Abstract
Nicotinamide treatment of plants and plant cell cultures has been shown to promote defense and decrease levels of DNA methylation. In the present study, we used RNA-seq technology to study overall changes in gene expression induced in roots of 3-month-old spruce (Picea abies) seedlings grown from nicotinamide-treated seeds to examine the molecular mechanisms underlying the defense promotion. Approximately 350 genes were identified as differentially expressed in roots after the seed treatment. Stress response genes, including transcription factors MYB77 and LHY and two chitinase enzymes, were generally upregulated, whereas genes thought to be involved in epigenetic regulation such as DDM1, known to promote DNA methylation, were present at high frequency among the downregulated genes. Across all samples, the expression of downregulated epigenetic-related genes was highly correlated with the nicotinamide treatment, indicating a common regulation. Our results support an earlier hypothesis regarding a potential role of nicotinamide as a defense-signal mediator.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses limit productivity in agriculture and forestry. Plants must adapt to biotic and abiotic stresses to survive. Norway spruce (Picea abies L. Karst.) is a significant element of European ecology and the forestry industry. Newly planted seedlings often suffer from attacks by pests such as pine weevils. A considerable proportion of the pesticides used in forestry are intended to reduce insect attacks so that newly planted young seedlings survive, but many of these chemical agents have broad ecotoxicity. An alternative approach is to activate and strengthen the plants’ own innate defense systems.
Systemic acquired resistance (SAR) refers to the induction of a strengthened defense response in the whole plant (Luna et al. 2012; Conrath et al. 2015). Both priming and long-distance hormonal signaling are parts of SAR. Priming is a procedure where plants are induced to become more stress-tolerant via early exposure to a stressor or a defense-promoting signal molecule. Subsequent re-exposures lead to stress responses that are far stronger than the first response (Conrath et al. 2015). The plants can acquire a “memory” that protects them not only from the specific stress they have been exposed to, but also to other stresses. UV-C exposure of lettuce, for instance, results in a more salt-tolerant plant (Ouhibi et al. 2014). An advantage of the priming technique is that it can be applied to seeds (Jisha et al. 2013; Worrall et al. 2012).
Stress memory in plants lasts only for a short time period when it is mediated via proteins, RNAs and metabolites. However, stress-induced epigenetic changes, which are considered to be one result of seed priming, can be maintained for longer durations and may even be inherited trans-generationally (He and Li 2018). Epigenetic regulation in plants involves methylation of cytosine in the sequence contexts CG, CHG and CHH in the DNA, as well as histone modifications and other chromatin rearrangements (Bartels et al. 2018). It has been demonstrated that the stress-induced DNA methylome is correlated with simultaneous differential gene expression (Dowen et al. 2012; Zogli and Libault 2017). Establishment of an epigenetic memory in spruce has previously been discussed around the influence of temperature during embryogenesis (Carneros et al. 2017). Other important factors influencing the epitype include the geography and climate of origin, as demonstrated for transcript patterns and DNA methylation in plants of Populus (Raj et al. 2011) and Arabidopsis thaliana (Kawakatsu et al. 2016) from different locations. Environmental factors in the habitat of ancestral plants might also pre-adapt some plants to biotic and abiotic stresses via inherited epigenetic patterns to confer an evolutionary advantage (Luna et al. 2012; Bräutigam et al. 2013).
Nicotinamide (NIC), a form of vitamin B3 (niacin), is a metabolite found in all cells. It can be released from NAD in the reaction where the enzyme poly(ADP-ribose) polymerase (PARP) is involved in repairing single-strand DNA breaks, which can be caused by oxidative stress (Rissel and Peiter 2019). NAD-dependent histone deacetylases, using NAD as a substrate, also release NIC (Briggs et al. 2017). Furthermore, NIC can inhibit PARP and NAD-dependent histone deacetylases, creating feedback loops (North and Verdin 2004; Rissel and Peiter 2019). Since increased histone acetylation promotes more open chromatin, NIC-inhibited deacetylation thereby has an explicit epigenetic effect, increasing histone acetylation and the resulting opportunity for transcription to occur (Avalos et al. 2005).
NIC has been hypothesized to play a role in eukaryotic cells as a stress signaling molecule when it is released by PARP as a consequence of DNA strand breaks (Berglund 1994). Earlier studies showed that NIC levels increased in stressed plant tissues (Kalbin et al. 1997) and that multiple defense pathways were activated by NIC (Berglund et al. 1993a, 1993b, 2017). NIC has also been shown to decrease the level of DNA methylation in plant tissue cultures (Berglund et al. 2017). A hypomethylating effect was also seen in spruce plants grown from treated seeds (Berglund et al. 2016).
In plant cells, NIC is metabolized to nicotinic acid (NIA), which in turn is further metabolized to NAD or N-methyl nicotinic acid (trigonelline) (Noctor et al. 2006). NIC is a PARP inhibitor (on protein level), but NIA and trigonelline are not. Both NIC and NIA improved stress tolerance to the heavy metals Cd, Zn and Cu in Salix viminalis (Ohlsson et al. 2008) and protect against DNA damage and cell leakage caused by oxidative stress (Berglund et al. 2017).
To our knowledge, there has so far been no large-scale gene expression study of NIC-treated plants. We thus explored the underlying molecular mechanisms behind NIC-induced changes in genes involved in defense and DNA methylation to form a general picture of the potential importance of NIC seed treatment for inducing stress tolerance in plants. In other studies of interactions between pests/pathogens and plant defense or stress response systems, leaves and needles have been the primary focus, leaving roots relatively unexplored. Here we examined changes in transcript abundances in roots from 3-month-old Norway spruce seedlings using RNA-seq technology to elucidate any changes in gene expression induced by NIC seed treatment.
Materials and methods
Plant material and seed treatment
Norway spruce seeds, collected in the orchard of Öhn (origin 57°00' N, alt. 55 m), were treated in water with 2.5 mM NIC and 1 µM Tween 80 for 4 h and gentle shaking at 23 °C in the dark. Control seeds were similarly treated in water-Tween 80. Directly after treatment, seeds were sown in wet peat and kept at 23 °C in the dark until germination started after 1 week. Plants were grown at 23 °C with an 18 h light/6 h dark cycle under fluorescent lighting tubes (150 µmol m–2 s–1). After 1 month, the seedlings were transplanted in fresh peat. Seedlings were harvested 3 months after sowing, snap frozen in liquid nitrogen, then stored at −80 °C. Roots were ground into powder in liquid nitrogen before RNA extraction.
RNA extraction
RNA was extracted using the Qiagen RNeasy Plant Mini Kit and the manufacturer’s protocol. RNA integrity and quantity were measured using an Agilent 2100 Bioanalyzer. The limit for RNA integrity was RIN > 8.0.
Library construction and RNA-seq analysis
Three biological replicates were created for each of the two experimental conditions. Library preparation and RNA sequencing were carried out at National Genomics Infrastructure Sweden at SciLifeLab in Stockholm. Libraries were constructed from 200 ng total RNA each, using the Illumina TruSeq Stranded mRNA Library Preparation Kit according to the manufacturer’s instructions. Sequencing was performed with the Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) using a standard protocol with read lengths of 2 × 100 bases. The average sequencing depth was 26 million reads (Table S1). The mapping frequency was 53%. The sequences are publicly available at EBI ArrayExpress (accession number E-MTAB-11007).
Data analyses
RNA-seq data were mapped using a pseudoalignment algorithm, which produces robust, accurate, and fast results compared to other strategies (Bray et al. 2016), in the software Kallisto (Bray et al. 2016). Kallisto uses transcripts per million (TPM), a normalized value for each transcript, making it easy to compare different samples. Differentially expressed genes (DEG) were identified using a method designed for downstream analysis of the mapped data. The algorithm takes into account the alignment statistics. It is implemented in the R package sleuth (Pimentel et al. 2017). The threshold for differential expression was set to p < 0.01. The reference sequence for mapping was high confidence coding sequences from the P. abies genome v1.0 (Nystedt et al. 2013). An online platform developed by the Norway spruce genome project, congenie.org, was used to download genomic data for annotation and enrichment analysis (Sundell et al. 2015). For biological interpretation of the data, the enriched DEGs were annotated for biological processes using tools available at congenie.org and the complete genome available as a reference data set. The p-values were adjusted for multiple-testing and represent false discovery rates. Data for an epigenetic regulating subset of genes were compared to expression data for the same genes in P. abies in other experiments and the corresponding genes in A. thaliana and Populus trichocarpa. The data were retrieved from congenie.org, atgenie.org and popgenie.org, respectively (Sundell et al. 2015).
Validation with qRT-PCR
Quantitative reverse transcription PCR (qRT-PCR) was performed for all six sequenced samples. First strand cDNA was synthesized using Superscript III according to the manufacturer’s instructions. For the qRT-PCR analyses, stress response genes CHIT4, ETR1 and MYB77, which were identified as differentially expressed in the RNA-seq analysis, were selected as suitable for the experimental conditions used in the qRT-PCR. QRT-PCR primers were designed using the online Primer3 software (Untergasser et al. 2012), using default parameters except for a primer melting temperature of 60 °C and a product length of 100–150 bp. A list of the primer sequences is available in Table S2. qRT-PCR was performed in a C1000 Thermocycler (Bio-Rad) using the SYBR green kit (Bio-Rad) with a reaction volume of 10 µL. All reactions were carried out in triplicate, and a no-template control was run for each primer-pair. Actin (PaACT) was used as the reference gene (Yakovlev et al. 2006). Quantification was performed using the 2−ΔΔCq method (Livak and Schmittgen 2001).
Results
Differentially expressed genes are associated with stress and epigenetics
Six RNA-seq libraries were generated from roots of six spruce seedlings, three from NIC-treated seeds, three control plants water-treated seeds. In the comparison of the number of sequences in the NIC-treated samples with that of the control group using a stringent cut off, 357 genes were found to be differentially expressed; 98 were upregulated and 259 downregulated (Tables S3 and S4). In the comparison of transcript abundances of up- and downregulated genes using the median tpm (transcripts per million) found in control samples (Fig. S1), the median expression of all upregulated genes was 15 TPM and 8.7 TPM for all downregulated genes. Sequence data are publicly available at EBI ArrayExpress (accession number E-MTAB-11007).
The gene ontology (GO) enrichment analysis confirmed enrichment for stress-related genes among the upregulated subset and for genes connected to epigenetic regulation among the downregulated subset (Fig. S2 A and B). A subset of differentially expressed stress- and epigenetics-related genes was then selected for further characterization, described next.
Upregulated stress-response genes
When all upregulated genes were screened for biological functions in stress pathways (Table 1), involvement in both abiotic and biotic stress responses was found.
Myb transcription factors comprise one of the largest groups of transcription factors in plants. They contain a highly conserved DNA-binding domain and regulate numerous stress-related genes (Li et al. 2015). In this study, two myb genes were upregulated: LHY and MYB77 (MA_115536g0010, MA_89683g0010).
Gene expression of several genes involved in defense against biotic stress were induced. Chitinases play a role in pathogen defense by breaking down chitin in insect exoskeletons and fungal cell walls (Nunes and Philipps-Wiemann 2018). Two of the genes with increased transcript levels belong to the chitinase family (MA_102538g0010, MA_10427514g0010). Other upregulated pathogen defense response genes included LECRK44 (L-type lectin-domain containing receptor kinase IV.4, MA_76027g0010) and PSKR2 (phytosulfokine-alpha receptor 2, MA_65838g0010) (Loivamäki et al. 2010; Wang et al. 2014a). LECRK44, a kinase in the plasma membrane, has been reported to be involved in plant immunity (Wang et al. 2014b). PSKR2 is a receptor for phytosulfokine, a hormone involved in pathogen infection signaling (Zhang et al. 2018).
Hormone receptors COI1 and ETR1 were also upregulated. COI1 is a mediator of wound- and jasmonate-signaling and is therefore potentially important for defense mechanisms (Devoto et al. 2005). ETR1 (ethylene receptor 1) is involved in ethylene-induced abiotic and biotic stress pathways (O’Donnell et al. 1996; Penninckx et al. 1998).
Downregulated genes involved in epigenetic response
Twelve of the downregulated transcripts of genes are thought to be involved in epigenetic gene regulation, according to the Gene Ontology annotation (Table 2). The most recognized gene in this group is DDM1, which is an important chromosomal factor for DNA methylation (Alvarez et al. 2010).
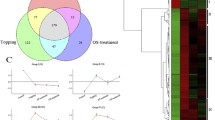
The correlation analysis of the set of downregulated genes with roles in epigenetic regulation revealed that their expression profiles were highly correlated (Fig. 1, Table 2). For example, ICU2 (INCURVATA2) (Hyun et al. 2013) has a Pearson correlation to DDM1 of r > 0.99. Moreover, the correlation of these genes was observed across conditions and tissues in other experiments in P. abies (Fig. S3, congenie.org). Furthermore, this correlation was found for A. thaliana and P. trichocarpa (Fig. S4, atgenie.org, popgenie.org).
Validation with qRT-PCR supports the methodology
The qRT-PCR was performed to validate the RNA-seq expression values for three differentially expressed genes, selected because of their strong relation to stress response (see section Upregulated stress response genes) The results of the comparison between the two methods are presented in Fig. S5. The Pearson’s r was 0.91 using the gene expression measurements for each sample, respectively (Fig. S5 A). Fig. S5 B displays the differences in log2 fold-changes for the tested genes after NIC treatment. This result shows that RNAseq and qRT-PCR produce concordant results.
Discussion
Long-term “memories” of stress signaling can be generated in plants through epigenetic changes to the DNA chromatin structure. In this study, we showed that treating Norway spruce seeds with NIC produces seedlings with a root gene expression profile showing several changes in expression levels of genes related to stress tolerance and epigenetics. Although analysis of epigenetic molecular hallmarks like histone modification and DNA methylation was beyond the scope of this investigation, our results indicate that NIC treatment of seeds may induce change at an epigenetic level, especially considering that our treatment at spruce embryo level (seed) is reflected in gene expression levels in months-old plants. Although further confirmation is needed, our results are in line with earlier studies that showed NIC can decrease global DNA methylation and influence an array of defense variables (Berglund et al. 1993a, 1993b, 2017). The transcriptome produced by NIC seed treatment may thereby predict a stronger defense against various kinds of stress.
The plants used in this study are not clones, since we wanted to use plant material that is typically used in forestry. Therefore, the individual plants may differ genetically to some degree. Still, our analyses reveal that NIC treatment resulted in similar responses in the samples tested, which may reveal a shared mechanism underlying changes in gene expression due to stress signals generated by NIC.
Expression of genes connected to epigenetics
DNA hypomethylation in plants is associated with various stresses such as cold, heat, bacteria, insects, salinity, and wounding (Alvarez et al. 2010; Santos et al. 2011; Lewandowska-Gnatowska et al. 2014). Logically, therefore, some genes involved in epigenetic regulation may have altered transcript abundances when stress signaling is activated. One of the downregulated genes in our study was DDM1, a key regulator of DNA methylation (Jeddeloh et al. 1999), which is required for DNA methylation of transposable elements (TEs); its loss results in strong activation of TEs (Lippman et al. 2004; Deniz et al. 2019). TEs are frequently found near genes that are upregulated by stress in maize and other plants (Makarevitch et al. 2015; Espinas et al. 2016).
In addition to DNA methylation, epigenetic changes also comprise histone modification (Ramirez-Prado et al. 2018). DDM1 also plays a role in histone methylation, which in turn could influence DNA methylation (Gendrel et al. 2002). Another gene affecting histone modification is ICU2, which plays a key role for the maintenance of repressive histone methylation (Hyun et al. 2013). Expression of ICU2 was downregulated here, as was the histone deacetylating HD2C (Histone deacetylase 2C). Downregulation of ICU2 and HD2C may lead to a chromatin state that can be reformed toward a more open configuration at some locations, allowing upregulation of certain other genes. The function of NIC as inhibitor of NAD-dependent histone deacetylases (North and Verdin 2004; Bond et al. 2009) is another way for NIC to promote histone acetylation. In addition, three transcripts encoding for histones are downregulated (MA_213048g0010, MA_1527811g0010, MA_74555g0010), although the exact function of the corresponding proteins are still unknown.
Besides DNA methylation and histone modifications, the chromatin state is also stabilized by other interactions. One important protein in this regard is the histone-binding FAS1, which has been linked to transcriptional gene silencing. FAS1 is downregulated in the NIC-treated group, and its expression is highly correlated to that of DDM1 (r = 0.97). In A. thaliana, mutation of FAS1 leads to expression of TEs (Ono et al. 2006). FAS1 is a subunit of chromatin assembly factor 1 (CAF-1), which cooperates with ASF1 and PCNA to contribute to chromatin assembly during replication. ASF1 and PCNA were also downregulated in this experiment and displayed a high correlation to DDM1 (r = 0.98, 0.93). Downregulation of the chromatin assembly-activity of CAF-1 can lead to reprogramming of embryonic stem cells (Ishiuchi et al. 2015; Kaufman 2015), and PCNA affects epigenetic inheritance (Shibahara and Stillman 1999; Zhang et al. 2000). CAF-1-PCNA-mediated chromatin assembly is also linked to single-strand DNA breaks (Moggs et al. 2000). PARP interacts with PCNA, which may be one mode of recruitment of PCNA to sites of DNA damage (Frouin et al. 2003), but might also have other consequences for chromatin remodeling.
It can be difficult to reliably compare data obtained across multiple studies of gene expression profiles in different tissues, species, young and old plants, monocots and eudicots, all exposed to different experimental stimuli. Therefore, it is remarkable that despite the range of experimental diversity mentioned, it is clear that there is a co-variation across experiments and species between the set of downregulated genes with putative roles in epigenetic regulation that were identified in this study. High correlation and coexpression of transcripts are often indicative of a common regulating mechanism, sometimes involving motifs in the sequence of a gene or in its genomic location (Bucher 1999). In our case, NIC might have a similar kind of general epigenetic regulatory role.
Taken together, the gene expression pattern in response to NIC treatment leads to less-condensed chromatin, promoting transcription.
Expression of genes connected to stress signaling
Increased expression of certain transcription factors may confer a more-tolerant epitype in which stress adaptation is more quickly induced (López-Galiano et al. 2018). In the present study, after the NIC seed treatment, the stress-response-inducing MYB transcription factors LHY and MYB77 were upregulated in 3-month-old plants (Table 1). These transcription factors are involved in many stress-signaling pathways and can also be induced by the hormones abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), and ethylene (ET) in A. thaliana (Shin et al. 2007; Jaradat et al. 2013; Grundy et al. 2015). LHY regulates circadian rhythm and reactive oxygen species (ROS) homeostasis (Lai et al. 2012); maximal stress tolerance with optimization of resources is achieved by circadian control. MYB77 has a role in maintaining homeostasis upon stress exposure and in the control of auxin-mediated signaling (Shin et al. 2007; Jaradat et al. 2013).
Many of the upregulated genes are known to specifically take part in biotic stress resistance. Examples of upregulated pathogen attack response genes are CHIT4, PSKR2, BAH1 and BAG5 (Hietala et al. 2004; Doukhanina et al. 2006; Yaeno and Iba 2008; Loivamäki et al. 2010; Yaqoob et al. 2012) (Tables 1; S3). Chitinases can be induced both by ET and JA (Rakwal et al. 2004), and two chitinase transcripts were upregulated in the present experiment. The expression of the ethylene receptor ETR1 is also upregulated after NIC treatment, which suggests that the response to ET should be faster and/or greater. ET and JA cooperatively activate stress tolerance under pathogen attack, cold, salinity, drought, and heat (Cheng et al. 2013; Kazan 2015; Vos et al. 2015). Furthermore, expression of COI1 (MA_263909), required for the JA response, was upregulated in our study. Another indicator that the ET/JA pathway was promoted compared to the SA pathway is that the two SA- and disease-resistance-inducing genes PAD4 and EDS1 (Rietz et al. 2011) were downregulated. PAD4 and EDS1 are repressors of the ET/JA pathway in A. thaliana (Straus et al. 2010). PAD4 and EDS1 are also important mediators of oxidative stress signaling (Straus et al. 2010) as discussed further below. Other well-known genes involved in epigenetic processes and stress responses, such as MET1, CMT3, CMT2, ROS1 and DME (Xu et al. 2018), were not differentially expressed in our study, perhaps because the seedlings analyzed were not exposed to stress as such. Rather, the seeds were treated with NIC, a putative signaling compound normally formed as a result of oxidative stress, 3 months before the analysis of the seedling roots.
Our gene expression results indicate that the ET/JA pathway is induced, whereas the SA pathway is suppressed.
Expression of genes related to oxidative stress and defense
Inhibition of PARP by exogenous chemical treatment leads to increased plant growth and to reduced production of anthocyanins and other stress-responsive molecules (Schulz et al. 2012). Decreased expression of oxidative stress-related genes is also a result of PARP deficiency, as shown in A. thaliana (Vanderauwera et al. 2007). In those cases, when PARP activity is inhibited, the NAD pool will be stable or increase, thus promoting growth in both stressed and unstressed plants. On the other hand, the lack of defense response observed may be due to the lack of NIC release via PARP activity, since NIC is known to stimulate several defense responses (Berglund and Ohlsson 1995).
NIC also has a general role as an antioxidant, decreasing cellular ROS content. NIC has an attenuating effect on the ROS content in human cells (Choi et al. 2015; Kwak et al. 2015) and increases glutathione levels in plant cells (Berglund et al. 1993a, 1993b, 2017). PARP is also important for basal plant defense in A. thaliana (Briggs et al. 2017). In addition, the PARP inhibitor 3-aminobenzamide prevents the increase in phenylalanine ammonia-lyase (PAL) activity caused by oxidative-stress-generating compounds (Berglund et al. 1996). The transcription factor LHY, which was upregulated after NIC seed treatments in our study, is also important for oxidative stress tolerance (Lai et al. 2012). Glutathione peroxidases (GPX) are involved in the antioxidant system and upregulated in response to various stresses (Rodriguez Milla et al. 2003). GPX6 (MA_125131g0010) was upregulated in our experiment as was OXS3 (oxidative stress 3, MA_9367190g0010), known for its ability to increase tolerance to oxidative stress. OXS3 may operate as a chromatin remodelling factor during stress responses (Blanvillain et al. 2009). ROS has an important influence on epigenetic modifications, resulting in a more open chromatin, and on stress responses (Kumar et al. 2020), but the mechanisms behind these effects are largely unknown.
An important ROS-induced process in woody plants is lignification (Barros et al. 2015), and genes encoding enzymes in the lignin biosynthesis pathway (e.g., cinnamyl alcohol dehydrogenase, dirigent-like protein and peroxidase) were downregulated, which may be linked to reduced ROS levels after the NIC seed treatment in our study.
Finally, there is evidence that NIC protects cells against DNA damage in yeast (Rössl et al. 2016), human (Surjana et al. 2010) and plant cells (Berglund et al. 2017), and we found that several DNA repair genes were downregulated in this experiment (e.g., BRCA1), which may imply that NIC reduces DNA damage. The ability of NIC to decrease oxidative stress may also partly explain the increased metal tolerance reported previously (Ohlsson et al. 2008).
The changes in gene expression in response to NIC treatment suggests that NIC has an antioxidative effect.
Negative correlation between stress and epigenetic related gene expression
The expression of several of the stress-related genes discussed in the present study was strongly inversely correlated with downregulated genes that have putative roles in epigenetic regulation (Table S5). MYB77, ETR1, COI1, CHIT4, BAH1, and PSKR2 all had a Pearson’s r < –0.85 with DDM1 (MA_89683g0010). The two genes with the strongest inverse correlation (r = −0.97, −0.94) both encode membrane proteins involved in defense response. Thus, transcription of these genes might be tightly controlled by a set of potential epigenetic regulators, and/or they are affected by as yet-unidentified mechanisms, yielding an indirect relationship that is discoverable at the transcriptional level.
These data suggest that the transcription of many of the upregulated genes in this experiment is affected by a change in the abundance of putative epigenetic regulators via mechanisms that are yet to be determined.
No trade-off between growth and defense mechanisms induced by NIC treatment?
In experiments on the development of stress-tolerant phenotypes, there is often a trade-off between stress responses and growth processes. Stronger defense mechanisms come at the cost of overall growth and other function as resources are reallocated for the plant to survive the stress (Huot et al. 2014). However, a positive effect of DNA hypomethylation on disease resistance in A. thaliana without negatively affecting growth was recently demonstrated (Furci et al. 2019). Similarly, we did not observe any negative effects on the growth and development of the plants originating from NIC-treated seeds. All spruce seedlings had the same size and growth rate based on visual observations and in agreement with a previous study (Berglund et al. 2016). Thus, NIC may have induced a protective state that did not affect the growth rate in our experimental conditions, perhaps because there is no decrease in NAD since NIC is added exogenously and independent of PARP activity. Nevertheless, general metabolism may be slightly modified, altering plant characteristics. Thus, for each particular application, functional differences should be further explored.
The increased levels of NIC, achieved independently of PARP activity, may promote defense activation without lowering the NAD content and, consequently, without restricting growth.
Conclusions
NIC seed treatment induced changes in the transcription of approximately 350 genes in the roots of 3-month-old spruce seedlings compared to the water-treated control group. Many of the upregulated genes encoded important stress signaling molecules, which can quickly mediate a strong life-saving response in plants upon exposure to a threatening stress, while several of the downregulated genes are thought to be involved in epigenetic regulation. This study increases our knowledge about NIC-induced expression of genes involved in defense and epigenetic processes, in line with NIC as a stress-signal mediator.
Abbreviations
- ABA:
-
Abscisic acid
- DEG:
-
Differentially expressed genes
- ET:
-
Ethylene
- GO:
-
Gene ontology
- JA:
-
Jasmonic acid
- NIC:
-
Nicotinamide
- PARP:
-
Poly-ADP(ribose) polymerase
- qRT-PCR:
-
Quantitative reverse transcription PCR
- ROS:
-
Reactive oxygen species
- SA:
-
Salicylic acid
- SAR:
-
Systemic acquired resistance
- TE:
-
Transposable elements
References
Alvarez ME, Nota F, Cambiagno DA (2010) Epigenetic control of plant immunity. Mol Plant Pathol 11:563–576. https://doi.org/10.1111/j.1364-3703.2010.00621.x
Avalos JL, Bever KM, Wolberger C (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD + cosubstrate specificity of a Sir2 enzyme. Mol Cell 17:855–868. https://doi.org/10.1016/j.molcel.2005.02.022
Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115:1053–1074
Bartels A, Han Q, Nair P, Stacy L, Gaynier H, Mosley M, Huang Q, Pearson J, Hsieh TF, An YQ, Xiao W (2018) Dynamic DNA methylation in plant growth and development. Int J Mol Sci 19:2144. https://doi.org/10.3390/ijms19072144
Berglund T (1994) Nicotinamide, a missing link in the early stress response in eukaryotic cells: a hypothesis with special reference to oxidative stress in plants. FEBS Lett 351:145–149. https://doi.org/10.1016/0014-5793(94)00850-7
Berglund T, Ohlsson AB (1995) Defensive and secondary metabolism in plant tissue cultures, with special reference to nicotinamide, glutathione and oxidative stress. Plant Cell Tiss Organ Cult 43:137–145
Berglund T, Ohlsson AB, Rydstrom J (1993a) Nicotinamide increases glutathione and anthocyanin in tissue culture of Catharanthus roseus. J Plant Physiol 141:596–600. https://doi.org/10.1016/S0176-1617(11)80462-1
Berglund T, Ohlsson AB, Rydström J, Jordan B, Strid Å (1993b) Effect of nicotinamide on gene expression and glutathione levels in tissue cultures of Pisum sativum. J Plant Physiol 142:676–684. https://doi.org/10.1016/S0176-1617(11)80901-6
Berglund T, Kalbin G, Strid Å, Rydström J, Ohlsson AB (1996) UV-B- and oxidative stress-induced increase in nicotinamide and trigonelline and inhibition of defensive metabolism induction by poly(ADP-ribose)polymerase inhibitor in plant tissue. FEBS Lett 380:188–193. https://doi.org/10.1016/0014-5793(96)00027-0
Berglund T, Lindström A, Aghelpasand H, Stattin E, Ohlsson AB (2016) Protection of spruce seedlings against pine weevil attacks by treatment of seeds or seedlings with nicotinamide, nicotinic acid and jasmonic acid. Forestry 89:127–135. https://doi.org/10.1093/forestry/cpv040
Berglund T, Wallström A, Nguyen TV, Laurell C, Ohlsson AB (2017) Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol Biochem 118:551–560. https://doi.org/10.1016/j.plaphy.2017.07.023
Blanvillain R, Kim JH, Wu S, Lima A, Ow D (2009) OXIDATIVE STRESS 3 is a chromatin-associated factor involved in tolerance to heavy metals and oxidative stress. Plant J 57:654–665. https://doi.org/10.1111/j.1365-313X.2008.03717.x
Bond DM, Dennis ES, Pogson BJ, Finnegan EJ (2009) Histone acetylation, vernalization insensitive 3, flowering locus C, and the vernalization response. Mol Plant 2:724–737. https://doi.org/10.1093/mp/ssp021
Bräutigam K, Vining KJ, Lafon-Placette C, Fossdal CG, Mirouze M, Marcos JG, Fluch S, Fraga MF, Guevara MÁ, Abarca D, Johnsen Ø, Maury S, Strauss SH, Campbell MM, Rohde A, Diaz-Sala C, Cervera MT (2013) Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol 3:399–415. https://doi.org/10.1002/ece3.461
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotech 34:525–527. https://doi.org/10.1038/nbt.3519
Briggs AG, Adams-Phillips LC, Keppler BD, Zebell SG, Arend KC, Apfelbaum AA, Smith JA, Bent AF (2017) A transcriptomics approach uncovers novel roles for poly(ADP-ribosyl)ation in the basal defense response in Arabidopsis thaliana. PLoS ONE 12:e0190268. https://doi.org/10.1371/journal.pone.0190268
Bucher P (1999) Regulatory elements and expression profiles. Curr Opin Struct Biol 9:400–407. https://doi.org/10.1016/S0959-440X(99)80054-2
Carneros E, Yakovlev I, Viejo M, Olsen JE, Fossdal CG (2017) The epigenetic memory of temperature during embryogenesis modifies the expression of bud burst-related genes in Norway spruce epitypes. Planta 246:553–566. https://doi.org/10.1007/s00425-017-2713-9
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582. https://doi.org/10.1104/pp.113.221911
Choi HJ, Jang S, Hwang ES (2015) High-dose nicotinamide suppresses ROS generation and augments population xxpansion during CD8+ T cell Activation. Mol Cells 38:918–924. https://doi.org/10.14348/molcells.2015.0168
Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53:97–119. https://doi.org/10.1146/annurev-phyto-080614-120132
Deniz Ö, Frost JM, Branco MR (2019) Regulation of transposable elements by DNA modifications. Nat Rev Genet 20:417–431. https://doi.org/10.1038/s41576-019-0106-6
Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58:497–513. https://doi.org/10.1007/s11103-005-7306-5
Doukhanina EV, Chen S, van der Zalm E, Godzik A, Reed J, Dickman M (2006) Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J Biol Chem 281:18793–18801. https://doi.org/10.1074/jbc.M511794200
Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A 109:E2183–E2191. https://doi.org/10.1073/pnas.1209329109
Espinas NA, Saze H, Saijo Y (2016) Epigenetic control of defense signaling and priming in plants. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01201
Frouin I, Maga G, Denegri M, Riva F, Savio M, Spadari S, Prosperi E, Scovassi AI (2003) Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1: a dynamic exchange of partners. J Biol Chem 278:39265–39268. https://doi.org/10.1074/jbc.C300098200
Furci L, Jain R, Stassen J, Berkowitz O, Whelan J, Roquis D, Baillet V, Colot V, Johannes F, Ton J (2019) Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. Elife. https://doi.org/10.7554/eLife.40655.001
Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 80(297):1871–1873. https://doi.org/10.1126/science.1074950
Grundy J, Stoker C, Carré IA (2015) Circadian regulation of abiotic stress tolerance in plants. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00648
He YH, Li ZC (2018) Epigenetic environmental memories in plants: establishment, maintenance, and reprogramming. Trends Genet 34:856–866. https://doi.org/10.1016/j.tig.2018.07.006
Hietala AM, Kvaalen H, Schmidt A, Jøhnk N, Solheim H, Fossdal CG (2004) Temporal and spatial profiles of chitinase expression by Norway spruce in response to bark colonization by Heterobasidion annosum. Appl Environ Microbiol 70:3948–3953. https://doi.org/10.1128/AEM.70.7.3948-3953.2004
Huot B, Yao J, Montgomery BL, He SY (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7:1267–1287
Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y (2013) The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140:156–166. https://doi.org/10.1242/dev.084624
Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovíc A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas TM, Torres-Padilla ME (2015) Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22:662–671
Jaradat MR, Feurtado JA, Huang DQ, Lu YQ, Cutler AJ (2013) Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol 13:192. https://doi.org/10.1186/1471-2229-13-192
Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22:94–97
Jisha KC, Vijayakumari K, Puthur JT (2013) Seed priming for abiotic stress tolerance: an overview. Acta Physiol Plant 35:1381–1396. https://doi.org/10.1007/s11738-012-1186-5
Kalbin G, Ohlsson AB, Berglund T, Rydström J, Strid Å (1997) Ultraviolet-B-radiation-induced changes in nicotinamide and glutathione metabolism and gene expression in plants. Eur J Biochem 249:465–472. https://doi.org/10.1111/j.1432-1033.1997.00465.x
Kaufman PD (2015) Want reprogramming? Cut back on the chromatin assembly! Nat Struct Mol Biol 22:648–650
Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, Chen H, Dubin M, Lee C-R, Wang C, Bemm F, Becker C, O’Neil R, O’Malley RC, Quarless DX, Consortium The 1001 Genomes, Schork NJ, Weigel D, Nordborg M, Ecker JR (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana Accessions. Cell 166:492–505. https://doi.org/10.1016/j.cell.2016.06.044
Kazan K (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20:219–229. https://doi.org/10.1016/j.tplants.2015.02.001
Kumar RMS, Wang YB, Zhang XP, Cheng H, Sun LR, He SB, Hao FS (2020). Redox components: Key regulators of epigenetic modifications in plants. Int J Mol Sci 21: undefined. https://doi.org/10.3390/ijms21041419
Kwak JY, Ham HJ, Kim CM, Hwang ES (2015) Nicotinamide exerts antioxidative effects on senescent cells. Mol Cells 38:229–235. https://doi.org/10.14348/molcells.2015.2253
Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schipper JHM, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci 109:17129–17134. https://doi.org/10.1073/pnas.1209148109
Lewandowska-Gnatowska E, Polkowska-Kowalczyk L, Szczegielniak J, Barciszewska M, Barciszewski J, Muszynska G (2014) Is DNA methylation modulated by wounding-induced oxidative burst in maize? Plant Physiol Biochem 82:202–208. https://doi.org/10.1016/j.plaphy.2014.06.003
Li C, Ng CKY, Fan LM (2015) MYB transcription factors; active players in abiotic stress signaling. Environ Exp Bot 114:80–91. https://doi.org/10.1016/j.envexpbot.2014.06.014
Lippman Z, Gendrel AV, Black M, Vaughn MV, Dedhia N, Richard McCombie W, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martinssen R (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471–476. https://doi.org/10.1038/nature02651
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Loivamäki M, Stührwohldt N, Deeken R, Steffens B, Roitsch T, Hedrich R, Sauter M (2010) A role for PSK signaling in wounding and microbial interactions in Arabidopsis. Physiol Plant 139:348–357. https://doi.org/10.1111/j.1399-3054.2010.01371.x
López-Galiano MJ, González-Hernández AI, Crespo-Salvador O, Rausell C, Real MD, Escamilla M, Camaes G, García-Agustín P, González-Bosch C, García-Robles I (2018) Epigenetic regulation of the expression of WRKY75 transcription factor in response to biotic and abiotic stresses in Solanaceae plants. Plant Cell Rep 37:167–176. https://doi.org/10.1007/s00299-017-2219-8
Luna E, Bruce TJA, Roberts MR, Flors V, Ton J (2012) Next-generation systemic acquired resistance. Plant Physiol 158:844–853. https://doi.org/10.1104/pp.111.187468
Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11:e1004915. https://doi.org/10.1371/journal.pgen.1004915
Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G (2000) A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol 20:1206–1218. https://doi.org/10.1128/MCB.20.4.1206-1218.2000
Noctor G, Queval G, Gakière B (2006) NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57:1603–1620. https://doi.org/10.1093/jxb/erj202
North BJ, Verdin E (2004) Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 5:224. https://doi.org/10.1186/gb-2004-5-5-224
Nunes CS, Philipps-Wiemann P (2018) Chitinases. In: Enzymes in human and animal nutrition: Principles and perspectives. Elsevier Inc., pp 361–378
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hällman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Käller M, Luthman J, Lysholm F, Nittylä T, Olson Å, Rilakovic M, Ritland C, Rosselló JA, Sena J, Svensson T, Talavera-López C, Theißen G, Tuominen H, Vanneste K, Wu Z-Q, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bosquet J, Garcia Gil R, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Lee Thompson S, Van de Peer Y, Andersson B, Nilsson O, Ingvarsson PK, Lundeberg J, Jansson S (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. https://doi.org/10.1038/nature12211
O’Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274:1914–1917. https://doi.org/10.1126/science.274.5294.1914
Ohlsson AB, Landberg T, Berglund T, Greger M (2008) Increased metal tolerance in Salix by nicotinamide and nicotinic acid. Plant Physiol Biochem 46:655–664. https://doi.org/10.1016/j.plaphy.2008.04.004
Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, Kato M, Kakutani T, Scheid OM, Araki T, Ki S (2006) Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11:153–162. https://doi.org/10.1111/j.1365-2443.2006.00928.x
Ouhibi C, Attia H, Rebah F, Msilini M, Chebbi M, Aarrouf J, Urban L, Lachaal M (2014) Salt stress mitigation by seed priming with UV-C in lettuce plants: Growth, antioxidant activity and phenolic compounds. Plant Physiol Biochem 83:126–133. https://doi.org/10.1016/j.plaphy.2014.07.019
Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10:2103–2113
Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14:687–690. https://doi.org/10.1038/nmeth.4324
Raj S, Bräutigam K, Hamanishi ET, Wilkins O, Thomas BR, Schroeder W, Mansfield SD, Plant AL, Campbell MM (2011) Clone history shapes Populus drought responses. Proc Natl Acad Sci U S A 108:12521–12526. https://doi.org/10.1073/pnas.1103341108
Rakwal R, Yang G, Komatsu S (2004) Chitinase induced by jasmonic acid, methyl jasmonate, ethylene and protein phosphatase inhibitors in rice. Mol Biol Rep 31:113–119. https://doi.org/10.1023/B:MOLE.0000031407.18708.95
Ramirez-Prado JS, Piquerez SJM, Bendahmane A, Hirt H, Raynaud C, Benhamed M (2018) Modify the histone to win the battle: Chromatin dynamics in plant – pathogen Interactions. Front Plant Sci 9:355. https://doi.org/10.3389/fpls.2018.00355
Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Corina Vlot A, Feys BJ, Niefind K, Parker JE (2011) Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol 191:107–119. https://doi.org/10.1111/j.1469-8137.2011.03675.x
Rissel D, Peiter E (2019) Poly(ADP-ribose) polymerases in plants and their human counterparts: parallels and peculiarities. Int J Mol Sci 20:1–29. https://doi.org/10.3390/ijms20071638
Rodriguez Milla MA, Maurer A, Huete AR, Gustafson JP (2003) Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J 36:602–615. https://doi.org/10.1046/j.1365-313X.2003.01901.x
Rössl A, Bentley-DeSousa A, Tseng YC, Nwosu C, Downey M (2016) Nicotinamide suppresses the DNA damage sensitivity of Saccharomyces cerevisiae independently of sirtuin deacetylases. Genetics 204:569–579. https://doi.org/10.1534/genetics.116.193524
Santos AP, Ferreira L, Maroco J, Oliveira MM (2011) Abiotic stress and induced DNA hypomethylation cause interphase chromatin structural changes in rice rDNA loci. Cytogenet Genome Res 132:297–303. https://doi.org/10.1159/000322287
Schulz P, Neukermans J, van der Kelen K, Mühlenbock P, van Breusegem F, Noctor G, Teige M, Metziaff M, Hannah MA (2012) Chemical PARP inhibition enhances growth of Arabidopsis and reduces anthocyanin accumulation and the activation of stress protective mechanisms. PLoS ONE 7(5):e37287. https://doi.org/10.1371/journal.pone.0037287
Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575–585. https://doi.org/10.1016/S0092-8674(00)80661-3
Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19:2440–2453. https://doi.org/10.1105/tpc.107.050963
Straus MR, Rietz S, Van Themaat E VL, Bartsch M, Parker JE (2010) Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 62:628–640. https://doi.org/10.1111/j.1365-313X.2010.04178.x
Sundell D, Mannapperuma C, Netotea S, Delhomme N, Lin YC, Sjödin A, Van de Peer Y, Jansson S, Hvidsten TR, Street NR (2015) The plant genome integrative explorer resource: plantgenie.org. New Phytol 208:1149–1156. https://doi.org/10.1111/nph.13557
Surjana D, Halliday GM, Damian DL (2010) Role of nicotinamide in DNA damage, mutagenesis, and DNA repair. J Nucleic Acids 2010:1–13. https://doi.org/10.4061/2010/157591
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40:e115–e115. https://doi.org/10.1093/nar/gks596
Vanderauwera S, De Block M, Van De Steene N, Van De Cotte B, Metziaff M, Breusegem FV (2007) Silencing of Poly(ADP-Ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci United States Am 104:15150–15155. https://doi.org/10.1073/pnas.0706668104
Vos IA, Moritz L, Pieterse CMJ, Van Wees SCM (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front Plant Sci 6:639. https://doi.org/10.3389/fpls.2015.00639
Wang M, Qin LM, Xie C, Li W, Yuan JR, Kong LN, Yu WL, Xia GM, Liu SW (2014a) Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol 55:1354–1365. https://doi.org/10.1093/pcp/pcu059
Wang Y, Bouwmeester K, Beseh P, Shan W, Govers F (2014b) Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol Plant Microbe Interact 27:1390–1402. https://doi.org/10.1094/MPMI-06-14-0191-R
Worrall D, Holroyd GH, Moore JP, Glowacz M, Croft P, Taylor JE, Paul ND, Roberts MR (2012) Treating seeds with activators of plant defence generates long-lasting priming of resistance to pests and pathogens. New Phytol 193:770–778. https://doi.org/10.1111/j.1469-8137.2011.03987.x
Xu JD, Zhou SS, Gong XQ, Song Y, van Nocker S, Ma FW, Guan QM (2018) Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol J 16:672–687. https://doi.org/10.1111/pbi.12820
Yaeno T, Iba K (2008) BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol 148:1032–1041. https://doi.org/10.1104/pp.108.124529
Yakovlev IA, Fossdal CG, Johnsen Ø, Junttila O, Skrøppa T (2006) Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genet Genomes 2:39–52. https://doi.org/10.1007/s11295-005-0031-z
Yaqoob N, Yakovlev IA, Krokene P, Kvaalen H, Solheim H, Fossdal CG (2012) Defence-related gene expression in bark and sapwood of Norway spruce in response to Heterobasidion parviporum and methyl jasmonate. Physiol Mol Plant Pathol 77:10–16. https://doi.org/10.1016/j.pmpp.2011.11.001
Zhang Z, Shibahara K, Stillman B (2000) PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408:221–225. https://doi.org/10.1038/35041601
Zhang H, Hu ZJ, Lei C, Zhen CF, Wang J, Shao SJ, Li X, Xia XJ, Cai XZ, Zhou J, Zhou YH, Yu JQ, Foyer CH, Shi K (2018) A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 30:652–667
Zogli P, Libault M (2017) Plant response to biotic stress: Is there a common epigenetic response during plant-pathogenic and symbiotic interactions? Plant Sci 263:89–93. https://doi.org/10.1016/j.plantsci.2017.07.008
Acknowledgements
The authors acknowledge support from Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, the National Genomics Infrastructure funded by the Swedish Research Council, and the Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. They thank Docent Henrik Aspeborg for comments on the manuscript and Docent Lauren McKee for manuscript editing.
Funding
Open access funding provided by Royal Institute of Technology.
Author information
Authors and Affiliations
Contributions
T. B. and A. O. conceptualized and directed the project; C. L., T. B. and A. O. designed the experiments; C. L. performed the analysis and interpreted the data, and prepared figures, tables, and references; C. L. wrote the original draft, and all authors revised the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: The work was supported by ÅForsk (https://aforsk.com) [15-416], Stiftelsen Tornspiran (http://stiftelsentornspiran.se), Anna och Nils Håkanssons Stiftelse (http://www.annaochnilshakanssonsstiftelse.se), Helge Ax:son Johnsons stiftelse (http://haxsonj.se/www/) [770721-0204], and Magnus Bergvalls Stiftelse (http://www.magnbergvallsstiftelse.nu) [2014-00501].
The online version is available at http://www.springerlink.com.
Corresponding editor: Tao Xu.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Laurell, C., Berglund, T. & Ohlsson, A.B. Transcriptome analysis shows nicotinamide seed treatment alters expression of genes involved in defense and epigenetic processes in roots of seedlings of Picea abies. J. For. Res. 33, 1365–1375 (2022). https://doi.org/10.1007/s11676-021-01410-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-021-01410-z