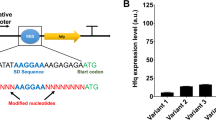
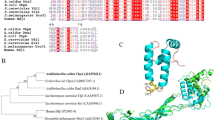
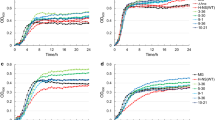
Abstract
Acidithiobacillus caldus is a typical extreme acidophile widely used in the biohydrometallurgical industry, which often experiences extreme environmental stress in its natural habitat. Hfq, an RNA-binding protein, typically functions as a global regulator involved in various cellular physiological processes. Yet, the biological functions of Hfq derived from such extreme acidophile have not been extensively investigated. In this study, the recombinant strain Δhfq/Achfq, constructed by CRISPR/Cas9-mediated chromosome integration, fully or partially restored the phenotypic defects caused by hfq deletion in Escherichia coli, including impaired growth performance, abnormal cell morphology, impaired swarming motility, decreased stress resistance, decreased intracellular ATP and free amino acid levels, and attenuated biofilm formation. Particularly noteworthy, the intracellular ATP level and biofilm production of the recombinant strain were increased by 12.2% and 7.0%, respectively, compared to the Δhfq mutant. Transcriptomic analysis revealed that even under heterologous expression, AcHfq exerted global regulatory effects on multiple cellular processes, including metabolism, environmental signal processing, and motility. Finally, we established a potential working model to illustrate the regulatory mechanism of AcHfq in bacterial resistance to environmental stress.







Similar content being viewed by others
Data availability
The data that support for the findings of this study are all contained in the manuscript and supplement materials.
References
Arce-Rodríguez A, Calles B, Nikel PI, de Lorenzo V (2016) The RNA chaperone hfq enables the environmental stress tolerance super-phenotype of Pseudomonas putida. Environ Microbiol 18(10):3309–3326. https://doi.org/10.1111/1462-2920.13052
Assis NG, Ribeiro RA, da Silva LG, Vicente AM, Hug I, Marques MV (2019) Identification of hfq-binding RNAs in Caulobacter crescentus. RNA Biol 16(6):719–726. https://doi.org/10.1080/15476286.2019.1593091
Attia AS, Sedillo JL, Wang W, Liu W, Brautigam CA, Winkler W, Hansen EJ (2008) Moraxella catarrhalis expresses an unusual hfq protein. Infect Immun 76(6):2520–2530. https://doi.org/10.1128/IAI.01652-07
Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE (2009) Cyanobacteria contain a structural homologue of the hfq protein with altered RNA-binding properties. FEBS J 276(14):3904–3915. https://doi.org/10.1111/j.1742-4658.2009.07104.x
Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, Dupuy B, Martin-Verstraete I, Soutourina O (2014) Pleiotropic role of the RNA chaperone protein hfq in the human pathogen Clostridium difficile. J Bacteriol 196(18):3234–3248. https://doi.org/10.1128/JB.01923-14
Brennan RG, Link TM (2007) Hfq structure, function and ligand binding. Curr Opin Microbiol 10(2):125–133. https://doi.org/10.1016/j.mib.2007.03.015
Bruinenberg PM, van Dijken JP, Scheffers WA (1983) An enzymic analysis of NADPH production and consumption in Candida utilis. J Gen Microbiol 129(4):965–971. https://doi.org/10.1099/00221287-129-4-965
Cai H, Roca J, Zhao YF, Woodson SA (2022) Dynamic refolding of OxyS sRNA by the Hfq RNA chaperone. J Mol Biol 434(18):167776. https://doi.org/10.1016/j.jmb.2022.167776
Caillet J, Gracia C, Fontaine F, Hajnsdorf E (2014) Clostridium difficile Hfq can replace Escherichia coli Hfq for most of its function. RNA 20(10):1567–1578. https://doi.org/10.1261/rna.043372.113
Chate AV, Redlawar AA, Bondle GM, Sarkate AP, Tiwari SV, Lokwani DK (2019) A new efficient domino approach for the synthesis of coumarin-pyrazolines as antimicrobial agents targeting bacterial D-alanine-D-alanine ligase. New J Chem 43(23):9002–9011. https://doi.org/10.1039/C9NJ00703B
Chen W, Westerhoff P, Leenheer JA, Booksh K (2003) Fluorescence excitation-emission matrix regional integration to quantify spectra for dissolved organic matter. Environ Sci Technol 37(24):5701–5710. https://doi.org/10.1021/es034354c
Das D, Kozbial P, Axelrod HL, Miller MD, McMullan D, Krishna SS, Abdubek P, Acosta C, Astakhova T, Burra P, Carlton D, Chen C, Chiu HJ, Clayton T, Deller MC, Duan L, Elias Y, Elsliger MA, Ernst D, Farr C, Feuerhelm J, Grzechnik A, Grzechnik SK, Hale J, Han GW, Jaroszewski L, Jin KK, Johnson HA, Klock HE, Knuth MW, Kumar A, Marciano D, Morse AT, Murphy KD, Nigoghossian E, Nopakun A, Okach L, Oommachen S, Paulsen J, Puckett C, Reyes R, Rife CL, Sefcovic N, Sudek S, Tien H, Trame C, Trout CV, van den Bedem H, Weekes D, White A, Xu Q, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA (2009) Crystal structure of a novel Sm-like protein of putative cyanophage origin at 2.60 a resolution. Proteins 75(2):296–307. https://doi.org/10.1002/prot.22360
Dendooven T, Sonnleitner E, Bläsi U, Luisi BF (2023) Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding. EMBO J 42(3):e111129. https://doi.org/10.15252/embj.2022111129
Dos Santos RF, Arraiano CM, Andrade JM (2019) New molecular interactions broaden the functions of the RNA chaperone hfq. Curr Genet 65(6):1313–1319. https://doi.org/10.1007/s00294-019-00990-y
Fantappiè L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I (2009) The RNA chaperone hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun 77(5):1842–1853. https://doi.org/10.1128/IAI.01216-08
Feng S, Qiu Y, Huang Z, Yin Y, Zhang H, Zhu D, Tong Y, Yang H (2021) The adaptation mechanisms of Acidithiobacillus caldus CCTCC M 2018054 to extreme acid stress: Bioleaching performance, physiology, and transcriptomics. Environ Res 199:111341. https://doi.org/10.1016/j.envres.2021.111341
Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14(9):563–575. https://doi.org/10.1038/nrmicro.2016.94
Franze de Fernandez MT, Eoyang L, August JT (1968) Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219(5154):588–590. https://doi.org/10.1038/219588a0
Granato LM, Picchi SC, Andrade Mde O, Takita MA, de Souza AA, Wang N, Machado MA (2016) The ATP-dependent RNA helicase HrpB plays an important role in motility and biofilm formation in Xanthomonas citri subsp. Citri. BMC Microbiol 16:55. https://doi.org/10.1186/s12866-016-0655-1
Hede N, Khandeparker L (2020) Extracellular polymeric substances mediate the coaggregation of aquatic biofilm-forming bacteria. Hydrobiologia 847:4249–4272. https://doi.org/10.1007/s10750-020-04411-x
Hou S, Tong Y, Yang H, Feng S (2021) Molecular insights into the copper-sensitive Operon Repressor in Acidithiobacillus caldus. Appl Environ Microbiol 87(16):e0066021. https://doi.org/10.1128/AEM.00660-21
Hu W, Feng S, Tong Y, Zhang H, Yang H (2020) Adaptive defensive mechanism of bioleaching microorganisms under extremely environmental acid stress: advances and perspectives. Biotechnol Adv 42:107580. https://doi.org/10.1016/j.biotechadv.2020.107580
Hu W, Tong Y, Liu J, Chen P, Yang H, Feng S (2023) Improving acid resistance of Escherichia coli base on the CfaS-mediated membrane engineering strategy derived from extreme acidophile. Front Bioeng Biotechnol 11:1158931. https://doi.org/10.3389/fbioe.2023.1158931
Huang Z, Feng S, Tong Y, Yang H (2019) Enhanced contact mechanism for interaction of extracellular polymeric substances with low-grade copper-bearing sulfide ore in bioleaching by moderately thermophilic Acidithiobacillus caldus. J Environ Manage 242:11–21. https://doi.org/10.1016/j.jenvman.2019.04.030
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81(7):2506–2514. https://doi.org/10.1128/AEM.04023-14
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589. https://doi.org/10.1038/s41586-021-03819-2
King KA, Benton AH, Caudill MT, Stoyanof ST, Kang L, Michalak P, Lahmers KK, Dunman PM, DeHart TG, Ahmad SS, Jutras BL, Poncin K, De Bolle X, Caswell CC (2024) Post-transcriptional control of the essential enzyme MurF by a small regulatory RNA in Brucella abortus. Mol Microbiol 121(1):129–141. https://doi.org/10.1111/mmi.15207
Kovac A, Konc J, Vehar B, Bostock JM, Chopra I, Janezic D, Gobec S (2008) Discovery of new inhibitors of D-alanine:D-alanine ligase by structure-based virtual screening. J Med Chem 51(23):7442–7448. https://doi.org/10.1021/jm800726b
Lekontseva NV, Stolboushkina EA, Nikulin AD (2021) Diversity of LSM family proteins: similarities and differences. Biochem (Mosc) 86(Suppl 1):S38–S49. https://doi.org/10.1134/S0006297921140042
Li Y, Dai F, Li Y, Liang W, Li C, Zhang W (2022) Hfq, a global regulator contributes to the virulence of Vibrio splendidus AJ01. Aquaculture 546:737416. https://doi.org/10.1016/j.aquaculture.2021.737416
Lin Z, Li J, Yan X, Yang J, Li X, Chen P, Yang X (2021) Engineering of the small noncoding RNA (sRNA) DsrA together with the sRNA chaperone hfq enhances the Acid Tolerance of Escherichia coli. Appl Environ Microbiol 87(10). https://doi.org/10.1128/AEM.02923-20
Liu H, Wang Q, Liu Q, Cao X, Shi C, Zhang Y (2011) Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl Microbiol Biotechnol 91(2):353–364. https://doi.org/10.1007/s00253-011-3286-3
Lu P, Ma D, Chen Y, Guo Y, Chen GQ, Deng H, Shi Y (2013) L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res 23(5):635–644. https://doi.org/10.1038/cr.2013.13
Ma X, Zhang S, Xu Z, Li H, Xiao Q, Qiu F, Zhang W, Long Y, Zheng D, Huang B, Chen C, Lu Y (2020) SdiA improves the Acid Tolerance of E. Coli by regulating GadW and GadY expression. Front Microbiol 11:1078. https://doi.org/10.3389/fmicb.2020.01078
Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, Nikulin A (2010) The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr Sect F Struct Biol Cryst Commun 66(Pt 7):760–764. https://doi.org/10.1107/S1744309110017331
Mura C, Randolph PS, Patterson J, Cozen AE (2013) Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on sm function. RNA Biol 10(4):636–651. https://doi.org/10.4161/rna.24538
Ngan JYG, Pasunooti S, Tse W, Meng W, Ngan SFC, Jia H, Lin JQ, Ng SW, Jaafa MT, Cho SLS, Lim J, Koh HQV, Abdul Ghani N, Pethe K, Sze SK, Lescar J, Alonso S (2021) HflX is a GTPase that controls hypoxia-induced replication arrest in slow-growing mycobacteria. Proc Natl Acad Sci U S A 118(12):e2006717118. https://doi.org/10.1073/pnas.2006717118
Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, Garber M, Nikonov S (2005) Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr D Biol Crystallogr 61(Pt 2):141–146. https://doi.org/10.1107/S0907444904030008
Pan X, Tang M, You J, Osire T, Sun C, Fu W, Yi G, Yang T, Yang ST, Rao Z (2022) PsrA is a novel regulator contributes to antibiotic synthesis, bacterial virulence, cell motility and extracellular polysaccharides production in Serratia marcescens. Nucleic Acids Res 50(1):127–148. https://doi.org/10.1093/nar/gkab1186
Park S, Prévost K, Heideman EM, Carrier MC, Azam MS, Reyer MA, Liu W, Massé E, Fei J (2021) Dynamic interactions between the RNA chaperone hfq, small regulatory RNAs, and mRNAs in live bacterial cells. Elife 10. https://doi.org/10.7554/eLife.64207
Qi J, Caiyin Q, Wu H, Tian K, Wang B, Li Y, Qiao J (2017) The novel sRNA s015 improves nisin yield by increasing acid tolerance of Lactococcus lactis F44. Appl Microbiol Biotechnol 101(16):6483–6493. https://doi.org/10.1007/s00253-017-8399-x
Qiao Y, Liu G, Leng C, Zhang Y, Lv X, Chen H, Sun J, Feng Z (2018) Metabolic profiles of cysteine, methionine, glutamate, glutamine, arginine, aspartate, asparagine, alanine and glutathione in Streptococcus thermophilus during pH-controlled batch fermentations. Sci Rep 8(1):12441. https://doi.org/10.1038/s41598-018-30272-5
Rudra P, Hurst-Hess KR, Cotten KL, Partida-Miranda A, Ghosh P (2020) Mycobacterial HflX is a ribosome splitting factor that mediates antibiotic resistance. Proc Natl Acad Sci U S A 117(1):629–634. https://doi.org/10.1073/pnas.1906748117
Sauer E (2013) Structure and RNA-binding properties of the bacterial LSm protein hfq. RNA Biol 10(4):610–618. https://doi.org/10.4161/rna.24201
Schachterle JK, Zeng Q, Sundin GW (2019) Three hfq-dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora. Mol Microbiol 111(6):1476–1492. https://doi.org/10.1111/mmi.14232
Seo SW, Kim D, O’Brien EJ, Szubin R, Palsson BO (2015) Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun 6:7970. https://doi.org/10.1038/ncomms8970
Shin GY, Schachterle JK, Shyntum DY, Moleleki LN, Coutinho TA, Sundin GW (2019) Functional characterization of a global virulence Regulator Hfq and Identification of Hfq-Dependent sRNAs in the Plant Pathogen Pantoea ananatis. Front Microbiol 10:2075. https://doi.org/10.3389/fmicb.2019.02075
Sonnleitner E, Moll I, Bläsi U (2002) Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiol (Reading) 148(Pt 3):883–891. https://doi.org/10.1099/00221287-148-3-883
Sun X, Zhulin I, Wartell RM (2002) Predicted structure and phyletic distribution of the RNA-binding protein hfq. Nucleic Acids Res 30(17):3662–3671. https://doi.org/10.1093/nar/gkf508
Sysoev M, Grötzinger SW, Renn D, Eppinger J, Rueping M, Karan R (2021) Bioprospecting of Novel Extremozymes from prokaryotes-the Advent of Culture-Independent methods. Front Microbiol 12:630013. https://doi.org/10.3389/fmicb.2021.630013
Tan Z, Fan J, He S, Zhang Z, Chu H (2023) sRNA21, a novel small RNA, protects Mycobacterium abscessus against oxidative stress. J Gene Med 25(6):e3492. https://doi.org/10.1002/jgm.3492
Trouillon J, Han K, Attrée I, Lory S (2022) The core and accessory Hfq interactomes across Pseudomonas aeruginosa lineages. Nat Commun 13(1):1258. https://doi.org/10.1038/s41467-022-28849-w
Tsui HC, Winkler ME (1994) Transcriptional patterns of the mutl-miaa superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie 76(12):1168–1177. https://doi.org/10.1016/0300-9084(94)90046-9
Tsui HC, Leung HC, Winkler ME (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13(1):35–49. https://doi.org/10.1111/j.1365-2958.1994.tb00400.x
Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9(8):578–589. https://doi.org/10.1038/nrmicro2615
Walsh IM, Bowman MA, Soto Santarriaga IF, Rodriguez A, Clark PL (2020) Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc Natl Acad Sci U S A 117(7):3528–3534. https://doi.org/10.1073/pnas.1907126117
Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136(4):615–628. https://doi.org/10.1016/j.cell.2009.01.043
Wilusz CJ, Wilusz J (2013) Lsm proteins and hfq: life at the 3’ end. RNA Biol 10(4):592–601. https://doi.org/10.4161/rna.23695
Wu H, Song S, Tian K, Zhou D, Wang B, Liu J, Zhu H, Qiao J (2018) A novel small RNA S042 increases acid tolerance in Lactococcus lactis F44. Biochem Biophys Res Commun 500(3):544–549. https://doi.org/10.1016/j.bbrc.2018.04.069
Wu C, Zhu G, Ding Q, Zhou P, Liu L, Chen X (2020) CgCmk1 activates CgRds2 to resist Low-pH stress in Candida Glabrata. Appl Environ Microbiol 86(11). https://doi.org/10.1128/AEM.00302-20
Yao Y, Wunier W, Morigen M (2015) Absence of the BaeR protein leads to the early initiation of DNA replication in Escherichia coli. Genet Mol Res 14(4):16888–16895. https://doi.org/10.4238/2015.December.14.16
Yin Y, Tong Y, Yang H, Feng S (2022) EpsRAc is a copper-sensing MarR family transcriptional repressor from Acidithiobacillus caldus. Appl Microbiol Biotechnol 106(9–10):3679–3689. https://doi.org/10.1007/s00253-022-11971-6
Yu L, Li W, Xue M, Li J, Chen X, Ni J, Shang F, Xue T (2020) Regulatory Role of the two-component system BasSR in the expression of the EmrD Multidrug Efflux in Escherichia coli. Microb Drug Resist 26(10):1163–1173. https://doi.org/10.1089/mdr.2019.0412
Zhao Y, Ren J, Jiang H, Chen X, Xu M, Li Y, Zhao J, Chen D, Zhang K, Li H (2021) Metabolomics and lipidomics analyses delineating hfq deletion-induced metabolic alterations in Vibrio alginolyticus. Aquaculture 535:736349. https://doi.org/10.1016/j.aquaculture.2021.736349
Zhu C, Chen J, Wang Y, Wang L, Guo X, Chen N, Zheng P, Sun J, Ma Y (2019) Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli. Biotechnol Bioeng 116(8):2018–2028. https://doi.org/10.1002/bit.26981
Acknowledgements
We are grateful to Linzhi Ding for his help in three-dimensional excitation emission matrix (3D-EEM) analysis.
Funding
This study was supported by grants from the National Key Research and Development Program of China (2022YFC3401304/001), the National Natural Science Foundation of China (No. 32371540; 21878128), the funding of Key Laboratory of Industrial Biotechnology, Ministry of Education (KLIBKF202005).
Author information
Authors and Affiliations
Contributions
WH and SF designed the research. WH, XH, HB, and ZC performed the experiments. WH and XH analyzed the data. WH wrote the first draft of the manuscript. SF, HY, and JZ contributed to manuscript revision, read, and approved the submitted version. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, W., Huo, X., Bai, H. et al. Insights into the complementation potential of the extreme acidophile’s orthologue in replacing Escherichia coli hfq gene—particularly in bacterial resistance to environmental stress. World J Microbiol Biotechnol 40, 105 (2024). https://doi.org/10.1007/s11274-024-03924-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-03924-0