Abstract
Growing evidence reveals that microorganisms in the gut are linked to metabolic health and disease risk in human beings to a considerable extent. The focus of research at this stage must tend to focus on cause-and-effect studies. In addition to being a component of DNA and RNA, purine metabolites can be involved in purine signalling in the body as chemical messengers. Abnormalities in purinergic signalling may lead to neuropathy, rheumatic immune diseases, inflammation, tumors, and a wide range of other diseases. It has proved that gut microbes are involved in purinergic signalling. The relationship between these gut-derived purinergic signalling molecules and host metabolism may be one of the important clues to our understanding of the mechanisms by which the microbiota affects host metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, multidisciplinary studies in epidemiology, pathology, histology, cells, and animals have uncovered a growing body of evidence that microbes in the gut are linked to metabolic health and disease risk in human beings to a considerable extent. Over the past 15 years, numerous descriptive studies have provided ample evidence for the theory that there is a bidirectional mechanism of influence between the gut microbiota and the metabolic homeostasis of the host, and that disordered gut flora contributes to the development of a variety of common metabolic diseases, including obesity, type 2 diabetes, nonalcoholic liver disease, metabolic heart disease, and malnutrition. To further understand the mechanisms of how the gut microbiota affects host metabolism, it is important to stand focus on cause-and-effect studies [1, 2].
Besides as a component of DNA and RNA, purine metabolites can also act as a chemical messenger to be involved in purine signalling in the organism, which can be cross-linked with other metabolic networks to coordinate many physiological processes such as cell proliferation, differentiation, migration, and apoptosis. Abnormalities in purinergic signalling can also lead to neuropathy, rheumatic immune diseases, inflammation, tumors, and many other diseases [3]. Adenosine 5′-triphosphate (ATP) is the most widespread purinergic signalling molecule, and this purinergic system exists in microorganisms in addition to mammals. Mitochondria are ATP-generating organelles, the endosymbiotic origin of which was a key event in the evolution of eukaryotic cells [4]; phylogenetic evidence suggests that alphaproteobacterial may be the ancestor of mitochondria, and mitochondria themselves are considered as a kind of alphaproteobacterial [5], although some studies have suggested that mitochondria evolved from a proteobacterial lineage that branched off before the divergence of all sampled [6]. However, evidence of microbial involvement in the purine system does exist, thus showing that microorganisms are inextricably linked to the purine system.
It has been shown that intestinal microorganisms can release extracellular ATP (eATP) into the gut or metabolize it to produce adenosine-like derivatives. The eATP from intestinal flora is able to bind to P2X7 receptors on small intestinal Tfh cells to maintain host immune homeostasis [7], and the adenosine metabolites from intestinal microbes can participate in the immune regulation via A2A receptors in the gut [8, 9]. The relationship between these gut-derived purinergic signalling molecules and host metabolism may be one of the important clues to our understanding of the mechanisms in which the microbiota affects host metabolism.
This paper aims to summarize the evidence that gut microbes are involved in the purinergic system and to discuss the possible causal relationships. The combining purinergic systems have provided a new way to study the gut microbiota. The studies in the metabolism of the gut microbes and purinergic systems have demonstrated their respective roles in a variety of metabolic diseases, immune regulation, tumors, and microbial-derived purinergic signalling. This provides the potential to serve as a bridge linking the microbiota, purine metabolism, and various physiological phenomena or disease occurrence.
Purinergic system
Cellular metabolism involves a highly regulated series of sequential biochemical reactions designed to produce the necessary substrates for essential cellular processes. Purines are one of the most abundant metabolites in mammalian cells. In addition to the production of DNA and RNA molecules, purine nucleotides such as adenosine 5′-triphosphate (ATP) and guanosine 5′-triphosphate (GTP) are essential for the provision of cellular energy and intracellular signalling, respectively. Purines can also be incorporated into more complex biomolecules and act as cofactors, such as nicotinamide adenine dinucleotide (NAD) and coenzyme A. Purine metabolites are involved in energy transfer and nucleotide metabolism in the body, providing essential energy and cofactors for cells, and are important basic metabolic compounds in the body.
The metabolic environment in which the intracellular purine metabolites are interconverted together constitutes the cellular purine pool. Intracellular adenosine can be converted to AMP by phosphorylation and thus reused by the body, and AMP and other purine nucleotides (i.e., inosine monophosphate (IMP), xanthosine monophosphate (XMP), and guanosine monophosphate (GMP)) can be interconverted to form intracellularly, resulting in a cellular purine pool. In addition, bases present in the extracellular matrix can be translocated into the cell to produce the corresponding nucleotides.
Uric acid (UA) salts are the end product of intracellular purine metabolism. The catabolism of purine nucleotides takes place mainly in the liver, small intestine, and kidney. The accumulation of UA can result in hyperuricemia (HUA) and the deposition of monosodium urate (MSU) in the joints can cause gout, which is associated with dysregulation of purine catabolism [10]. Diets rich in purines (including seafood, meat, animal offal, and alcohol) can lead to excessive UA production [11]. The metabolism of alcohol and fructose consumes a large amount of ATP, which leads to the accumulation of AMP and thus accelerates the breakdown of nucleotides and increases the production of UA [12]. UA is produced through the oxidation of xanthine by xanthine oxidase; the use of xanthine oxidase inhibitors can alleviate this metabolic abnormality [13]. The kidneys and intestines are responsible for the UA excretion and about 25% of UA is excreted into the intestines [14]. Recent evidence suggests that the pathogenesis and progression of HUA are inextricably linked to gut microbes [15]. The intestinal bacteria such as Escherichia coli, Clostridium, and Pseudomonas can be involved in the metabolism of UA in the intestine [16, 17]. Enrichment of Bacteroides caccae, Bacteroides xylanisolvens, and other flora could be detected in the feces of gout patients, while there was a decrease in the abundance of genera such as Faecalibacterium prausnitzii and Bifidobacterium pseudoatenulatum [18]. Meanwhile, the transplantation of feces from HUA mice into normal mice resulted in an increase in UA levels in normal mice [19]. All of this evidence suggests a link between purine metabolism and gut microbes.
Possible targets of gut microbiota involved in purinergic signalling
In addition to direct involvement in purine metabolism, gut microbes can also participate in the regulation of metabolism in vivo through purinergic signalling. This regulatory effect is mainly mediated through purinergic receptors in the intestine and the release of extracellular ATP (eATP) and nucleoside metabolites from gut microbes, which is also influenced by extracellular nucleotidases.
Apart from participating in the synthesis of DNA and RNA, purine metabolites can also bind to purine receptors directly or as ligands for purinergic reactions, allowing purine metabolites to participate in cellular communication as a signalling molecule. Extracellular nucleotides (e.g., ATP, UTP, ADP, NAD) and their derived nucleosides (e.g., adenosine, inosine) are initiators and major components of purinergic reactions [20]. Purinergic signalling can regulate many aspects of cell behavior, such as proliferation, differentiation, migration, apoptosis, and other important cellular physiological processes, through synergistic interactions with other transmitter networks. Purinergic receptors on intestinal epithelial cells as well as immune cells inevitably have a complex connection with the gut microbiota. The microbial ecosystem is an important player in the treatment and intervention of inflammatory bowel disease and some autoimmune diseases, which makes the link between gut microbes and purinergic signalling potentially harboring important physiological implications.
Purinergic receptors are divided into two subfamilies: P1 receptors and P2 receptors. The P1 receptor is a G protein–coupled receptor, and adenosine can act as an endogenous ligand to bind to the P1 receptor and participate in physiological responses such as cardiac rhythm regulation [21]. The P1 receptor family has four main members: A1, A2A, A2B, and A3. A1 receptors are mainly expressed in the nervous system, while the other three subtypes are widely expressed in the nervous system, spleen, colon, testis, and other tissues [22].
The pathophysiological function of P1 receptors is extremely complex. A1 and A3 receptors can bind to Gi proteins and reduce intracellular cyclic AMP (cAMP) levels by inhibiting adenylate cyclase (AC) activity. In contrast, A2A and A2B bind to Gs proteins and activate AC to upregulate intracellular cAMP levels [23]. CD8+ T cells predominantly express A2A and A2B receptors. Activation of A2A receptors increases intracellular cAMP levels, inhibits Th1/Th2 differentiation, and increases intracellular cAMP levels, exerting an immunosuppressive effect during immune regulation [24]. In addition, A1 and A2A receptors (A2AR) can form A1-A2A complexes that bind to both Gi and Gs proteins and can exert opposite regulatory effects depending on the ligand concentration [25, 26]. The use of A2AR activators reduces symptoms in mice with colitis, which clarifies the link between A2AR and colitis [27]. Gut microbiota–mediated modulation of A2AR signalling alleviates DSS-induced intestinal inflammation model and simultaneously detects an increase in plasma inosine concentration in mice [28]. This suggests that the activation of A2AR may be a pathway to reduce the symptoms of colitis and that intestinal microbes are involved in this metabolic process.
The P2 nucleotide receptor family is divided into two subfamilies: P2X and P2Y [29]. P2 receptors can be activated by ATP and its analogs to regulate cellular metabolism and influence inflammatory and immune responses. Extracellular ATP can directly regulate T cell responses and induce intestinal inflammation by binding to P2X receptors [30]. Apart from the release of extracellular ATP from the damaged cells, intact cells also release ATP for the regulation of various immune cell functions such as the maturation of dendritic cells (DCs) and the activation of B and T cells under normal conditions [31]. P2 receptor has received a great deal of attention as a therapeutic target for colonic inflammation. It was found that P2RX1 knockout (P2rx1−/−) mice were less affected by DSS compared to WT mice. Meanwhile, the indole alkaloid biogenesis pathway was upregulated, and indole-producing microbes such as Clostridiaceae, Bacteroidaceae, Rikenellaceae, and Lachnospiraceae families were enriched in the intestine [32]. Indole alkaloids have been shown to induce regeneration of the mucosal barrier and to play a protective role in colitis [33]. Such results suggest that P2RX1 knockout–induced changes in gut microbiota could provide stronger gut protection. The cause of this phenomenon needs to be further confirmed.
Extracellular ATP can be catabolized by extracellular nucleotidases (e.g., CD39, CD73) to ADP and AMP, eventually generating adenosine (ADO) to transmit purinergic signals between cells. CD39 is the prototype of the ectonucleoside triphosphate diphosphate hydrolase (ENTPDase) family. ENTPDase1/CD39 can be expressed in immune cells and regulate the immune response by downregulating ATP levels. ENTPDase7 is the major ectonucleoside triphosphate diphosphate hydrolase in the intestine. The Entpd7 gene, which encodes ENTPDase7, is highly expressed in all types of intestinal epithelial cells (EC) in the small intestine. The expression of Entpd7 was not altered in the small intestine of mice treated with oral antibiotics, indicating that the microbiota did not directly regulate the expression of Enptd7 [34].
ENTPDase8 is highly expressed in large intestinal epithelial cells and can regulate eATP concentration in the large intestine [35]. In fecal microbiota of P2rx4−/− mice, the relative abundance of Bacteroidetes was greater than that in WT mice, while the relative abundance of Proteobacteria was lower [35]. These results suggest that P2X4R contributes to the maintenance of the gut microbial community. It has also been found that ENTPDase8 can similarly reduce intestinal inflammation by limiting P2Y6 receptor activation [36]. But additional studies on the P2Y6 receptor have found that P2Y6−/− mice are more susceptible to DSS-induced inflammation than WT mice, and P2Y6 deficiency can affect the secretion of a protective mucus layer in the gut or lead to abnormal activation of Th17/Th1 lymphocytes [37, 38]. This change may be related to the alteration of intestinal microbiota after P2 receptor deletion, and high-affinity IgA may maintain the balance of intestinal ecology by limiting the growth of certain bacteria. More studies are needed to clarify this overall regulatory effect.
Extracellular ATP is usually considered to be a pro-inflammatory signal, but extracellular adenosine produced by ATP/ADP/AMP degradation exhibits immunosuppressive effects. Thus, production by adenosine can often exert a regulatory effect on the eATP-induced inflammatory response [39]. Some evidence suggests that the gut microbiota can regulate the purinergic signalling by releasing eATP or by participating in nucleotide metabolism in which this connection may play an important role in purinergic signalling–mediated immunomodulatory effects [32, 40,41,42,43,44]. In the following, we will further discuss the possible mechanisms of this moderating effect.
Possible mechanisms of extracellular ATP from commensal bacteria involved in purinergic signalling in the intestine
The intestinal microbiota is involved in the formation of the intestinal mucosal barrier to maintain the homeostasis of the intestinal environment and can participate in purinergic signalling in the gut by releasing eATP to bind to P2X7 receptors in the intestine. This mechanism can maintain the homeostasis of microflora in the intestine through immunomodulatory effects. Several studies focusing on the P2X7 receptor (P2RX7) on T follicular helper (Tfh) cells in Peyer’s patches (PPs) have shown that the microbiota in the gut can release ATP via mechanosensitive channels and the eATP may be involved in purinergic signalling through the gut, affecting a range of inflammatory responses and immune regulation [45, 46].
Microbes in the gut can stimulate the development of gut-associated lymphoid tissue [47], Tfh cells in PPs, a subset of CD4+ T cells, promote germinal center (GC) responses and IgA release, thereby limiting the colonization of invasive bacteria and the entry of microbial-associated inflammatory factors from the intestinal lumen, constituting an effective mucosal defense mechanism in the intestine [48, 49]. High-affinity IgA protects the intestine by limiting bacterial growth [50], and the regulation of the high-affinity IgA response by T follicle–regulated (Tfr) cells promotes intestinal flora diversification and influences the composition of the intestinal flora [51]. It has been found that ATP released by bacteria can penetrate the intestinal epithelium and affect intestinal homeostasis by inducing apoptosis of the cells that regulate high-affinity secretory IgA in the intestine. High concentrations of ATP can be found in ileal and hepatic portal blood. Vancomycin/ampicillin/metronidazole (VAM) treatment leads to a significant increase in ATP concentrations in ileal and hepatic portal blood and induces massive apoptosis of the cells, a phenomenon that suggests that antibiotic treatment may harm the induction of this intestinal immunity [52]. In contrast, depletion of eATP in the intestine with adenosine triphosphate bisphosphatase (apyrase) enhances the sIgA response. In addition, the addition of apyrase to oral vaccine formulations against bacteria to counteract eATP in the intestine has been used to boost IgA titers in serum [52]. All these phenomena demonstrate the association of eATP with gut microbiota homeostasis.
The P2RX7 purinergic receptor is an ATP-gated cation channel to which high concentrations of extracellular ATP bind to activate the NLRP3 inflammasome pathway, ultimately leading to the maturation and release of the pro-inflammatory cytokines IL-1β and IL-18 [53, 54]. P2RX7 plays a key role in the innate immune response and regulation of the T cell population [55, 56]. In the intestines, the activation of P2RX7 downregulates the number of Tfh cells in PPs to promote host-microbiota symbiosis under homeostatic conditions [40], and drives Th1 cell differentiation and controls follicular helper T cell populations to prevent Plasmodium malaria [57]. Mice lacking P2X7 (P2rx7−/−) exhibited a significant increase in Tfh cells due to impaired P2XY receptor function, resulting in resistance to eATP-induced apoptosis. The alteration of Tfh cells in these mice resulted in an enhanced secretory IgA response [40]. This suggests that beyond its singular pro-inflammatory role, P2RX7 regulates the intestinal environment in a more complex manner.
Hierarchical clustering of mouse cecum microbiota revealed an increase of Lachnospiraceae and Helicobacteraceae in the intestine of P2rx7−/− mice [7]. In contrast, enrichment of Paraprevotellaceae and Caulobacteraceae occurred in wild-type mice [7]. The increase in Lachnospiraceae may be associated with obesity [58]. Many species of this genus have been shown to produce butyrate [59]. The occurrence of these microbiome changes may be due to the deficiency of P2RX7 [7]. This shows that blocked purinergic signalling can directly affect the composition of intestinal microbes, which demonstrates the complex link between purinergic signalling and intestinal microbes in the intestine. Lack of P2RX7 leads to an increase in PP Tfh cells, but not in the cellular abundance of Tfr. Dysregulation of Tfh cell activation may lead to a loss of controlled diversity of stimulatory bacteria that play an important role in the regulation of gut microbial homeostasis [51].
In addition, the intestinal microbiota can assist in the constitutive development of Th17 cells in the intestinal lamina propria (LP). The segmented filamentous bacteria (SFB) are among the most potent Th17 cell inducers [60]. SFB are a mucosa-associated symbiont in the intestine composed of a group of spore-forming Gram-positive bacteria. Close adhesion to small intestinal epithelial cells (EC) is a distinctive feature of SFB [61], and the colonization of SFB can trigger unique signalling pathways in the intestine to produce a favorable environment for Th17. There is evidence that microbiota-mediated Th17 cell development is independent of the major innate immune receptors [62]. SFB as a component of the gut microbiota can promote autoimmune arthritis by inducing PP Tfh cells, and SFB-containing mouse feces can exacerbate the inflammatory response in SPF K/BxN mice, but there is evidence that the immune response induced by SFB is not directly linked to the role of P2RX7 [63]. Notably, microbial-host interactions were also observed in P2RX7 deficiency concerning host-influenced microbiota. For example, it has been previously shown that P2RX7 deficiency triggers an enhanced PP Tfh cell response, leading to increased IgA production by B cells to inhibit SFB colonization [40].
Adenosine derivative in the intestine involved in immunomodulatory effects
Gut microbes can also regulate the activity of A2AR by affecting the level of inosine, thus participating in a complex immunomodulatory process. Inosine is formed from adenosine in a reaction catalyzed by adenosine deaminase in the intracellular and extracellular spaces. IMP is dephosphorylated intracellularly to inosine by 5′-nucleotidase and shunted to the extracellular space via the membrane nucleoside transporter. Inosine is degraded intracellularly by purine nucleoside phosphorylase to hypoxanthine and ribulose 1-phosphate. Hypoxanthine is subsequently converted to xanthine catalyzed by xanthine oxidase and eventually converted to UA [64].
In the past, inosine was generally thought to be an inactive breakdown product of adenosine, but it has since been shown that inosine can also bind to adenosine receptors and initiate intracellular signalling. Among the four AR subtypes, A2AR is the most effective in downregulating inflammation by regulating intracellular cAMP levels, but inosine is thought to be a functional agonist for A1R and A3R. Recent evidence has also supported the active effect of inosine against A2AR [65]. The theoretical basis for the belief that inosine is not an effective agonist of A2AR is mostly derived from in vitro experiments [66, 67]. In contrast, in experiments in vivo, inosine tends to exert A2AR-dependent anti-inflammatory and immunomodulatory effects [67]. With the advancement of gut microbial studies and probiotic-related studies, the link between the biological activity of inosine and microorganisms provides more perspectives on the biological significance of inosine.
Lactobacillus reuteri strain DSM 17,938 (LR) is a probiotic originally isolated from breast milk [68]. LR has been shown to have anti-inflammatory and immunomodulatory effects by inhibiting the Toll-like receptor 4-mediated NF-κB pathway [69]. In recent years, it has been found that its mechanism of action is related to purinergic signalling and inosine is a key effector molecule in Lactobacillus roxellanae therapy. Studies have found that LR can alleviate autoimmune diseases caused by defective regulatory T (Treg) cells by restoring inosine levels in the intestine [41, 70]. Forkhead box protein 3 (Foxp3) is a major transcription factor associated with T reg cell development and function. Mutations or deletions of the Foxp3 gene cause human IPEX syndrome (immune dysregulation, polyendocrinopathy, and enteropathy, with X-linked inheritance), an autoimmune disease associated with Treg deficiency. Scurfy (SF) mice with mutations in the Foxp3 gene can exhibit a similar clinical phenotype [71].
Treg deficiency led to an increase in the number of IFN-γ-producing CD4+ T cells (Th1) and IL-4-producing CD4+ T cells (Th2) in mouse spleen MLNs and significantly reduced the alpha diversity of the intestinal flora in SF mice, while the relative abundance of Lactobacillus was significantly reduced and the relative abundance of Bacteroides was significantly increased in the feces of SF mice [8].
In contrast, oral administration of LR reversely reduced the plasma levels of IFN-γ and IL-4 in SF mice and prolonged the survival time of SF mice. Meanwhile, the relative abundance of Lactobacillus, Oscillospira, and Firmicutes was increased while that of Tenericutes and Bacteroides was decreased in the feces of SF mice treated with LR [8], but the effects of these alterations in the metabolic process still need to be confirmed by more studies. The in vitro fermentation showed that LR could not metabolize inosine directly, but LR treatment can restore normal plasma levels of inosine, which may be related to the upregulation of expression of equilibrative nucleoside transporter 1 (ENT1) and concentrative nucleoside transporter 2 (CNT2) in the intestine, assisting the entry of inosine in the intestinal lumen into the intestinal epithelium [72, 73]; the altered gut microbiota composition may also have influenced this process, but the exact mechanism of this action needs to be identified by additional studies. Undeniably, A2A receptors (A2AR) play a key role in the process of remodeling the gut microbiota and suppressing autoimmune responses that occur due to Treg cell defects; the use of A2AR antagonists directly counteracts the immune-protective effects induced by LR [74].
Inosine has immunomodulatory effects on immune cells such as macrophages [64] and also functions as an immunosuppressive agent by binding to adenosine A2AR, thereby stimulating the release of intracellular cyclic adenosine monophosphate (cAMP) and inhibiting Th1/Th2 differentiation [8, 64]. Thus, the microbiota-inosine-A2AR axis can be an alternative approach to perform immunomodulatory functions similar to those of Treg cells when Treg cells are functionally deficient and unable to play a normal immunomodulatory role, which provides a potential pathway for the gut microbiota to adjust the immune function of the organism.
Besides, several intestinal bacteria were found to be associated with the efficacy of immune checkpoint blockade (ICB) [9, 44]. By using colorectal cancer (CRC) animal models to identify specific ICB-promoting bacteria, the researchers found that inosine produced by specific gut bacteria Bifidobacterium pseudolongum (B. pseudolongum) or Akkermansia muciniphila (A. muciniphila) and Lactobacillus johnsoni (L. johnsonii) can combine with adenosine A2AR to enhance the immunotherapeutic response and reduce the incidence and the size of inflammation-induced colon cancers in mice [9].
Oral supplementation of B. pseudolongum to germ-free mice with MC38 colon cancer enhanced the effect of immunotherapy blocked by CTLA-4 or PD-1, the process in which enhanced activation of Th1/Tc1 cells and their corresponding effector functions were observed. Further investigation of the mechanism of this phenomenon revealed that inosine produced by bacterial metabolism and A2AR on T cells are critical factors in this process. Compared with germ-free mice, mice treated with CTLA-4 blocker plus B. pseudolongum had increased serum inosine and its degradation factors xanthine and hypoxanthine, and inosine concentrations were highest in the duodenum and gradually decreased along the small and large intestine. This antitumor activity can be mimicked by the use of dibutyryl cAMP (Bucladesine, db-cAMP), which was depleted by either blocking A2AR or purging inosine [9].
A. muciniphila can modulate PD-1 blockade and produce inosine in vitro in an interleukin 12–dependent way by increasing the recruitment of CCR9+ CXCR3+ CD4+ T lymphocytes to the mice tumor bed [75]. The potentiation of ICB efficacy by A. muciniphila is also based on the A2AR expression of T cells [9, 75]. L. johnsonii promotes the antitumor efficacy of anti-CTLA-4, and its metabolite hypoxanthine also acts as a ligand binding to A2AR on the surface of T cells, though the ICB-promoting effect induced by L. johnsonii is relatively weak. This effect of enhanced immune activity is in contradiction with the function of A2AR in ordinary physiological conditions, which, as mentioned above, exerts immunosuppressive effects on inhibiting Th1 differentiation through the release of cAMP [9].
Novel immune checkpoint inhibitors based on the immunosuppressive effects of adenosine and A2AR signalling are also available, such as monoclonal antibodies (MAb) targeting CD73, CD39, and CD38, as well as pharmacological antagonists of A2AR, many of which are currently in clinical trials [76]. However, some studies have shown that inosine analogs can play a pro-inflammatory role, and A2AR can maintain Th1/anti-tumor immunity in mice [77,78,79].
This phenomenon may be related to the influence of the specific environment on which inosine is dependent when it is engaged in immune regulation.
In vitro experiments showed that inosine promotes Th1 differentiation of T cells when exogenous IFN-γ is present and, conversely, inhibits Th1 differentiation when IFN-γ is absent. cAMP is a downstream signalling molecule of A2AR. In the condition of blocking A2AR, db-cAMP can restore Th1 differentiation circumventing the requirements of inosine [9]. Moreover, even in the absence of exogenous IFN-γ, inosine could equally enhance Th1 differentiation when naïve T cells are triggered by anti-CD3/anti-CD28. Thus, the regulatory impact of the inosine-A2AR pathway on Th1 is affected by the specific microenvironment in which it is exposed, acting either as a promoter or as a suppressor [9].
The combination of inosine compounds coupled with CTLA-4 blockade alone was unable to exert antitumor activity in mice, while the addition of the immune adjuvant synthetic oligodeoxynucleotides that contain unmethylated CG dinucleotides (CpG ODN) resulted in the recurrence of the antitumor activity exhibited by B. pseudolongum plus CTLA-4 blockade exhibited reproducible antitumor activity, demonstrating the reliability of A2AR on the microenvironment [9]. Gut microbes are linked to the body’s immune system through complex and diverse pathways, and studies about immunomodulation through optimization of the gut microbiota remain to be deepened, and that these discoveries based on purinergic signalling may provide brand new insights for future studies.
Dietary and herbal interventions associated with gut microbial–mediated purinergic signalling
The effects of diet can also affect purine metabolism and purinergic signalling through the involvement of gut microbes. It has been found in past studies that inappropriate dietary habits can lead to disturbances in purine metabolism, often culminating in the accumulation of purine metabolites leading to diseases such as HUA and gout [10], accompanying changes in gut microbiology [80]. High fructose intake exacerbates purine degradation and intestinal ecological dysbiosis [81,82,83], and a high-purine diet can also lead to disturbances in the intestinal ecology [19, 84]. Dietary and herbal interventions can play a useful part in the early prevention or treatment of disease, and if their mechanisms of action are clarified, they can also advance the development of targeted drugs in the future.
There has been much evidence that natural products can alleviate HUA by modulating the gut microbiota, alleviating chronic inflammation, and promoting UA excretion [85], as well as inducing microbial-associated purinergic signalling to improve the symptoms of related diseases. Rhein is the main component of several traditional Chinese medicines, including Rhubarb, Aloe, and Sennae Folium. Rhein treatment alleviated DSS-induced colitis, altered gut microbiota composition, and increased Lactobacillus level. It also alters the purine metabolism level to restore the concentration of UA. As Rhein is unable to metabolize UA directly, this regulatory effect may be related to the enrichment of Lactobacillus [86]. Sunflower head enzymatic hydrolysate (SHEH) can ultimately alleviate HUA by restoring disturbances in the composition of the gut microbiota and reducing circulatory LPS levels [87], a Dendrobium candidum leaf extract was also reported to have a similar function [88].
There is already some evidence that certain active ingredients from dietary fiber or Chinese herbs can participate in purinergic signalling regulation via the gut microbiota. Cordycepin (CCS) is a major bioactive component separated from Cordyceps militaris. Its chemical identity is 3-deoxyadenosine. CCS was found by improving gut microbiology in high-fat diet (HFD) mice and regulating body weight by upregulating adenosine A1 receptor expression and inhibiting prolactin secretion [89, 90]. But its specific targets of action need to be further defined. Barley leaf (BL) is the young grass of the crop barley (Hordeum vulgare L.). Its antioxidant properties have been reported in previous studies [91]. As a traditional Chinese medicine, it has been used to protect the function of the intestinal tract. The study found that, in addition to improving dysbiosis of the intestinal microbiota induced by dextran sodium sulfate (DSS), BL significantly affected the levels of purine metabolites in serum and colonic contents, with the most significant increases in inosine and guanosine in vitro. The fermentation experiments revealed that BL could be used by upregulating the relative abundance of Firmicutes and Lactobacillus and that enriched Lactobacillus was positively correlated with the concentrations of inosine and guanosine. Subsequently, further studies revealed that inosine could exert a protective effect against colitis through A2AR/PPARγ signalling [28]. This provides a basis for the involvement of dietary interventions in the regulation of purinergic signalling through gut microbes.
Inosine and guanosine are valuable food additives which can be manufactured by microbial fermentation techniques in industrial production. By using genetic engineering, several bacterial strains such as Bacillus subtilis [92], Corynebacterium ammoniagenes [93], and Escherichia coli [94] have been used to increase the production of these purine nucleotides.
In recent years, engineered yeasts developed through biosynthetic techniques have also found novel applications in the regulation of purinergic signalling mediated by the gut microbiota [95, 96]. Based on an inflammatory therapeutic strategy that regulates eATP-adenosine homeostasis, Benjamin M. et al. developed a novel engineered yeast; this engineered yeast can secrete apyrase in response to the metabolite eATP produced in the inflamed gut, thereby depleting pro-inflammatory eATP and promoting the production of immunosuppressive adenosine [43]. The use of engineered yeast probiotics to modulate the pro-inflammatory-anti-inflammatory balance in the gut may provide a more flexible treatment for intestinal inflammation and inflammatory diseases targeting tissues other than the intestinal system. All of this evidence suggests that probiotics can indeed act on the gut microbiota and that natural medicines and probiotics can indeed be involved in metabolic or disease processes through inflammatory and immune responses by participating in the metabolism of gut microbes.
Conclusion
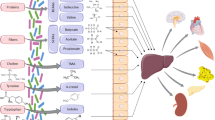
Over the last fifteen years, great progress has been made in the human microbiota field, yet difficulty lies in figuring out the specific functions performed by each microbial species in the prognosis or treatment of human diseases and in identifying the effective biomarkers that play key roles. By summarizing the existing studies, we suggest that ATP and adenosine derived from gut microbes can transmit purinergic signals in vivo by entering the gut as well as the bloodstream and binding to the corresponding purinergic receptors (Fig. 1).
The release of eATP from the gut microbiota can induce apoptosis of Tfh cells and regulate IgA release by binding to P2X7 receptors on Tfh cells in Peyer’s patches (PPs). Also, the intestinal commensal SFB can induce the differentiation of Th17 and Tfh and stimulate the release of IgA with other cytokines. When the intestinal environment is stable, IgA produced in PPs can play a protective role in the intestinal environment. In contrast, once the homeostasis of the intestinal environment gets disrupted, SFB induces a strong autoimmune response. Inosine (INO), which is metabolized by some microorganisms in the gut, can act as a purinergic signalling factor binding to A2AR on the surface of CD4+ T cells, inhibiting the differentiation of helper T cells to play a Treg-like role. However, in a tumor environment or the presence of other inflammatory factors infiltration, the inosine-A2AR axis can play the opposite role and promote T cell differentiation
Gut microbes can either actively release ATP into the gut or passively release ATP into the gut lumen after being disrupted. These extracellular ATPs can induce apoptosis of Tfh cells by binding to P2X7 receptors in Peyer’s patches (PPs), thus exerting immunosuppressive effects. And the activation of Tfh cells stimulates plasma cell differentiation and production of high-affinity SIgA to kill invasive pathogens in the intestine. This immune regulation modulated by microbial-derived eATP can maintain the stability of the gut microbiome that shows a form of co-existence between host and microorganism. Evidence suggests that immune responses mediated by SFB may also be related to this mechanism. The SFB, as a gut symbiont, contributes to immune homeostasis by inducing Th17 cells in normal physiological metabolism and inducing a strong autoimmune response when the gut microbiota is in an ecological imbalance. SFB can invoke Th17 cells to differentiate and produce multiple immune factors, leading to many autoimmune diseases, including rheumatoid arthritis (RA). Conducting more studies linking alterations in gut microbes with eATP and related purinergic receptors could provide more evidence for SFB-related metabolic mechanisms. Infection or immunity induces Tfh cells to produce complex immune responses, the disruption of Tfh cells can also alter the intestinal commensal community, and dysbiosis of the microbiota in the gut can stimulate Tfh cells to participate in immune regulation, emphasizing the importance played by Tfh cells in intestinal immune homeostasis. Immunomodulatory signals from the gut play a different role, and there may be additional applications for the immunosuppression exerted by P2X7 receptors in intestinal immunity beyond the multiple immune stimuli. Aiming to clarify the specific mechanism of this immunoregulation, more in vitro experiments on P2X7 receptors are needed.
In addition to eATP signalling, the adenosine derivatives inosine and hypoxanthine produced by microbial metabolism can also combine to A2AR on T cells and induce or inhibit Th cell differentiation in different contexts, thereby participating in the bidirectional regulation of the immune response. When Treg cell functionality is defective, the microbiota-inosine-A 2A receptor axis can exert immunomodulatory functions similar to those of Treg cells. For example, including B. pseudolongum, several bacteria promote the mouse immune system in tumor environment by inducing Th1 differentiation by the combination of their inosine-metabolites and A2AR, thereby enhancing the antitumor activity of ICB therapy. This phenomenon contradicts the immune-suppressive activity previously demonstrated by A2AR. In vitro experiments confirm that A2AR requires the coactivation of IFN-γ or CpG to exert this immune-promoting effect. The context-dependent feature shows the complex mechanisms behind the purinergic signalling–related immune regulation and highlights the necessity of a multi-omics study on it. Adenosine is the main ligand of A2AR, but metabolomic studies revealed extremely low levels of adenosine in the intestinal tract, while high concentrations of inosine are involved in purinergic signalling, which may be implicated in the metabolic reaction occurring in the intestine. To determine whether adenosine is a gut-derived purinergic signalling molecule required more microbiome as well as metabolomic studies. LR can improve plasma inosine concentrations but does not produce inosine in the in vitro experiments, and its direct link to metabolites needs to be further explored. This similar regulatory mechanism may also not be specific to LR, and there is much room for exploration of the link between microbial and purinergic signalling.
Different gut microbes can play a role in facilitating or inhibiting the immune response in the gut through diverse pathways. The maintenance or restoration of this homeostasis could be of great value in the development of gut flora–based therapeutic regimens. Existing studies have shown a close link between the purinergic system and gut microbial metabolism, but further microbiomic and metabolomic studies need to be combined with multifaceted analyses to clarify this underlying link. In addition to affecting the metabolism of uric acid, these effects also play an important role in the regulation of purinergic signalling. The discovery of more specific links between gut microbes and purinergic signalling could provide a more theoretical basis for the development of microbiota-based therapeutic approaches, as well as a detailed explanation of the physiological functions of purinergic signalling.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Abbreviations
- ATP:
-
Adenosine 5′-triphosphate
- eATP:
-
Extracellular Adenosine triphosphate
- GTP:
-
5′-Triphosphate
- NAD:
-
Nicotinamide adenine dinucleotide
- IMP:
-
Inosine monophosphate
- XMP:
-
Xanthosine monophosphate
- GMP:
-
Guanosine monophosphate
- HPRT:
-
Hypoxanthine–guanine phosphoribosyltransferase
- APRT:
-
Adenine phosphoribosyltransferase
- PRPP:
-
Phosphoribosyl pyrophosphate
- PRA:
-
5-Phosphoribosylamine
- cAMP:
-
Cyclic adenosine monophosphate
- ADO:
-
Adenosine
- P2RX7:
-
P2X7 receptor
- ENTPDase:
-
Ectonucleoside triphosphate diphosphate hydrolase
- VAM:
-
Vancomycin/ampicillin/metronidazole
- PPs:
-
Peyer’s patches
- Tfh:
-
T follicle-helper
- Tfr:
-
T follicle-regulated
- SFB:
-
Segmented filamentous bacteria
- Foxp3:
-
Forkhead box protein 3
- IPEX:
-
Immune dysregulation, polyendocrinopathy, and enteropathy, with X-linked inheritance
- ENT1:
-
Equilibrative nucleoside transporter 1
- CNT2:
-
Concentrative nucleoside transporter 2
- ICB:
-
Immune checkpoint blockade
References
Lynch SV, Pedersen O (2016) The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. https://doi.org/10.1056/NEJMra1600266
Fan Y, Pedersen O (2021) Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19:55–71. https://doi.org/10.1038/s41579-020-0433-9
Huang Z, Xie N, Illes P, Di Virgilio F, Ulrich H, Semyanov A, Verkhratsky A, Sperlagh B, Yu SG, Huang C, Tang Y (2021) From purines to purinergic signalling: molecular functions and human diseases. Signal Transduct Target Ther 6:162. https://doi.org/10.1038/s41392-021-00553-z
Embley TM, Martin W (2006) Eukaryotic evolution, changes and challenges. Nature 440:623–630. https://doi.org/10.1038/nature04546
Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283:1476–1481. https://doi.org/10.1126/science.283.5407.1476
Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG (2018) Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557:101–105. https://doi.org/10.1038/s41586-018-0059-5
Perruzza L, Gargari G, Proietti M, Fosso B, D’Erchia AM, Faliti CE, Rezzonico-Jost T, Scribano D, Mauri L, Colombo D, Pellegrini G, Moregola A, Mooser C, Pesole G, Nicoletti M, Norata GD, Geuking MB, McCoy KD, Guglielmetti S, Grassi F (2017) T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota-derived extracellular ATP. Cell Rep 18:2566–2575. https://doi.org/10.1016/j.celrep.2017.02.061
He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, Luo M, Tran DQ, Zhou J, Tatevian N, Luo F, Molina JG, Blackburn MR, Gomez TH, Roos S, Rhoads JM, Liu Y (2017) Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med 214:107–123. https://doi.org/10.1084/jem.20160961
Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, Lewis IA, Geuking MB, McCoy KD (2020) Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369:1481–1489. https://doi.org/10.1126/science.abc3421
Choi HK (2010) A prescription for lifestyle change in patients with hyperuricemia and gout. Curr Opin Rheumatol 22:165–172. https://doi.org/10.1097/BOR.0b013e328335ef38
Richette P, Bardin T (2010) Gout Lancet 375:318–328. https://doi.org/10.1016/S0140-6736(09)60883-7
Choi HK, Willett W, Curhan G (2010) Fructose-rich beverages and risk of gout in women. JAMA 304:2270–2278. https://doi.org/10.1001/jama.2010.1638
Richette P, Doherty M, Pascual E, Barskova V, Becce F, Castaneda-Sanabria J, Coyfish M, Guillo S, Jansen TL, Janssens H, Liote F, Mallen C, Nuki G, Perez-Ruiz F, Pimentao J, Punzi L, Pywell T, So A, Tausche AK, Uhlig T, Zavada J, Zhang W, Tubach F, Bardin T (2017) 2016 updated EULAR evidence-based recommendations for the management of gout. Ann Rheum Dis 76:29–42. https://doi.org/10.1136/annrheumdis-2016-209707
de Oliveira EP, Burini RC (2012) High plasma uric acid concentration: causes and consequences. Diabetol Metab Syndr 4:12. https://doi.org/10.1186/1758-5996-4-12
Pascart T, Liote F (2019) Gout: state of the art after a decade of developments. Rheumatology (Oxford) 58:27–44. https://doi.org/10.1093/rheumatology/key002
Crane JK (2013) Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Gut Microbes 4:388–391. https://doi.org/10.4161/gmic.25584
Crane JK, Mongiardo KM (2014) Pro-inflammatory effects of uric acid in the gastrointestinal tract. Immunol Invest 43:255–266. https://doi.org/10.3109/08820139.2013.864667
Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, Su X, Qiao J, Zheng Y, Wang L, Koh E, Danliang H, Xu J, Lee YK, Zhang H (2016) Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep 6:20602. https://doi.org/10.1038/srep20602
Liu X, Lv Q, Ren H, Gao L, Zhao P, Yang X, Yang G, Xu D, Wang G, Yang W, Wang P, Wang Z, Xing S (2020) The altered gut microbiota of high-purine-induced hyperuricemia rats and its correlation with hyperuricemia. PeerJ 8:e8664. https://doi.org/10.7717/peerj.8664
Luthje J (1989) Origin, metabolism and function of extracellular adenine nucleotides in the blood. Klin Wochenschr 67:317–327. https://doi.org/10.1007/BF01741386
Mustafa S J, Morrison R R, Teng B, and Pelleg A (2009) Adenosine receptors and the heart: role in regulation of coronary blood flow and cardiac electrophysiology. Handb Exp Pharmacol:161–88. https://doi.org/10.1007/978-3-540-89615-9_6
Fredholm BB, IJzerman AP, Jacobson KA, Klotz KN, Linden J (2001) International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev 53:527–52
Sachdeva S, Gupta M (2013) Adenosine and its receptors as therapeutic targets: an overview. Saudi Pharm J 21:245–253. https://doi.org/10.1016/j.jsps.2012.05.011
Sitkovsky M, Lukashev D, Deaglio S, Dwyer K, Robson SC, Ohta A (2008) Adenosine A2A receptor antagonists: blockade of adenosinergic effects and T regulatory cells. Br J Pharmacol 153(Suppl 1):S457–S464. https://doi.org/10.1038/bjp.2008.23
Brugarolas M, Navarro G, Martinez-Pinilla E, Angelats E, Casado V, Lanciego JL, Franco R (2014) G-protein-coupled receptor heteromers as key players in the molecular architecture of the central nervous system. CNS Neurosci Ther 20:703–709. https://doi.org/10.1111/cns.12277
Navarro G, Cordomi A, Zelman-Femiak M, Brugarolas M, Moreno E, Aguinaga D, Perez-Benito L, Cortes A, Casado V, Mallol J, Canela EI, Lluis C, Pardo L, Garcia-Saez AJ, McCormick PJ, Franco R (2016) Quaternary structure of a G-protein-coupled receptor heterotetramer in complex with Gi and Gs. BMC Biol 14:26. https://doi.org/10.1186/s12915-016-0247-4
Pallio G, Bitto A, Pizzino G, Galfo F, Irrera N, Squadrito F, Squadrito G, Pallio S, Anastasi GP, Cutroneo G, Macri A, Altavilla D (2016) Adenosine receptor stimulation by polydeoxyribonucleotide improves tissue repair and symptomology in experimental colitis. Front Pharmacol 7:273. https://doi.org/10.3389/fphar.2016.00273
Li D, Feng Y, Tian M, Ji J, Hu X, Chen F (2021) Gut microbiota-derived inosine from dietary barley leaf supplementation attenuates colitis through PPARgamma signaling activation. Microbiome 9:83. https://doi.org/10.1186/s40168-021-01028-7
Burnstock G (2018) Purine and purinergic receptors. Brain Neurosci Adv 2:2398212818817494. https://doi.org/10.1177/2398212818817494
Schenk U, Westendorf AM, Radaelli E, Casati A, Ferro M, Fumagalli M, Verderio C, Buer J, Scanziani E, Grassi F (2008) Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci Signal 1:ra6. https://doi.org/10.1126/scisignal.1160583
Trautmann A (2009) Extracellular ATP in the immune system: more than just a “danger signal.” Sci Signal 2:pe6. https://doi.org/10.1126/scisignal.256pe6
Wang X, Yuan X, Su Y, Hu J, Ji Q, Fu S, Li R, Hu L, Dai C (2021) Targeting purinergic receptor P2RX1 modulates intestinal microbiota and alleviates inflammation in colitis. Front Immunol 12:696766. https://doi.org/10.3389/fimmu.2021.696766
Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, Stockinger B (2018) The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity 49(353–362):e5. https://doi.org/10.1016/j.immuni.2018.07.010
Kusu T, Kayama H, Kinoshita M, Jeon SG, Ueda Y, Goto Y, Okumura R, Saiga H, Kurakawa T, Ikeda K, Maeda Y, Nishimura J, Arima Y, Atarashi K, Honda K, Murakami M, Kunisawa J, Kiyono H, Okumura M, Yamamoto M, Takeda K (2013) Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J Immunol 190:774–783. https://doi.org/10.4049/jimmunol.1103067
Tani H, Li B, Kusu T, Okumura R, Nishimura J, Okuzaki D, Motooka D, Arakawa S, Mori A, Yoshihara T, Ogino T, Tsai S H, Furuta Y, Muneta M, Nakamura S, Fukusaki E, Yamamoto K, Yagita H, Kayama H, and Takeda K (2021) The ATP-hydrolyzing ectoenzyme E-NTPD8 attenuates colitis through modulation of P2X4 receptor-dependent metabolism in myeloid cells. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2100594118
Salem M, Lecka J, Pelletier J, Gomes Marconato D, Dumas A, Vallieres L, Brochu G, Robaye B, Jobin C, Sevigny J (2022) NTPDase8 protects mice from intestinal inflammation by limiting P2Y6 receptor activation: identification of a new pathway of inflammation for the potential treatment of IBD. Gut 71:43–54. https://doi.org/10.1136/gutjnl-2020-320937
Salem M, El Azreq MA, Pelletier J, Robaye B, Aoudjit F, Sevigny J (2019) Exacerbated intestinal inflammation in P2Y6 deficient mice is associated with Th17 activation. Biochim Biophys Acta Mol Basis Dis 1865:2595–2605. https://doi.org/10.1016/j.bbadis.2019.06.019
Placet M, Molle CM, Arguin G, Geha S, Gendron FP (2021) The expression of P2Y6 receptor promotes the quality of mucus in colitic mice. FEBS J 288:5459–5473. https://doi.org/10.1111/febs.15819
Beldi G, Enjyoji K, Wu Y, Miller L, Banz Y, Sun X, Robson SC (2008) The role of purinergic signaling in the liver and in transplantation: effects of extracellular nucleotides on hepatic graft vascular injury, rejection and metabolism. Front Biosci 13:2588–2603. https://doi.org/10.2741/2868
Proietti M, Cornacchione V, Rezzonico Jost T, Romagnani A, Faliti CE, Perruzza L, Rigoni R, Radaelli E, Caprioli F, Preziuso S, Brannetti B, Thelen M, McCoy KD, Slack E, Traggiai E, Grassi F (2014) ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer’s patches to promote host-microbiota mutualism. Immunity 41:789–801. https://doi.org/10.1016/j.immuni.2014.10.010
Liu Y, Tian X, He B, Hoang TK, Taylor CM, Blanchard E, Freeborn J, Park S, Luo M, Couturier J, Tran DQ, Roos S, Wu G, Rhoads JM (2019) Lactobacillus reuteri DSM 17938 feeding of healthy newborn mice regulates immune responses while modulating gut microbiota and boosting beneficial metabolites. Am J Physiol Gastrointest Liver Physiol 317:G824–G838. https://doi.org/10.1152/ajpgi.00107.2019
Bahreyni A, Samani SS, Khazaei M, Ryzhikov M, Avan A, Hassanian SM (2018) Therapeutic potentials of adenosine receptors agonists and antagonists in colitis; current status and perspectives. J Cell Physiol 233:2733–2740. https://doi.org/10.1002/jcp.26073
Scott BM, Gutierrez-Vazquez C, Sanmarco LM, da Silva PJA, Li Z, Plasencia A, Hewson P, Cox LM, O’Brien M, Chen SK, Moraes-Vieira PM, Chang BSW, Peisajovich SG, Quintana FJ (2021) Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nat Med 27:1212–1222. https://doi.org/10.1038/s41591-021-01390-x
Luo B, Zhang Y, Zhang C, Liu X, Shi C (2021) Intestinal microbiota: a potential target for enhancing the antitumor efficacy and reducing the toxicity of immune checkpoint inhibitors. Cancer Lett 509:53–62. https://doi.org/10.1016/j.canlet.2021.04.001
Berrier C, Coulombe A, Szabo I, Zoratti M, Ghazi A (1992) Gadolinium ion inhibits loss of metabolites induced by osmotic shock and large stretch-activated channels in bacteria. Eur J Biochem 206:559–565. https://doi.org/10.1111/j.1432-1033.1992.tb16960.x
Booth IR, Edwards MD, Black S, Schumann U, Miller S (2007) Mechanosensitive channels in bacteria: signs of closure? Nat Rev Microbiol 5:431–440. https://doi.org/10.1038/nrmicro1659
Macpherson AJ, Harris NL (2004) Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 4:478–485. https://doi.org/10.1038/nri1373
Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T (2002) Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298:1424–1427. https://doi.org/10.1126/science.1077336
Mantis NJ, Rol N, Corthesy B (2011) Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 4:603–611. https://doi.org/10.1038/mi.2011.41
Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, Voller T, Minola A, Fernandez-Rodriguez B, Agatic G, Barbieri S, Piccoli L, Casiraghi C, Corti D, Lanzavecchia A, Regoes RR, Loverdo C, Stocker R, Brumley DR, Hardt WD, Slack E (2017) High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544:498–502. https://doi.org/10.1038/nature22058
Kawamoto S, Maruya M, Kato L M, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, Hattori M, and Fagarasan S (2014) Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41:152–65. https://doi.org/10.1016/j.immuni.2014.05.016
Proietti M, Perruzza L, Scribano D, Pellegrini G, D’Antuono R, Strati F, Raffaelli M, Gonzalez SF, Thelen M, Hardt WD, Slack E, Nicoletti M, Grassi F (2019) ATP released by intestinal bacteria limits the generation of protective IgA against enteropathogens. Nat Commun 10:250. https://doi.org/10.1038/s41467-018-08156-z
Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S (2017) The P2X7 receptor in infection and inflammation. Immunity 47:15–31. https://doi.org/10.1016/j.immuni.2017.06.020
Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, Perfettini JL, Schlemmer F, Tasdemir E, Uhl M, Genin P, Civas A, Ryffel B, Kanellopoulos J, Tschopp J, Andre F, Lidereau R, McLaughlin NM, Haynes NM, Smyth MJ, Kroemer G, Zitvogel L (2009) Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med 15:1170–1178. https://doi.org/10.1038/nm.2028
Bartlett R, Stokes L, Sluyter R (2014) The P2X7 receptor channel: recent developments and the use of P2X7 antagonists in models of disease. Pharmacol Rev 66:638–675. https://doi.org/10.1124/pr.113.008003
Bernardazzi C, Castelo-Branco M T L, Pego B, Ribeiro B E, Rosas S L B, Santana P T, Machado J C, Leal C, Thompson F, Coutinho-Silva R, and de Souza H S P (2022) The P2X7 receptor promotes colorectal inflammation and tumorigenesis by modulating gut microbiota and the inflammasome. Int J Mol Sci 23. https://doi.org/10.3390/ijms23094616
Salles EM, Menezes MN, Siqueira R, Borges da Silva H, Amaral EP, Castillo-Mendez SI, Cunha I, Cassado ADA, Vieira FS, Olivieri DN, Tadokoro CE, Alvarez JM, Coutinho-Silva R, D’Imperio-Lima MR (2017) P2X7 receptor drives Th1 cell differentiation and controls the follicular helper T cell population to protect against Plasmodium chabaudi malaria. PLoS Pathog 13:e1006595. https://doi.org/10.1371/journal.ppat.1006595
Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626. https://doi.org/10.1038/nature11400
Meehan CJ, Beiko RG (2014) A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 6:703–713. https://doi.org/10.1093/gbe/evu050
Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K (2008) ATP drives lamina propria T(H)17 cell differentiation. Nature 455:808–812. https://doi.org/10.1038/nature07240
Davis CP, Savage DC (1974) Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun 10:948–956. https://doi.org/10.1128/iai.10.4.948-956.1974
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. https://doi.org/10.1016/j.cell.2009.09.033
Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, Wu HJ (2016) Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 44:875–888. https://doi.org/10.1016/j.immuni.2016.03.013
Hasko G, Sitkovsky MV, Szabo C (2004) Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol Sci 25:152–157. https://doi.org/10.1016/j.tips.2004.01.006
da Rocha LF, de Oliveira AP, Accetturi BG, de Oliveira MI, Domingos HV, de Almeida CD, de Lima WT, Santos AR (2013) Anti-inflammatory effects of inosine in allergic lung inflammation in mice: evidence for the participation of adenosine A2A and A 3 receptors. Purinergic Signal 9:325–336. https://doi.org/10.1007/s11302-013-9351-x
Fredholm BB, Irenius E, Kull B, Schulte G (2001) Comparison of the potency of adenosine as an agonist at human adenosine receptors expressed in Chinese hamster ovary cells. Biochem Pharmacol 61:443–448. https://doi.org/10.1016/s0006-2952(00)00570-0
Welihinda AA, Kaur M, Greene K, Zhai Y, Amento EP (2016) The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cell Signal 28:552–560. https://doi.org/10.1016/j.cellsig.2016.02.010
Walter J, Britton RA, Roos S (2011) Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A 108(Suppl 1):4645–4652. https://doi.org/10.1073/pnas.1000099107
Liu Y, Tran DQ, Fatheree NY, Marc Rhoads J (2014) Lactobacillus reuteri DSM 17938 differentially modulates effector memory T cells and Foxp3+ regulatory T cells in a mouse model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 307:G177–G186. https://doi.org/10.1152/ajpgi.00038.2014
Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO (2010) Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol 11:618–627. https://doi.org/10.1038/ni.1884
Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F (2001) Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27:68–73. https://doi.org/10.1038/83784
Ward JL, Sherali A, Mo ZP, Tse CM (2000) Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J Biol Chem 275:8375–8381. https://doi.org/10.1074/jbc.275.12.8375
Okada M, Suzuki K, Nakashima M, Nakanishi T, Fujioka N (2006) The nucleotide derivatives inosine and inosinic acid inhibit intestinal absorption of mizoribine in rats. Eur J Pharmacol 531:140–144. https://doi.org/10.1016/j.ejphar.2005.12.013
He B, Hoang TK, Tran DQ, Rhoads JM, Liu Y (2017) Adenosine A2A receptor deletion blocks the beneficial effects of Lactobacillus reuteri in regulatory T-deficient scurfy mice. Front Immunol 8:1680. https://doi.org/10.3389/fimmu.2017.01680
Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragon L, Jacquelot N, Qu B, Ferrere G, Clemenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359:91–97. https://doi.org/10.1126/science.aan3706
Csoka B, Himer L, Selmeczy Z, Vizi ES, Pacher P, Ledent C, Deitch EA, Spolarics Z, Nemeth ZH, Hasko G (2008) Adenosine A2A receptor activation inhibits T helper 1 and T helper 2 cell development and effector function. FASEB J 22:3491–3499. https://doi.org/10.1096/fj.08-107458
Cekic C, Linden J (2014) Adenosine A2A receptors intrinsically regulate CD8+ T cells in the tumor microenvironment. Cancer Res 74:7239–7249. https://doi.org/10.1158/0008-5472.CAN-13-3581
Lasek W, Janyst M, Wolny R, Zapala L, Bocian K, Drela N (2015) Immunomodulatory effects of inosine pranobex on cytokine production by human lymphocytes. Acta Pharm 65:171–180. https://doi.org/10.1515/acph-2015-0015
Lioux T, Mauny MA, Lamoureux A, Bascoul N, Hays M, Vernejoul F, Baudru AS, Boularan C, Lopes-Vicente J, Qushair G, Tiraby G (2016) Design, synthesis, and biological evaluation of novel cyclic adenosine-inosine monophosphate (cAIMP) Analogs that activate stimulator of interferon genes (STING). J Med Chem 59:10253–10267. https://doi.org/10.1021/acs.jmedchem.6b01300
Chu Y, Sun S, Huang Y, Gao Q, Xie X, Wang P, Li J, Liang L, He X, Jiang Y, Wang M, Yang J, Chen X, Zhou C, Zhao Y, Ding F, Zhang Y, Wu X, Bai X, Wu J, Wei X, Chen X, Yue Z, Fang X, Huang Q, Wang Z, Huang R (2021) Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes 7:66. https://doi.org/10.1038/s41522-021-00235-2
Zhao S, Jang C, Liu J, Uehara K, Gilbert M, Izzo L, Zeng X, Trefely S, Fernandez S, Carrer A, Miller KD, Schug ZT, Snyder NW, Gade TP, Titchenell PM, Rabinowitz JD, Wellen KE (2020) Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature 579:586–591. https://doi.org/10.1038/s41586-020-2101-7
Silva-Veiga FM, Miranda CS, Martins FF, Daleprane JB, Mandarim-de-Lacerda CA, Souza-Mello V (2020) Gut-liver axis modulation in fructose-fed mice: a role for PPAR-alpha and linagliptin. J Endocrinol 247:11–24. https://doi.org/10.1530/JOE-20-0139
Brosh S, Boer P, Sperling O (1982) Effects of fructose on synthesis and degradation of purine nucleotides in isolated rat hepatocytes. Biochim Biophys Acta 717:459–464. https://doi.org/10.1016/0304-4165(82)90288-4
Liu R, Han C, Wu D, Xia X, Gu J, Guan H, Shan Z, Teng W (2015) Prevalence of hyperuricemia and gout in mainland China from 2000 to 2014: a systematic review and meta-analysis. Biomed Res Int 2015:762820. https://doi.org/10.1155/2015/762820
Wang J, Chen Y, Zhong H, Chen F, Regenstein J, Hu X, Cai L, and Feng F (2021) The gut microbiota as a target to control hyperuricemia pathogenesis: potential mechanisms and therapeutic strategies. Crit Rev Food Sci Nutr:1–11. https://doi.org/10.1080/10408398.2021.1874287
Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, Wang A, Chen W, Sun Z, Lu Y (2020) Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics 10:10665–10679. https://doi.org/10.7150/thno.43528
Liu G, Chen X, Lu X, Zhao J, Li X (2020) Sunflower head enzymatic hydrolysate relives hyperuricemia by inhibiting crucial proteins (xanthine oxidase, adenosine deaminase, uric acid transporter1) and restoring gut microbiota in mice. J Funct Foods 72:104055. https://doi.org/10.1016/j.jff.2020.104055
Lou XJ, Wang YZ, Lei SS, He X, Lu TT, Zhan LH, Chen X, Chen YH, Li B, Zheng X, Lv GY, Chen SH (2020) Beneficial effects of macroporous resin extract of dendrobium candidum leaves in rats with hyperuricemia induced by a high-purine diet. Evid Based Complement Alternat Med 2020:3086106. https://doi.org/10.1155/2020/3086106
An Y, Li Y, Wang X, Chen Z, Xu H, Wu L, Li S, Wang C, Luan W, Wang X, Liu M, Tang X, Yu L (2018) Cordycepin reduces weight through regulating gut microbiota in high-fat diet-induced obese rats. Lipids Health Dis 17:276. https://doi.org/10.1186/s12944-018-0910-6
Li Y, Li Y, Wang X, Xu H, Wang C, An Y, Luan W, Wang X, Li S, Ma F, Ni L, Liu M, Tang X, Yu L (2018) Cordycepin modulates body weight by reducing prolactin via an adenosine A1 receptor. Curr Pharm Des 24:3240–3249. https://doi.org/10.2174/1381612824666180820144917
Yu YM, Chang WC, Chang CT, Hsieh CL, Tsai CE (2002) Effects of young barley leaf extract and antioxidative vitamins on LDL oxidation and free radical scavenging activities in type 2 diabetes. Diabetes Metab 28:107–114
Sheremet AS, Gronskiy SV, Akhmadyshin RA, Novikova AE, Livshits VA, Shakulov RS, Zakataeva NP (2011) Enhancement of extracellular purine nucleoside accumulation by Bacillus strains through genetic modifications of genes involved in nucleoside export. J Ind Microbiol Biotechnol 38:65–70. https://doi.org/10.1007/s10295-010-0829-z
Peifer S, Barduhn T, Zimmet S, Volmer DA, Heinzle E, Schneider K (2012) Metabolic engineering of the purine biosynthetic pathway in Corynebacterium glutamicum results in increased intracellular pool sizes of IMP and hypoxanthine. Microb Cell Fact 11:138. https://doi.org/10.1186/1475-2859-11-138
Shimaoka M, Takenaka Y, Kurahashi O, Kawasaki H, Matsui H (2007) Effect of amplification of desensitized purF and prs on inosine accumulation in Escherichia coli. J Biosci Bioeng 103:255–261. https://doi.org/10.1263/jbb.103.255
Mimee M, Nagler CR (2021) Engineered yeast tune down gut inflammation. Nat Med 27:1150–1151. https://doi.org/10.1038/s41591-021-01420-8
Tahir AH, Tang Y (2022) Engineered yeast-based eATP precisely controlled system for the treatment of inflammatory bowel disease. Purinergic Signal 18:9–11. https://doi.org/10.1007/s11302-022-09846-6
Funding
Partial financial support was received from Sichuan Science and Technology Program(2019YFS0160),
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Bowen Liu, Rong Li, and Ping Yang. The first draft of the manuscript was written by Mingjian Li. YH and PL provided many updated resources to enhance the quality of this work. All authors read and approved the final manuscript. all authors commented on previous versions of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflicts of interest
There is no conflict of interest in the article.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Liu, B., Li, R. et al. Exploration of the link between gut microbiota and purinergic signalling. Purinergic Signalling 19, 315–327 (2023). https://doi.org/10.1007/s11302-022-09891-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11302-022-09891-1