Abstract
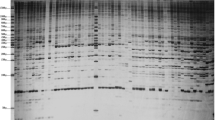
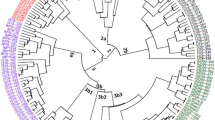
This study examined 63 tree peony specimens, consisting of 3 wild species and 63 cultivars, using sequence-related amplified polymorphism (SRAP) markers for the purpose of detecting genomic polymorphisms. Bulk DNA samples from each specimen were evaluated with 23 SRAP primer pairs. Among the 296 different amplicons, 262 were polymorphic. The maximum parsimony, neighbor-joining, and unweighted pair-group method using arithmetic average trees were largely in congruence. In the three trees, the wild species Paeonia ludlowii and P. delavayi formed separate clusters with strong bootstrap support, and P. ostii was closely related to all cultivars. The cultivars were divided into groups with various corresponding bootstrap values. The genetic similarity among the genotypes ranged from 0.02 to 0.73. These results demonstrate that SRAP markers are effective in detecting genomic polymorphisms in the tree peony and should be useful for linkage map construction and molecular marker assisted selection breeding.



Similar content being viewed by others
References
Budak H, Shearman RC, Parmaksiz I, Gaussoin RE, Riordan TP, Dweikat I (2004a) Molecular characterization of buffalograss germplasm using sequence-related amplified polymorphism markers. Theor Appl Genet 108:328–334
Budak H, Shearman RC, Parmaksiz I, Riordan TP, Dweikat I (2004b) Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor Appl Genet 109:280–288
Chen XM, Zheng GS, Zhang SW (2001) RAPD analysis of tree peony cultivars. Acta Hort Sin 28:370–372
Dice LR (1945) Measures of the amount of ecological association between species. Ecology 26:297–302
Ferriol M, Pico B, Nuze F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP) a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Li JJ (1999) Chinese tree peony and herbaceous peony. Chinese Forestry Press, Beijing, p 17
Li JJ (2005) Chinese tree peony (in English). China Forestry Publishing House, Beijing, pp 36–42
Liu P, Wang ZC, Shang F (2006a) AFLP analysis of genetic diversity of Paeonia suffruticosa cultivars in Henan Province. Acta Hort Sin 33:1369–1372
Liu YG, Teng YY, Pan C, Han YC, Zhou MQ, Hu ZL (2006b) Cluster analysis of Nelumbo based on SRAP markers. Amino Acids & Biotic Res 28:29–32
Mega K (1983) Tree peony and paeonia. In: Somei T (ed) NHK publisher, Japan, p 131
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Pei YL, Zou YP, Yin Z, Wang XQ, Zhang ZX, Hong DY (1995) Preliminary report of RAPD analysis in Paeonia suffruticosa subsp. spontanea and Paeonia rockii. Acta Phytotax Sin 33:350–356
Su X, Zhang H, Dong LN, Zhang JQ, Zhu XT, Sun K (2006) RAPD classification and identification of Paeonia rockii varieties planted in Gansu Province. Acta Bot Boreal–Occident Sin 26:696–701
Suo ZL, Zhou SL, Zhang HJ (2004) DNA molecular evidences of the inter-specific hybrids between Paeonia ostii and P. suffruticosa based on ISSR markers. Forest Res 17:700 –705
Suo ZL, Zhang HJ, Zhang ZM, Chen FF, Chen FH (2005) DNA molecular evidences of the inter-specific hybrids between Paeonia rockii and P. suffruticosa based on ISSR markers. Acta Bot Yunnan 27:42–48
Swofford DL (1998) PAUP, phylogenetic analysis using parsimony (and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Templeton AR (1983) Phylogenetic inference from restriction endoclease cleavagesite maps with particular reference to the evolution of human and apes. Evolution 37:221–224
Wang LS, Shiraishi A, Hashimoto F, Aoki N, Shimizu K, Sakata Y (2001) Analysis of petal anthocyanins to investigate flower coloration of Zhongyuan (Chinese) and Daikon Island (Japanese) tree peony cultivars. J Plant Res 114:33–43
Wang LS, Hashimoto F, Shiraishi A, Aoki N, Li JJ, Sakata Y (2004) Chemical taxonomy of the Xibei tree peony from China by floral pigmentation. J Plant Res 117:47–55
Zhao X, Zhou ZQ, Lin QB, Pan KY, Hong DY (2004) Molecular evidence for the interspecific relationships in Paeonia sect. Moutan: PCR-RFLP and sequence analysis of glycerol-3-phosphate acyltransferase (GPAT) gene. Acta Phytotax Sin 42:236–244
Zhou ZQ, Pan KY, Hong DY (2003) Advances in studies on relationships among wild tree peony species and the origin of cultivated tree peonies. Acta Hort Sin 30:751–757
Zou YP, Cai ML, Zhang ZX (1999a) The genetic diversity and protect strategy of Paeonia. Jishanensis. The Natural Sci Headway 9:468–472
Zou YP, Cai ML, Wang ZP (1999b) Systematic studies on Paeonia sect. Moutan DC. based on RAPD analysis. Acta Phytotax Sin 37:220–227
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program) (Grant No. 2006AA100109), the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2–YW-Z-064), and the Pilot Research Program of the Institute of Botany, CAS. The authors thank Prof. Hong-Jie Li for his kind help. The authors also thank Xia Tao and Cheng Li-Bao for their technical instruction. The authors also thank the members of the Physiology and Genetic Breeding of Ornamental Plants Research Group, IBCAS, for their kind help.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10528_2007_9140_MOESM1_ESM.xls
S1. Mean genetic similarity (Nei and Li 1979) among 66 tree peonies based on SRAP analysis. 1 ‘Fang Ji’, 2 ‘Wu Da Zhou’, 3 ‘Hua Jing’, 4 ‘Shou An Hong’, 5 ‘Hu Chuan Han’, 6 ‘Jin Yu Jiao Zhang’, 7 ‘Hua Wang’, 8 ‘Yao Huang’,9 ‘Jin Huang’, 10 ‘Yu Ban Bai’, 11 ‘Yu Pan Tuo Jin’, 12 ‘Feng Wei’, 13 ‘P. ludlowii’, 14 ‘P. delavayi’, 15 ‘Feng Dan’, 16 ‘Shu Hua Zi’, 17 ‘Chu Wu’, 18 ‘Zhao Fen’, 19 ‘Yin Bai He’, 20 ‘Yao Chi Jiu Nu’, 21 ‘Mu Ai’, 22 ‘Que Hao’, 23 ‘P. ostii’, 24 ‘Bai Wang Shi Zi’, 25 ‘Jing Yu’, 26 ‘Bai He Liang Chi’, 27 ‘Xue Zhong Song Tan’, 28 ‘Yu Ban Xiu Qiu’, 29 ‘Chun Hong Jiao Yan’, 30 ‘Zhong Chuan Fen’, 31 ‘Qing Chun’,32 ‘Hong Lian’, 33 ‘Han Ying Shi Zi’, 34 ‘Zui Xi Shi’, 35 ‘Shao Nu Qun’, 36 ‘Gui Fei Cha Cui’, 37 ‘Shan Hua Lan Man’, 38 ‘Fu Gui Hong’, 39 ‘Bai Yuan Chun’, 40 ‘Yu Yi Huang’, 41 ‘Huang Hua Kui’, 42 ‘Jin Dao1’, 43 ‘Tai Ping Hong Dan’, 44 ‘Tai Ping Hong Chong’, 45 ‘Jin Dao2’, 46 ‘Jin Zhi’, 47 ‘Ge Jin Zi’, 48 ‘Pan Zhong Qu Guo’, 49 ‘Jiu Zui Yang Fei’, 50 ‘Qi Bao Dian’, 51 ‘Guan Shi Mo Yu’, 52 ‘Ru Hua Si Yu’, 53 ‘Hu Lan’, 54 ‘Dao Da Chen’, 55 ‘Bai Yu Bing’, 56 ‘Hai Huang’, 57 ‘Jin Ge’, 58 ‘Zhu Sha Lei’, 59 ‘Yan Long Zi’, 60 ‘Huo Lian Jin Dan’, 61 ‘Hong Zhu Nu’, 62 ‘Jia Ge Jin Zi’, 63 ‘Zi Guang Ge’, 64 ‘Qing Luo’, 65 ‘Luo Yang Hong’, 66 ‘Tai Yang’. (XLS 36 kb)
10528_2007_9140_MOESM2_ESM.doc
S2. UPGMA dendrogram of genetic relationships among 66 tree peony genotypes, calculated on the basis of genetic similarity analysis by means of 23 SRAP primer pairs. Bootstrap values over 50 are indicated above the branch; based on 1,000 resamplings of the data set. (DOC 183 kb)
10528_2007_9140_MOESM3_ESM.doc
S3. A maximum parsimony tree (one of 1,934 trees) among 66 tree peony genotypes, calculated on PAUP 4.0b10 by means of 23 SRAP primer pairs. Bootstrap values over 50 are indicated above the branch; based on 1,000 resamplings of the data set. (DOC 181 kb)
Rights and permissions
About this article
Cite this article
Han, X.Y., Wang, L.S., Shu, Q.Y. et al. Molecular Characterization of Tree Peony Germplasm Using Sequence-Related Amplified Polymorphism Markers. Biochem Genet 46, 162–179 (2008). https://doi.org/10.1007/s10528-007-9140-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-007-9140-8