Abstract
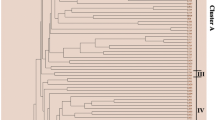
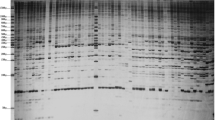
Chinese fir (Cunninghamia lanceolata (Lamb.) Hook) is one of the most important coniferous tree species used for timber production in China. Here, we conducted a sequence-related amplified polymorphism (SRAP) primer screening assay with a total of 594 primer combinations, using 22 forward and 27 reverse primers on four representative Chinese fir genotypes. The obtained results indicated that Chinese fir genomic DNA has a notable amplification bias on the employed forward or reverse primer nucleotides (3′ selection bases). Out of the tested primer sets, 35 primer combinations with clearly distinguished bands, stable amplification, and rich polymorphism were selected and identified as optimal primer sets. These optimal primer pairs gave a total of 379 scorable bands, including 265 polymorphic bands, with an average of 10.8 bands and 7.6 polymorphic bands per primer combination. The produced band number for each optimal primer set ranged from 7 to 14 with a percentage of polymorphic bands spanning from 33.3 to 100.0 %. These primer combinations could facilitate the next SRAP analysis assays in Chinese fir.


Similar content being viewed by others
References
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 27(4):617–631
Ahmad R, Potter D, Southwick SM (2004) Genotyping of peach and nectarine cultivars with SSR and SRAP molecular markers. J Am Soc Hortic Sci 129(2):204–210
Ai PF, Zhen ZJ, Jin ZZ (2011) Genetic diversity and relationships within sweet kernel apricot and related Armeniaca species based on sequence-related amplified polymorphism markers. Biochem Syst Ecol 39(4–6):694–699
Baloch FS, Kurt C, Arioğlu H, Ozkan H (2010) Assaying of diversity among soybean (Glycin max (L.) Merr.) and peanut (Arachis hypogaea L.) genotypes at DNA level. Turk J Agric For 34:285–301
Bracci T, Busconi M, Fogher C, Sebastiani L (2011) Molecular studies in olive (Olea europaea L.): overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep 30(4):449–462
Budak H, Shearman RC, Gaussoin RE, Dweikat I (2004) Application of sequence-related amplified polymorphism markers for characterization of turfgrass species. HortScience 39:955–958
Castonguay Y, Cloutier J, Bertrand A, Michaud R, Laberge S (2010) SRAP polymorphisms associated with superior freezing tolerance in alfalfa (Medicago sativa spp. sativa). Theor Appl Genet 120(8):1611–1619
Chen YQ, Ye BY, Zhu JM, Zhuang ZH, Pan DR, Chen RK (2001) Analysis of genetic relationship among Chinese fir (Cuninghamia lanceolata Hook) provenances by RAPD. Appl Environ Biol 7(2):l30–l133 (In Chinese)
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B Biol Sci 363(1491):557–572
Feng FJ, Chen MM, Zhang DD, Sui X, Han SJ (2009) Application of SRAP in the genetic diversity of Pinus koraiensis of different provenances. Afr J Biotechnol 8(6):1000–1008
Ferriol M, Picó B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107(2):271–282
Guo DL, Luo ZR (2006) Genetic relationships of some PCNA persimmons (Diospyros kaki Thunb.) from China and Japan revealed by SRAP analysis. Genet Resour Crop Evol 53:1597–1603
Han XY, Wang LS, Shu QY, Liu ZA, Xu SX, Tetsumura T (2008) Molecular characterization of tree peony germplasm using sequence-related amplified polymorphism markers. Biochem Genet 46:162–179
He ZX, Shi JS, Yin ZF, Chen XC, Yu RZ (2000) Detection of genetic markers associated with growth traits in Chinese fir. J Zhejiang For Coll 17(4):350–354 (In Chinese)
Huang HH, Xu LL, Tong ZK, Lin EP, Liu QP, Cheng LJ, Zhu MY (2012) De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom 13:648
Jing ZB, Wang XP, Cheng JM (2013) Analysis of genetic diversity among Chinese wild Vitis species revealed with SSR and SRAP markers. Genet Mol Res 12(2):1962–1973
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Li M, Shi J, Li F, Gan S (2007) Molecular characterization of elite genotypes with in a second-generation Chinese Fir (Cunninghamia lanceolata) breeding population using RAPD markers. Scientia Silvae Sinicae 43(12):50–55
Mishra MK, Nishani S, Jayarama (2011) Genetic relationship among indigenous coffee species from India using RAPD ISSR and SRAP markers. Biharean Biologist 5(1):17–24
Müller-Starck G, Liu YQ (1988) Genetics of Cuninghamia lanceolata HOOK. 1 genetic analysis. Silvae Genetica 37:236–243
Nei M, Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 79:5269–5273
Ouyang L, Chen JH, Zheng RH, Xu Y, Lin YF, Huang JH, Ye DQ, Fang YH, Shi JS (2014) Genetic diversity among the germplasm collections of the Chinese fir in 1st breeding population upon SSR markers. J Nanjing For Univ (Nat Sci Edn) 38(1):21–26 (In Chinese)
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9(1):6
Rana MK, Arora K, Singh S, Singh AK (2013) Multi-locus DNA fingerprinting and genetic diversity in jute (Corchorus spp.) based on sequence-related amplified polymorphism. J Plant Biochem Biotechnol 22(1):1–8
Shen AH, Li HB, Wang K, Ding HM, Zhang X, Fan L, Jiang B (2011) Sequence characterized amplified region (SCAR) markers-based rapid molecular typing and identification of Cunninghamia lanceolata. Afr J Biotechnol 10(82):19066–19074
Shi J, Zhen Y, Zheng RH (2010) Proteome profiling of early seed development in Cunninghamia lanceolata (Lamb.) Hook. J Exp Bot 61(9):2367–2381
Smutkupt S, Peyachoknagul S, Kowitwanich K, Onto S, Thanananta N, Julsrigival S, Kunkaew W, Punsupa V (2006) Varietal determination and genetic relationship analysis of highland legumes using SRAP markers. SABRAO J Breed Genet 38(1):19–27
Soleimani MH, Talebi M, Sayed-Tabatabaei BE (2012) Use of SRAP markers to assess genetic diversity and population structure of wild, cultivated, and ornamental pomegranates (Punica granatum L.) in different regions of Iran. Plant Syst Evol 298:1141–1149
Tong CF, Shi JS (2004) Constructing genetic linkage maps in Chinese fir using F1 progeny. Acta Genetica Sinica 31(10):1149–1156 (In Chinese)
Uzun A, Yesiloglu T, Aka-Kacar Y, Tuzcu O, Gulsen O (2009) Genetic diversity and relationships within Citrus and related genera based on sequence related amplified polymorphism markers (SRAPs). Sci Hortic 121:306–312
Valdez-Ojeda R, Hernandez-Stefanoni JL, Aguilar-Espinosa M, Rivera-Madrid R, Ortiz R, Quiros CF (2008) Assessing morphological and genetic variation in Annatto (Bixa orellana L.) by sequence-related amplified polymorphism and cluster analysis. HortScience 43(7):2013–2017
Walter C, Carson SD, Menzies MI, Richardson T, Carson M (1998) Review: application of biotechnology to forestry -molecular biology of conifers. World J Microbiol Biotechnol 14(3):321–330
Wang G, Gao Y, Yang L, Shi J (2007) Identification and analysis of differentially expressed genes in differentiating xylem of Chinese fir (Cunninghamia lanceolata) by suppression subtractive hybridization. Genome 50(12):1141–1155
Wang G, Gao Y, Wang J, Yang L, Song R, Li X, Shi J (2011) Overexpression of two cambium-abundant Chinese fir (Cunninghamia lanceolata) α-expansin genes ClEXPA1 and ClEXPA2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol J 9(4):486–502
Wang Z, Chen J, Liu W, Luo Z, Wang P, Zhang Y, Zheng R, Shi J (2013) Transcriptome characteristics and six alternative expressed genes positively correlated with the phase transition of annual cambial activities in Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook). PLoS One 8(8):e71562
Yang YL, Ma XQ, Zhang MQ (2009) Molecular polymorphic analysis for different geographic provenances of Chinese Fir. J Trop Subtrop Biol 17(2):183–189 (In Chinese)
Yeh FC, Shi J, Yang R, Hong J, Ye Z (1994) Genetic diversity and multilocus associations in Cunninghamia lanceolata (Lamb.) Hook from The People’s Republic of China. Theor Appl Genet 88:465–471
You Y, Hong JS (1998) Application of RAPD marker to genetic variation of Chinese fir provenances. Scientia Silvae Sinicae 34(4):32–38 (In Chinese)
Youssef M, James AC, Rivera-Madrid R, Ortiz R, Escobedo-GraciaMedrano RM (2011) Musa genetic diversity revealed by SRAP and AFLP. Mol Biotechnol 47(3):189–199
Zhao WG, Fang RJ, Pan YL, Yang YH, Chung JW, Chung IM, Park YJ (2009) Analysis of genetic relationships of mulberry (Morus L.) germplasm using sequence-related amplified polymorphism (SRAP) markers. Afr J Biotechnol 8(11):2604–2610
Acknowledgments
This research was supported by the National Natural Sciences Foundation of China (No. 31200506) and the Special Fund for Forest Scientific Research in the Public Welfare (No. 201404127).
Author information
Authors and Affiliations
Corresponding author
Additional information
Huiquan Zheng and Hongjing Duan contributed equally to this work.
The online version is available at http://www.springerlink.com.
Rights and permissions
About this article
Cite this article
Zheng, H., Duan, H., Hu, D. et al. Sequence-related amplified polymorphism primer screening on Chinese fir (Cunninghamia lanceolata (Lamb.) Hook). J. For. Res. 26, 101–106 (2015). https://doi.org/10.1007/s11676-015-0025-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-015-0025-0