturvedi MM (2007) Modulation of transcription factors by curcumin. In: The molecular targets and therapeutic uses of curcumin in health and disease. Springer US, Boston, MA, pp 127–148
Shivakumar D et al (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6(5):1509–1519
Skehan P et al (1990) New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI J Nati Cancer Inst 82(13):1107–1112
Srivastava R, Dikshit M, Srimal RC, Dhawan BN (1985) Anti-thrombotic effect of curcumin. Thromb Res 40(3):413–417
Vichai V, Kirtikara K (2006) Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 1(3):1112–1116
Vogel HA, Pelletier J (1815) Curcumin-biological and medicinal properties. J Pharm 1:289
Vyas A, Dandawate P, Padhye S, Ahmad A, Sarkarb F (2013) Perspectives on new synthetic curcumin analogs and their potential anticancer properties. Current Pharmaceutical Design Pharm Des 19(11):2047–2069
Xu Zhijie et al (2019) Identification of Aloperine as an Anti-Apoptotic Bcl2 Protein Inhibitor in Glioma Cells. PeerJ 7:e7652
Yip KW, Reed JC (2008) Bcl-2 Family proteins and cancer. Oncogene 27(50):6398–6406
Yoshino Y, Ishioka C (2015) Inhibition of glycogen synthase kinase-3 beta induces apoptosis and mitotic catastrophe by disrupting centrosome regulation in cancer cells. Sci Rep 5(1):13249
Zaidi D, Singh N, Ahmad IZ, Sharma R, Balapure AK (2011) Antiproliferative Effects of Curcumin plus Centchroman in MCF-7 and MDA MB-231 Cells. Int J Pharm Pharm Sci 3(2):212–216
Zeng, Jing et al. (2014) GSK3β Overexpression Indicates Poor Prognosis and Its Inhibition Reduces Cell Proliferation and Survival of Non-Small Cell Lung Cancer Cells. In: Srikumar P. Chellappan (Ed.) PLoS ONE. 9(3): e91231
Zhang H-Z, Zhao Z-L, Zhou C-H (2018) Recent advance in oxazole-based medicinal chemistry. Eur J Med Chem 144:444–492
Zorofchian Moghadamtousi S et al (2014) A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Res Int 2014:1–12
Acknowledgements
The authors wish to thank the Department of Chemistry, MIT-Manipal, and the Department of Pharmacology, MCOPS-Manipal, for rendering their laboratory facilities.
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. One of the authors wishes to acknowledge the Dr. TMA Pai PhD Fellowship and KSTePS, DST, Govt. of Karnataka, for the fellowship and continuity of funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no conflicts of interest to declare.
Ethical approval
This article does not include any studies with animal or human subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rodrigues, F.C., Kumar, N.V.A., Hari, G. et al. The inhibitory potency of isoxazole-curcumin analogue for the management of breast cancer: A comparative in vitro and molecular modeling investigation. Chem. Pap. 75, 5995–6008 (2021). https://doi.org/10.1007/s11696-021-01775-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11696-021-01775-9