Abstract
Hyperhydricity (HH) is one of the most important physiological disorders that negatively affects various plant tissue culture techniques. The objective of this study was to characterize optical features to allow an automated detection of HH. For this purpose, HH was induced in two plant species, apple and Arabidopsis thaliana, and the severity was quantified based on visual scoring and determination of apoplastic liquid volume. The comparison between the HH score and the apoplastic liquid volume revealed a significant correlation, but different response dynamics. Corresponding leaf reflectance spectra were collected and different approaches of spectral analyses were evaluated for their ability to identify HH-specific wavelengths. Statistical analysis of raw spectra showed significantly lower reflection of hyperhydric leaves in the VIS, NIR and SWIR region. Application of the continuum removal hull method to raw spectra identified HH-specific absorption features over time and major absorption peaks at 980 nm, 1150 nm, 1400 nm, 1520 nm, 1780 nm and 1930 nm for the various conducted experiments. Machine learning (ML) model spot checking specified the support vector machine to be most suited for classification of hyperhydric explants, with a test accuracy of 85% outperforming traditional classification via vegetation index with 63% test accuracy and the other ML models tested. Investigations on the predictor importance revealed 1950 nm, 1445 nm in SWIR region and 415 nm in the VIS region to be most important for classification. The validity of the developed spectral classifier was tested on an available hyperspectral image acquisition in the SWIR-region.
Key message
This study provides an approach that paves the way to automatic detection of hyperhydricity by identifying the key spectral features of this phenomenon.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyperhydricity (HH) represents one of the major challenges for increasing the efficiency of plant in vitro propagation as it limits plant quality, adventitious root formation and ex vitro survival rate, in particular when using liquid culture or bioreactor systems (Cardoso et al. 2018; Debergh et al. 1992; Gribble 1999). According to Kemat et al. (2020), at least 200 species are sensitive to HH and around 150 species can be affected seriously by HH emphasizing the relevance for commercial micropropagation. HH not only restricts the propagation of in vitro plants, but also affects the efficiency of genetic transformation mediated by Agrobacterium (van Altvorst et al. 1996) and the conservation of important species in germplasm banks (Lizárraga et al. 2017).
HH is a physiological disorder occurring under the specific conditions of plant tissue culture such as high humidity, high supplementation of sucrose, impaired gaseous exchange capacity and consequently low photosynthetic activity (George et al. 2008; Ziv 1991). The work of van den Dries et al. (2013) and Rojas-Martínez et al. (2010) provided strong evidence that the underlying mechanism of the HH etiology is the flooding of the apoplast, resulting in hypoxia and causing oxidative stress. This in turn leads to the macroscopic symptoms of water-soaked, wrinkled, curled, brittle and translucent tissue. The occurrence of HH was shown to be increased when the water availability for the in vitro explant was increased (Smith and Spomer 1995), e.g., by reduced concentration of the gelling agent (Ivanova and Van Staden 2011), or by the type of the gelling agent used (Pasqualetto et al. 1988; Tsay et al. 2006). The gelling agent gelrite induced HH in a wide range of plant genera (e.g., Arabidopsis sp., van den Dries et al. 2013, Malus sp. Pasqualetto et al. 1988, Prunus sp. Franck et al. 1998), even though the same gel strength as agar was used.
In addition to the anatomical changes of hyperhydric tissue which include larger intercellular spaces in the mesophyll and a drastically reduced number of palisade cells (Vieitez et al. 1985), several biochemical changes of hyperhydric tissue such as decreased chlorophyll contents (Phan and Letouze 1983; Franck et al. 1998), hypolignification (Kevers et al. 1987; Kemat et al. 2021), and high apoplastic water volume (Dries et al. 2013; Tian et al. 2015; de Klerk and Pramanik 2017) were reported. Paques et al. (1985) refer to HH as an inducible and reversible phenomenon and demonstrated that Malus sp. ‘M26’ plantlets could return to non-hyperhydric state if the induction phase in liquid culture did not exceed five days or if the symptoms of HH were not too severe. Recently, there were reports that hyperhydricity can be reversed by supplementation of agents to media such as silver nitrate and trichloroacetate (Gao et al. 2017; de Klerk and Pramanik 2017) or by controlling the environmental conditions in addition to media optimization (Mohamed et al. 2023), but no general countermeasure has been derived up to now. In commercial in vitro laboratories visual monitoring for contaminations and disorders are part of the routine work and therefore a costly and time-consuming repetitive matter (Mestre et al. 2017).
Nowadays, digitalization enters the horticultural sector, driven by digital solutions to increasingly complex work processes achieved through technological advances in sensors, automation and robotization, as well as data analysis through classical and advanced machine learning (ML) techniques. Automation of processes offers great economic potential for micropropagation laboratories since 60–70% of total costs of a micropropagated plant is due to manual labor (Chen 2016). An increasing number of reports on automating micropropagation processes such as explant cutting (Huang and Lee 2010), the commercial laser-based robotic cut and transplanting system RoBo®Cut (Bock Biosciences GmbH 2018), monitoring of cultures (Dhondt et al. 2014, Bethge et al. 2023) and transplanting of explants (Lee et al. 2019) were published within the recent years. In addition, there are several studies on the application of computer vision to micropropagation (Smith et al. 1989; Aynalem et al. 2006; Dhondt et al. 2014; Gupta and Karmakar 2017; Mestre et al. 2017) with imaging sensors being the crucial technology. Imaging sensors used in horticulture consist of affordable RGB cameras, multispectral cameras, thermal cameras, expensive hyperspectral imaging (HSI) systems, ToF (Time of Flight), LIDAR systems (Light Detection and Ranging) and more. The different sensor systems can be discriminated by their operating spectral range (UV, VIS, NIR, SWIR, MWIR, LWIR/Thermal-IR), spectral resolution from one (monochrome) to > 100 (hyperspectral) channels and cost of purchase. For example, the price of silicon (Si)-based hyperspectral cameras rise by a factor of 2 to 20 when switching the operating spectral range from VIS/NIR (400–1000 nm) to SWIR (900–1700 nm) with an Indium-Galium-Asenide (InGaAs) camera chip (Tisserand 2021). This needs to be considered, when selecting the appropriate spectral range and corresponding imaging technology. While computer vision coupled with ML offers already great potential to solve complex detection task in agriculture (reviewed in Patrício and Rieder 2018), for application in plant tissue culture only few reports are available up to now (reviewed in Prasad and Gupta 2008; Hesami and Jones 2020). However, these are limited in terms of live-monitoring, since they followed the “object to sensor” approach for plantlet clustering (Mahendra et al. 2004), classification of somatic embryos (Zhang et al. 1999) and estimation of shoot length (Honda et al. 1997).
The visual appearance of plants, and in particular leaf pigments, can be estimated by spectroscopic approaches based on their interaction with electromagnetic radiation. Single biochemical plant metabolites can be associated with specific wavelengths based on their major absorption peaks (Table 1).
Univariate data analysis, e.g., spectral indices or multivariate data analyses like partial least square (PLS), allows the prediction of leaf pigments’ concentrations and can be used for classification. These techniques also enable the discrimination of different plant species or the identification of growth anomalies by specific spectral features (Shaw and Kelley 2005). According to Hesami and Jones (2020), ML techniques applied to plant tissue culture problems will help in future to solve classification and regression problems and can be employed for automation and mechanization of in vitro propagation, genetic engineering and genome editing technologies. In addition, Nezami-Alanagh et al. (2019) demonstrated the positive impact of ML models in optimizing culture media in terms of time, cost and the occurrence of physiological disorders in the propagation of pistachio rootstocks. Prasad and Gupta (2008) proposed that an automated decision-making system based on computer vision coupled with ML models and combined with a robotic system will result in the mechanization of commercial mass propagation and help in evaluating various aspects of plant quality such as HH status, which might be difficult to determine by human visual inspection. To our knowledge, the spectral properties of HH have not yet been studied or used as a distinguishing feature for ML classification of in vitro cultured explants.
The objective of this study was to investigate the spectral fingerprints of hyperhydric tissue in two different plant species (Malus sp. and Arabidopsis thaliana) after forced induction of the growth anomaly and subsequent spectral analysis of the explants. Here, we selected Malus as a representative of classical in vitro shoot cultures and Arabidopsis as a model plant for the underlying mechanism of HH. A novel phenotyping system was tested to monitor the morphological characteristics of hyperhydric explants in time-series image data. Furthermore, we aimed at identifying specific absorption features of hyperhydric tissues that are sufficient for discrimination by ML techniques and to locate them within in the electromagnetic radiation spectrum. Putative discriminating models should be validated and discussed in terms of their feasibility in plant tissue culture. The findings of this study should pave the way for an automatic detection of HH by live-monitoring of in vitro cultures.
Material and methods
Plant material and experimental setup
Morphological characteristics of hyperhydricity
From in vitro apple shoot cultures (Malus sp. ‘G214’) uniform shoots of 10–15 mm length were prepared and cultivated on modified MS medium (Murashige and Skoog 1962) containing 2.2 µM 6-benzylaminopurine (BAP), 0.5 µM indole-3-butyric acid (IBA), 3% (w/v) sucrose and solidified with either 0.8% (w/v) agar (Plant agar, Duchefa, Haarlem, The Netherlands) for the control variant (“MS + agar”) or with 0.25% (w/v) gelrite (Duchefa, Haarlem, The Netherlands) for the HH induction variant (“MS + gelrite”). The pH of the medium was adjusted to 5.8 prior to autoclaving at 121 °C for 15 min.
Arabidopsis thaliana ‘Col-0’ seeds which had been stored at 4 °C, were surface-disinfected using 70% (v/v) isopropanol for 30 s, followed by 2% (v/v) sodium hypochlorite plus Tween 20 for 5 min and then rinsed thoroughly three times using sterile deionized water. The seeds were germinated for 10 days at 24 °C in 9 cm-Petri dishes (polystyrene) on modified plant growth regulator-free B5 medium (Gamborg et al. 1968), containing 1.5% (w/v) sucrose with 0.8% (w/v) Plant agar and pH 5.8. Uniform 10 day-old seedlings were selected and five seedlings per 500 mL-vessel were transferred to modified plant growth regulator-free B5 medium (Gamborg et al. 1968), containing 1.5% (w/v) sucrose and either 0.8% (w/v) Plant agar for the control variant (“B5 + agar”) or 0.25% (w/v) gelrite (“B5 + gelrite”) to induce HH. The pH of the medium was adjusted to 5.8 prior to autoclaving at 121 °C for 15 min.
Ten 500 mL polypropylene vessels were prepared for Experiment I (Table 2) and Experiment II, each with four plantlets and containing ~ 80 mL of one of the two different media (“B5/MS + agar”/“B5/MS + gelrite” supplemented with 1 g L-1 titanium dioxide). Titanium dioxide (food dye; Ruth GmbH & Co.KG, Bochum, Germany) was used to add a white color to the medium, because this enabled the height measurements of the robot system due to increased reflection of the culture media. A plastic film (PVC system foil; Klarsichtpackung GmbH, Hofheim, Germany) sealed the containers as a substitution of the lid of the containers to provide a fully transparent view while ensuring the aseptic condition of the cultures. These cultures were cultivated at 22 °C with a 16 h photoperiod and under a PPFD (Photosynthetic Photon Flux Density) of 35–40 \(\mathrm{\mu mol}{\mathrm{ m}}^{-2}{\mathrm{s}}^{-1}\), provided by two tubular fluorescent lamps (Philips MASTER TL-D 58W/865). The lab’s bottom-cooling system—provided by water-cooled plastic tubes below the shelf—prevented water condensation due a local shift of dew point. Room temperature ranged from 19 (night) to 25 °C (day) with an average of 22 °C over 24 h, while the average surface temperature of the cooled cultivation area ranged from 19 (night) to 24 °C (day) with an average of 21 °C over 24 h. In addition to the non-destructive monitoring approach (Exp. I & II), three experiments (Exp. III, Exp. IV, Exp. V; Table 2) were conducted with different evaluation time points. The evaluation time points were chosen based on the key events in the dynamic etiology of hyperhydricity during a culture passage (~ 4–5 weeks for Malus). Important morphological changes were observed during the first two weeks, so Exp. III and V covered this time span, while measurements in Exp. IV were undertaken to cover the second half of the culture passage.
Hyperhydricity induction
For Malus shoot cultures, 500 mL polypropylene containers containing 80 mL of the two different media were used and each container was inoculated with five shoots. Cultivation took place for 20 days (Table 2; Exp. III), 28 days (Exp. IV) and 16 days (Exp. V) at 22 °C (room temperature ranged from 19 (night) to 25 °C (day) with an average of 22 °C over 24 h) with a 16 h photoperiod and under a PPFD (Photosynthetic Photon Flux Density) of 35–40 µmol m−2 s−1, provided by tubular fluorescent lamps (Philips MASTER TL-D 58W/865). Arabidopsis plantlets were cultivated as described above for 20 days (Exp. III).
Evaluations
Morphological characteristics of hyperhydricity via image analysis
For visualization of the etiology of HH, the multisensory robot system “Phenomenon” (Bethge et al. 2023) was used. RGB images were captured in Exp. I and Exp. II every 4 h with a 12.3-megapixel RGB camera (Raspberry Pi Camera HQ, Raspberry Pi Foundation, Cambridge, UK) equipped with a 6 mm fixed focal length low-distortion lens (Edmund Optics: 6 mm wide angle lens, f/1.2, high resolution = 120 lp mm−1 (lp = line pairs), low distortion < 0.5%) and with the following camera parameters: resolution = 4054 px × 3040 px, shutter speed = 2000 ms, iso = 100, autowhite-balance = off and a fixed gain of 3.3, 1.5 (red, blue).
Sensor data from the multisensory robot system “Phenomenon” (Bethge et al. 2023) were processed, segmented, and various parameters were calculated. RGB image analysis was performed in Python (Van Rossum and Drake 2009), using the following packages: OpenCV v3.4.9 (Bradski 2000), NumPy v1.20.2 (Van Der Walt et al. 2011) and PlantCv v3.11.0 (Gehan et al. 2017) and the Software toolkit Ilastik v1.3.3 (Sommer et al. 2011) headless integrated in the Python script. RGB image analysis included a histogram stretching for normalization, segmentation via a trained random forest classifier, normalization to the day 0 plant area and calculation of projected plant area (37.7 px = 1 mm). Shape analyses were performed on the four largest objects by area and limited to the first nine days to avoid errors from overlapping explants. We used the installed shape function of PlantCv to calculate solidity (measure of density as the ratio between object area and area of the convex hull of the object) and eccentricity (measure of deviation of an ellipse to a circle (eccentricity = 0) as the ratio between major and minor axis).
Depth data were acquired once per day for each culture container with the point-measuring laser distance sensor as a spatial scan by sequential readout of the sensor while shifting the detector head of the “Phenomenon” robot system in xy direction, according to the scan pattern (100 mm × 100 mm; with a resolution of 1 mm × 1 mm). The laser distance sensor (OD-Mini OB1-B100, Sick AG, Waldkirch, Germany) used in this setup was specified by the manufacturer with a power consumption of < 1.92 W, laser emission wavelength of 655 nm, max. output of 390 µW (laser class 1), a measuring range of 50 to 150 mm and a linearity of ± 100 µm as well as spot size of 700 µm × 600 µm at a measuring distance of 100 mm. The analog output of the laser distance sensor (10 V) was connected via a small voltage divider circuit to a high precision 16-bit A/D-converter (ADS 1115), which communicated via Inter-Integral Circuit (I2C) with a microcontroller board (Wemos D1 Mini). Each distance measurement consisted of a up to ten single readouts and averaging (excluding default sensor values), to achieve a robust and low-noise measurement. A detailed description of the robot system “Phenomenon” can be found in Bethge et al. (2023).
Depth data of explants were obtained by measuring 10,000 data points of each culture vessel once a day with a scanning laser distance sensor. The depth data processing pipeline included the segmentation of culture media by a RANSAC (random sample consensus, Fischler and Bolles 1981) segmentation approach, subtraction of RANSAC plane, normalization to the day 0 plant height with the Python libraries: Open3D v0.15.1 (Zhou et al. 2018) and Pyvista v0.34.0 (Sullivan et al. 2019). Pipelines construction is described in detail in Bethge et al. (2023).
Statistical analysis of repeated measures data was performed using R software. Data were transformed, if necessary, with the R package bestNormalize v1.8.2 (Peterson and Peterson 2020). Different linear mixed-effect models from nlme v3.1-153 package (Pinheiro et al. 2017) were fitted to the data with different covariance structures: scaled identity, first-order autoregressive, first-order heterogeneous autoregressive, compound symmetry, Toeplitz and heterogenous Toeplitz. The mean model consisted of the fixed effects treatment/medium type and time and their interaction terms. An extra random effect was included in the model to account for the dependencies between measurements from the same culture container or in SI. 1 for shape analysis from the same explant (as nested random effect). We also included linear models with random intercept (CulturecontainerID) and random slope (Time). The respective best model (Fig. 1A: linear mixed model with scaled identity covariance structure and random slope; Fig. 1B and SI. 1A: linear mixed model with heterogenous Toeplitz covariance structure and random slope; SI. 1B: linear mixed model with heterogenous Toeplitz covariance structure and random intercept) was selected based on the Akaike Information Criterion (AIC, Sakamoto et al. 1986) values and residual analysis (QQ-plot). Pairwise comparisons using Tukey’s HSD test at p < 0.05 was performed and show significant differences between treatments within a time point.
Morphological differences in growth patterns of explants of A. thaliana Col-0 and Malus ‘G214’ cultivated on either agar or gelrite solidified media (Mean ± SD). A The curve for the increase in the projected plant area was calculated from the analysis of the segmented RGB images normalized to the plant area of day 0 and presented as projected plant area [cm2]. Since flower initiation started at later time points for A. thaliana and thus an error in the estimation of projected plant area might occur, the analysis of growth curves was limited to the first ten days. B The relative increase in mean canopy height resulted from analysis of segmented depth data collected with a scanning laser distance sensor and normalized to day 0 plant height. Yellows lines indicates cultivation on standard media formulation on Gamborg-B5 (A. thaliana) and MS-Medium (Malus) solidified with 0.8% agar (w/v), while dark gray lines display the cultivation on induction media containing 0.25% (w/v) gelrite, inducing HH. C Representative images at the endpoint of the experiments. Sample number (n) indicates the individual culture containers. Significance stars indicate comparisons of treatments within a time point (day) with *p < 0.05, **p < 0.01, ***p < 0.001. RGB and depth data were acquired with the multisensory robot system “Phenomenon” (Bethge et al. 2023). (Color figure online)
Visual scoring of hyperhydricity severity level
In the Experiments III to V, the severity of HH was assessed for each explant and at every time point (Exp. III: 0, 5, 10, 15, 20 days; Exp. IV: 14, 21, 28 days; Exp. V: 0, 4, 8, 12, 16 days) according to Tian et al. (2015) with minor modifications (Table 3). The starting plant material cultivated on control media represented the samples of 0 days after transfer (DAT 0).
Determination of apoplastic liquid volume
Per time point at least 10 samples per treatment were collected for the determination of apoplastic liquid volume, with DAT 0 samples representing the starting material. Apoplastic liquid was extracted from leaf tissue by mild centrifugation according to van den Dries et al. (2013) and Terry and Bonner (1980): Leaves (50–150 mg FM) from a single explant were excised, weighed, and placed into a 2 mL tube microcentrifuge filter without membrane (ClearLine®; Kisker Biotech GmbH & Co, Steinfurt, Germany). Samples were centrifuged at 3000 g for 20 min at 4 °C. Immediately after centrifugation, the leaves were reweighed to determine the apoplastic liquid volume (VAL) in µL g−1 fresh mass (FM) using the Eq. 1.
where FM = fresh mass of leaves in mg, Mac = mass of leaves after centrifugation and ρH2O = water density (the water density was taken as equal to 1 g mL−1 assuming the apoplastic liquid is mainly water and has a temperature of 4 °C).
Spectral data acquisition and analysis
Prior to the quantification of apoplastic liquid volume, one fully expanded leaf per explant under study was collected. The leaf was then placed in a 3D printed sample holder (SI. 2) in an adaxial position that allowed for flat clamping without exerting too much pressure on the leaf (with a cavity of 1 mm). The curled hyperhydric leaves were handled with care to obtain reflection spectra from a planar surface. The leaf reflectance spectra were examined with a Perkin-Elmer Lambda 900 UV–VIS-NIR-SWIR spectrometer (Perkin-Elmer Instruments, Norwalk, USA) equipped with 150 mm Indium-Gallium-Arsenide (InGaAs) integrating sphere. The reflectance intensity was measured in steps of 1 nm in the wavelength range between 200 and 2000 nm, and the reflectance was calculated using the reflection spectrum of the white reference standard Spectralon®. Raw spectra were pre-processed in R v4.1.2 using Rstudio (RStudio Team 2015) with the hsdar v1.0.4 package (Lehnert et al. 2018) allowing the cleaning of device errors, trimming to spectral range of 400 mn to 2000 nm and smoothing with the Savitzky-Golay filter at a window size of 25 data points of third-degree polynomials to remove noise from data.
Spectra of leaves obtained from three experiments (Exp. III: 147, Exp. IV: 39 spectra, Exp. V: 51) were divided into two groups based on the significance level of the apoplastic liquid volume and the HH score of the whole explant was assessed by visual observation. Here, the explants with a HH score of 0 and 1 were classified as normal explants while the explants with a HH score of 2 to 4 represented hyperhydric explants. This resulted in 100 and 137 spectra of normal and hyperhydric leaves, respectively, covering the two plant species Malus ‘G214’ (187 spectra) and A. thaliana (50 spectra). For visualization and isolation of the HH-specific absorption features, leaf spectra were further processed with the segmented upper hull continuum removal method described in detail in Lehnert et al. (2018). This normalization method allowed a comparison of individual absorption features on a common baseline formed by a segmented upper hull of local maxima and resulted in absorption features spectra. In addition, difference spectra of absorption feature spectra were calculated by subtracting normal leaf spectra from hyperhydric leaf spectra.
In addition, we defined three spectral ranges based on the sensitivity of the state-of-the-art sensor technologies such as standard RGB camera systems with silicon sensor chips (3 channels: B: 400 nm to 500 nm, G: 500 nm to 600 nm and R: 600 nm to 700 nm), multispectral camera systems with silicon sensor chips (4 channels: B: 400 nm to 500 nm, G: 500 nm to 600 nm, R: 600 nm to 700 nm and NIR: 750 nm to 850 nm) and SWIR-HSI camera systems with Indium-Gallium-Arsenide (InGaAs) sensor chips (SWIR: 900 nm to 1700 nm). This division was made as a decision support for assessing the potential of the candidate detection systems to detect HH based on their spectral sensitivity range and considering their affordability.
Identification of hyperhydricity-specific absorption features
Different ML models were trained with the caret v6.0-90 package (Kuhn 2008) in the R software to identify the key absorption features that discriminate between normal and hyperhydric explant leaf spectra. Here, pre-processed spectral data sets (237) were centered and scaled and divided into a training set (178 spectra; Malus: 143, A. thaliana: 35, with 103 normal and 75 hyperhydric explants, in total) and a test set (59 spectra; Malus: 44, A. thaliana: 15, with a total of 34 normal and 25 hyperhydric explants). All classification models were trained with the same resampling procedure consisting of a 10 times tenfold repeated cross validation (CV). The tenfold repeated CV divides the training data into 10 equal parts (10 subsamples with a size of 178/10). These parts are iterated 10 times, during each iteration, 9 of the 10 parts serve as training data, and the remaining 10th part as the validation set to calculate model performance metrics. In 10 times repeated tenfold CV this process is repeated 10 times; therefore, performance of training was validated on 100 validation subsamples consisting of 17–18 individual spectra.
In the confusion metrics, correctly classified normal and hyperhydric leaves formed the true-positive (TP) and the true-negative (TN) class, while false classified ones constituted the false-positive (FP) and false-negative (FN) class, respectively. For evaluation of model validation performance, the sensitivity (Eq. 2; TPR: true positive rate) and the specificity (Eq. 3; TNR: true negative rate) were calculated with normal explants as the positive class and the area under the curve (AUC) of the receiver-operator-characteristics (Eq. 4; AUCROC), while for evaluation of model test performance, the accuracy (Eq. 5) was determined. Here, misclassifications are described by the false negative rate (FNR) and false positive rate (FPR). Balanced accuracy (Eq. 6) and F1 score (Eq. 7) were calculated to account for putative class imbalances. To find the best suitable model for discriminating between normal and hyperhydric leaf spectra, different ML model structures were tested, including a neuronal net with the maximum allowable number of weights set to 2000 (“nnet” from nnet v7.3-16 package; Ripley et al. 2016), a linear discriminate analysis (“lda” from caret package), a supported vector machine (“svmLinear” from caret package), a random forest (“rf” from caret package), a high dimensional discriminate analysis (“hdda” from caret package) as well as a linear discriminate analysis (“lda” from caret package with PCA-preprocessed data set) with an upstream principal component analysis (PCA). Based on their resampled performance metrics, the best model was selected to identify its most relevant features/wavelengths on the basis of the underlying variable importance in the model.
Automated hyperhydricity detection
To test the validity of the developed spectral classifier, an HSI-system operating in the shortwave infrared (SWIR) region was used to acquire a single HSI data cube from a culture vessel containing a HH-sensitive apple genotype (Malus ‘Selection 4’). The imaging system that was developed and described by Thiel (2018) consisted of an EVK Helios Core NIR Line-scan camera (240 px × 1 px and 252 spectral channels in the wavelength region of 900 nm to 1700 nm), two 65 W halogen spot lights and a conveyer-belt system to move the sample. Image acquisition was performed in closed polypropylene culture vessels, so that sterile conditions could be maintained inside the vessel and water condensation was prevented by heat radiation from the halogen lamps. Since only one HSI data cube could be acquired, these results were considered to be an exemplary and preliminary validation test.
The developed spectral classifier was retrained with a reduced number of features to match the spectral channels of the imaging system (Features/wavelengths: 252 channels in the range between 900 and 1700 nm). Due to the binary classification output of the classifier, most of the background pixels were removed by creation of a binary mask with simple thresholding of the image slice at a wavelength of 1000 nm. Then each pixel of the segmented hyperspectral data cube was inserted as an input to the spectral classifier and class membership was predicted.
As a more affordable approach and as a proof of concept, an object detection model based on annotated RGB images acquired by the robot system was trained. Therefore, 250 images were randomly selected and annotated with the graphical user interface Roboflow© (Dwyer et al. 2022). The image data set consisted of 200 annotated images of eight culture containers from Experiment II and 50 images from a comparable experiment to increase variance in the number of explants, background colour, and colour of culture media. A total of 504 normal explants and 545 hyperhydric explants were included. The image data set was divided into 175 images as training set, 50 images as validation set and 25 images as test set. Data augmentation of annotated bounding boxes increased the training set to 1800 images and included: horizontal and vertical flip, rotation by 90° (clockwise, counter-clockwise, upside down), rotation by ± 5°, brightness by ± 10%, exposure by ± 7%, blur with 2px and noise with 2% of pixels. The data set (Bethge 2023) is publicly accessible via Roboflow© universe. Time series images of two culture vessels from Experiment II were retained and used to visualize the trained model. Object detection models perform attempts to identify and locate objects in images while assigning them to the appropriate classes. We selected YOLOv8 (Jocher et al. 2023) architecture as the latest versions of the YOLO (“You only look once”, Redmond et al. 2016) family. YOLO is a single-stage object detector, consisting of three parts in its architecture: backbone, neck and head. The backbone is defined by several convolutional layers which extract key features from the images, the neck uses the features and forms the feature pyramid by fully connected layers and the head is the final output layer for prediction of bounding boxes and classification. The training process was performed in the Google Colaboratory (Colab/Colab Pro) environment on a NVIDIA A100-SXM4-40 GB graphical processing unit (GPU) serviced by Google. In addition, the model was trained with the following parameters: epochs = 250 (early stopping occurred after 188 epochs), batch size = 16 images, image size = 640 px, patience = 100 epochs, learning rate = 0.01, momentum = 0.94, intersection over union (IoU) = 0.7. We let Roboflow train two object detection models, one from scratch and one with weights from a previously trained model (additional 125 images from the same experiment) to see the full potential of the dataset with the optimized pipeline. Evaluation of model performance was based on precision (Eq. 8), recall (Eq. 9), average precision (AP; Eq. 10) and mean average precision (mAP; Eq. 11) of the validation set. Here true positive (TP) indicate a correct detection and classification, false negative (FN) describes cases where the prediction missed the detection contained in the ground truth data, while in a false positive (FP) case a bounding box was predicted on a location not contained in the ground truth data. Thereby, AP represents the area under the precision-recall-curve across a range of probability confidence threshold values from 0 to 1. The mAP is the sum of AP of each class (k) divided by the number of classes (n) at a given intersection over union (IuO) threshold of 0.5. Intersection over union is defined as ratio between the overlap area to the united area of the predicted and ground truth bounding box. After the training process predications were obtained using the Python library roboflow v0.2.25 (Dwyer et al. 2021) with IuO threshold and confidence threshold set to 0.5.
Morphological characteristics of hyperhydricity via image analysis
Studying the morphology of the shoots of the two treatments revealed major differences in horizontal and vertical growth. Significantly stronger growth, quantified as projected plant area, was observed for the gelrite treatment at early time points (5 days) for both plant species (Fig. 1A). After 4 weeks of cultivation, shoots of Malus in culture vessels with gelrite medium had with 24.5 cm2 a 2.4 times greater increase in projected plant area than shoots in vessels with agar medium with 10.3 cm2. Here, 65% of the explants of Malus had a HH score > 2 in the gelrite treatment compared to 0% for agar treatment. For A. thaliana we evaluated the projected plant area only until day 10 to avoid distorting effects on projected plant area due to flower initiation starting at day 12. Shape analysis of single explants showed significant differences in solidity at day 3 and in eccentricity at day 6 for A. thaliana, whereas the shape differences of Malus explants were not significant (SI. 1). Vertical growth analysis, quantified as mean canopy height (Fig. 1B) and maximum shoot height as mean of upper 10th percentile (SI. 1), showed a significantly higher mean canopy height of Malus for the gelrite treatment at day 18 and of A. thaliana at day 16. An even earlier distinction was recorded for the maximum shoot height, i.e. at day 11 and 14 for A. thaliana and Malus, respectively.
Hyperhydricity induction
Visual scoring of HH revealed the dynamics of HH induction using gelrite in the two plant species under investigation. Anthocyanin accumulation was noted within the first 4 days in both treatments for Malus. However, it persisted only in the gelrite treatment until the end of the experiment in most explants. In Malus, severe symptoms of HH were induced even on the agar control medium in 12.5% of the shoots (Fig. 2A). In two experiments, significant differences in the HH score and the occurrence of severe symptoms (curled, thickened and translucent leaves = level 4 of the HH score) between the agar control and the gelrite induction treatment were identified 10 days (Fig. 2A) and 8 days (SI. 3) after transfer. When performing this experiment under a novel phenotyping system, time-lapse videos were taken. They confirmed these observations and visualized the temporal development of HH in the two plant species (Malus: SI. 4 and A. thaliana: SI. 5). Three of four apple shoots turned into a hyperhydric status and formed first hyperhydric leaves (SI. 4: arrows) on gelrite at 5 DAT (SI. 4: 100 h). They started to curl at 8 DAT and also became much larger than those on agar. After 27 days, severe symptoms appeared on dark green to reddish explants that exhibited compact growth with curled, epinastic, and brittle leaves.
Visual scoring of hyperhydricity of A Malus ‘G214’ and B A. thaliana Col-0 in vitro cultures over 20 days (DAT, Days After Treatment). Samples from 0 days after transfer (DAT 0) represent the starting plant material cultured on control media. Yellows bars indicate cultivation on standard media formulation A MS-Medium, B Gamborg-B5 solidified with 0.8% (w/v) agar, while gray bars display the cultivation on induction media containing 0.25% (w/v) gelrite. Dashed lines represent the medians of each histogram. Sample number (n) indicates the individual explants. The different sample numbers result from the combined evaluation with different methods (apoplastic liquid evaluation, reflection spectroscopy) of the same samples. Different letters resulting from Kruskal–Wallis test followed by Fisher’s LSD (p < 0.05) indicate significant differences between histograms. Kruskal–Wallis effect size could be determined to be very strong with A η2 = 0.62 and B η2 = 0.65. (Color figure online)
For A. thaliana, there was already a significant increase in the HH score after 5 days of treatment (Fig. 2B). Furthermore, decolorization of leaves was the predominating symptom of HH in A. thaliana on gelrite induction medium. For A. thaliana seedlings, first signs of HH (SI. 5: arrows) became visible at 5 DAT (SI. 5: 100 h) on gelrite-solidified medium and shoots developed longer petioles and much larger leaves with severe HH symptoms.
The apoplastic liquid volume increased steadily for Malus (Fig. 3A) until 15 DAT, while Experiment IV (SI. 3) demonstrated a decrease at later time points: 21 days and 28 days. A significant difference in apoplastic liquid content in both plant species was detected at the earliest time point: 4 DAT (SI. 3) and 5 DAT (Fig. 3A and B), where the apoplastic liquid volumes of explants on gelrite induction media were already twice as high as those of explants on agar control media. In Malus, three independent experiments (Fig. 3A and SI. 3) allowed us to confine the time of peak in apoplastic liquid volume at 12 to 16 DAT. Apparently, up to this timepoint, quantification of apoplastic liquid volume reflected the HH score well—even the occurrence of some hyperhydric explants on the agar control medium was also reflected in the increase in apoplastic water volume (Fig. 3A vs. SI. 3). However, at later time points, the severity of HH symptoms steadily increased, while apoplastic liquid volume stayed constant.
Apoplastic liquid volume of A Malus ‘G214’ and B A. thaliana Col-0 in vitro cultures over time (DAT, Days After Treatment). Samples from 0 days after transfer (DAT 0) represent the starting plant material cultured on control media. Yellow lines indicate cultures on standard media A MS-Medium, B Gamborg-B5 solidified with 0.8% (w/v) agar, while gray dashed lines display the cultures on HH induction media containing 0.25% (w/v) gelrite (Mean ± SD). The values of A. thaliana at DAT 0 B were masked in gray to indicate the authors’ uncertainty, because the plants were very small when the apoplastic water volume was determined at this time, and therefore a large influence of adhering water could not be excluded. Replicate number (n) indicates the individual explants. Different letters resulted from Tukey’s HSD test at p < 0.05 and indicate significant differences when comparing time points within one treatment, while asterisks indicate comparisons of treatments within a time point with * = p < 0.05, ** = p < 0.01, *** = p < 0.001. (Color figure online)
To prove the relation between the objective quantification of apoplastic liquid volume and the HH score determined by visual scoring, data pairs of a total of 349 measurements from both treatments of Malus were used (Fig. 4). We found the highest correlation between HH score and apoplastic volume to be ρ(18) = 0.83 (p < 0.001) 12 DAT using spearman’s rank correlation for all acquired time points for Malus. Interestingly, only three groups could be distinguished significantly by apoplastic liquid volume. Explants with a HH score of two had more than > 50% curled leaves and in average a double amount of apoplastic liquid volume. We therefore restricted the three significant groups to two classes (HH score 0–1: normal explants and HH score 2–4: hyperhydric explants) in further analysis, regardless of the treatment in order to exclude treatment-depended effects on the spectral analysis.
Relation between visual scoring of hyperhydricity and apoplastic liquid volume of Malus ’G214’ (Mean ± SD). Data obtained from three different induction experiments (Exp. III-V) covering time points from 0 to 28 DAT. Replicate number (n) indicates the individual explants. Different letters resulted from Tukey’s HSD test at p < 0.05 and show significant differences between score levels
Spectral analysis of hyperhydricity
The evaluation of leaf explants via UV–VIS-NIR-SWIR spectroscopy (Fig. 5) revealed the first major difference in significantly reduced reflectance in the RGB (400 nm to700 nm) region of 6.5 ± 3.2% for the hyperhydric explants compared to 8.7 ± 3.9% for normal explants. The largest difference in reflectance was recorded for NIR (750 nm to 850 nm) region with a reflectance of 20.5 ± 9.0% hyperhydric explants and normal explants with 28.3 ± 9.5%. Also, for the SWIR (950 nm to 1700 nm) region the overall reflectance was lower in hyperhydric explants (13.8 ± 8.2% for hyperhydric and 21.4 ± 8.7% for normal explants). Differences in average reflectance were most significant in the blue (p< 2.2e-16) region followed by SWIR (p< 4.3e-14), green (p< 1.1e-12), red (p< 2.5e-11) and the NIR region (p< 2.0e-09) according to the results of a Mann–Whitney test.
Raw reflectance spectra of Malus ‘G214’ and A. thaliana Col-0 in vitro leaves. Mean (solid) ± SD (dashed) spectra of normal leaves (N, in green) and hyperhydric leaves (HH, in blue). The distinction was based on visual scoring of HH (N: 0–1 HH score; HH: 2–4 HH score). Wavebands represent different spectral regions, defined by the sensitivity of silicon-based (Si) cameras, such as affordable RGB and RGB-NIR multispectral cameras, and a more expensive Indium-Galium-Asenide-based (InGaAs) detection sensor. Reflectance spectra were measured with an UV–VIS-NIR-SWIR spectrometer (PerkinElmer Lambda 950) in a wavelength range of 200 nm to 2000 nm and at a resolution of 1 nm. (Color figure online)
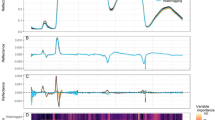
The emergence of HH-specific absorption features over time was recorded applying the continuum removal method to pre-processed spectra of Experiment V and the formation of difference spectra of the isolated band depth spectra, where the absorption features spectra of normal explants were subtracted from absorption features spectra of hyperhydric explants (Fig. 6). Greater absorption of hyperhydric explants was observed as early as 8 DAT, with a maximum at 1402 nm and a full width at half maximum of 157 nm. At later time points the difference in absorbance at around 980 nm, 1150 nm, 1400 nm, 1520 nm and 1780 nm increased negatively, while at around 1930 nm the difference positively increased. In the VIS region, two further local maxima arose at 460 nm and 695 nm at DAT 10, which were also detected at the later time points. However, these peaks can be considered as artefacts of the reduced reflection in the green region due to the continuum removal method based on connection of local maxima. In addition, a consistent positive peak indicating less absorption or higher reflection of hyperhydric explants was found with a maximum around 1930 nm, besides the two local minima at 1400 nm and 1520 nm. When combining data from all experiments (SI. 6), including different time points and the two different plant species, we identified reliable minima (arrows) at 980 nm, 1150 nm, 1400 nm, 1520 nm, and 1780 nm, indicating stronger absorption of the hyperhydric explants, and a reliable maximum at 1930 nm.
Spectral contrasting reflectance of Malus ‘G214’ normal and hyperhydric explants over time (DAT, Days After Treatment). The spectral data used originate from Experiment V. Upper row: Raw reflection spectra; middle row: extracted absorption features after segmented convex-hull removal of raw spectra; bottom row: difference spectrum of the absorption peaks, where the absorption features spectra of normal explants were subtracted from absorption features spectra of hyperhydric explants. Mean (solid) ± SD (dashed) spectra of normal explant leaves (N, in green) and hyperhydric explant leaves (HH, in blue). Distinction of N and HH was based on visual scoring of HH (N: 0–1 HH score; HH: 2–4 HH score). Arrows indicate putative major biochemical compounds absorbing in the given wavelength region, according to Curran (1989). Colored arrows represent: "Chl" = chlorophyll (dark green), "Antho" = anthocyanin (red), "H20" = water (dark blue), "L" = lignin (dark red), "P" = protein (green), "S" = sugar (yellow). Reflection spectra were measured with an UV–VIS-NIR-SWIR spectrometer (PerkinElmer Lambda 950) in a wavelength range of 200 mn to 2000 and at a resolution of 1 nm. Absorption features spectra of the other conducted experiments, showing similar results, can be found in SI. 6. (Color figure online)
To demonstrate whether the observed differences in reflectance spectra are sufficient to reliably discriminate between hyperhydric and non-hyperhydric explants, while generalizing plant species and time points, we performed a model spot checking for several ML models with whole spectral data sets as input (Table 4). The models spot check based on AUCROC metrics identified partial least square (PLS) and linear discriminate analysis with upstream principal component analysis (PCA.LD) with 0.94 to be superior in the training step when classifying the explants against the other models, while supported vector machine (SVM), neutral net (NNET) with 0.93 and random forest model (RF) with 0.92 performed only slightly worse. High dimensional linear discriminate analysis (HD.LD) showed the lowest performance and was therefore excluded. Furthermore, SVM was best in classifying normal explants as expressed in the sensitivity metrics with 0.91 ± 0.08, while NNET reached with 0.93 ± 0.13 the highest specificity indicating the best performance in identifying hyperhydric explants. On the test set consisting of 59 unseen spectra, SVM outperformed the other models with the highest accuracy with 0.85, the highest balanced accuracy with 0.84, the highest sensitivity with 0.91 and the highest F1 score of 0.87 and was therefore selected as final model, besides for its low training time and its better human interpretability. As a reference of a classical approach, we checked the classification performance of a two-band normalized difference ratio index using a threshold of 0.35, which resulted in a low accuracy of 0.63.
The evaluation of the predictor importance based on ROC-curve importance of SVM revealed the most important wavelength for classification (Fig. 7). The most relevant wavelength for classification was found at 1949 nm, followed by the peak at 1445 nm in the SWIR region, 424 nm in the blue region and 676 nm in the red region. The wavelength region from 700 to 900 nm, including the NIR region, contained the least essential information for the classification. In the green region, 500 nm was most important, while in the SWIR region two further peaks were identified at 975 nm and 1202 nm.
Variable importance of spectral classification of hyperhydricity using a support vector machine approach. Classification classes consisted of reflection spectra from either normal or hyperhydric leaves based on visual scoring of HH (N: 0–1 HH score; HH: 2–4 HH score). Wavebands representing different spectral regions, defined by the sensitivity of silicon-based (Si) cameras, such as affordable RGB and RGB-NIR multispectral cameras, and more expensive Indium-Galium-Asenide-based (InGaAs) sensors as candidates for detection
The 237 acquired spectra of the two species were further used to simulate three in literature stated HH-affected leaf compounds over time (anthocyanin, water, lignin) via described vegetation indices (Fig. 8; mARI, Gitelson et al. 2006; NDWI, Gao 1996; NDLI, Serrano et al. 2002). Hyperhydric explants of A. thaliana showed a relatively small increase in ARI, high increase in NDWI and notable reduction in NDLI compared to normal explants. All three vegetation indices simulated using spectra of Malus “G214” classified as hyperhydric, revealed a strong change over time compared to normal spectra.
Selection of contrasting vegetation indices to hyperhydricity inducing cultivation of Malus 'G214' and A. thaliana 'Col-0'. Vegetation indices were calculated from spectra from three different experiments (Exp. III-V). Data points from normal explants are indicated in green (N: 0–1 HH score), while the blue color represents data from hyperhydric tissue (HH: 2–4 HH score). Estimated 95% confidence interval was colorized in light gray, while lines illustrate the locally weighted data trend by 2nd order polynomial regression. ARI/mARI defined according to Gitelson et al. (2006), NDWI from Gao (1996) and NDLI according to Serrano et al. (2002). (Color figure online)
Automated detection of hyperhydricity
To test the validity of the SWIR region of the trained spectral classifier as spectral region with high importance for discrimination of HH and to see the generalization to a new domain, the classifier was applied on a previously acquired SWIR-HSI data set from culture vessels containing normal (N, green) and hyperhydric explants (HH, blue) of Malus ‘Selection 4’ (Fig. 9A). From the spectral signatures (Fig. 9B), a normalized difference ratio index (NDRI, Fig. 9C–E) as a two-band index with a HH-insensitive wavelength at 1086 nm (Fig. 9C) and a HH-responsive wavelength at 1432 nm (Fig. 9D) was derived. Hyperhydric explants became almost invisible due to their high absorption/ reduced reflection (R) at 1432 nm (Fig. 9D and SI. 7). Based on the acquired spectral signature a normalized difference ratio index (NDRI) could be derived (Eq. 12), which is formed by two wavelengths, an HH-insensitive correction wavelength at 1086 nm and a HH-sensitive at 1432 mn.
Validation test of major absorption features by hyperspectral imaging. A Reference RGB image of a Malus ‘Selection 4’ vessel with normal (N, green) and hyperhydric (HH, blue) explants used for hyperspectral imaging of the SWIR region with the EVK Helios Core NIR Line-scan camera (240 px × 1 px and 252 spectral channels in the wavelength region of 900 nm to 1700 nm, according to Thiel 2018). B SWIR reflectance spectra of normal leaf, hyperhydric leaf and culture media (CM) pixels (green, blue and orange) with the dashed vertical lines indicating spectral locations of selected wavelengths. C and D False-color images of selected wavelengths at 1086 nm and 1432 nm. E Normalized difference ratio of selected wavelengths used to illustrate classical discriminating approach via F segmentation of plant pixels and G binarization by thresholding. Proposed discriminating approach by application of H ML model “Support vector machine“ (ML-SVM) on segmented plant pixels of the SWIR-Hyperspectral-Image-Cube (SWIR-HSI-Cube). ML-SVM was laboratory-trained with single leaf reflection spectra and is presented as the predicted probability images of plant pixels. Hyperspectral imaging was performed through the lid of the culture vessel. (Color figure online)
The NDRI image (Fig. 9E) was segmented with a mask for plant pixels (Fig. 9F) and a threshold was applied to produce the classification image (Fig. 9G). For the ML approach that included the application of the spectral classifier (Fig. 9H), some modifications were made to the trained spectral classifier, such as spectral resampling to fit the spectral sensor channels and segmentation to limit the task to a two-class problem (see Materials and Methods section).
As a more affordable approach of HH detection—SWIR camera systems can cost hundred to thousand times more than an RGB camera system—three different object detection models were trained based on RGB image time series data sets to determine if the information contained in the three spectral channels of the RGB images (in addition to the observed morphological differences in the shape of the explants) was sufficient to correctly classify the hyperhydric explants. With all three trained models (Table 5) a high mAP of > 88% was observed for the validation set, indicating a high accuracy in localization and correct classification of the explants in the images. Highest precision of 86.8% in validation set was reached with the model PCTOC_V2. For this model, we used the Roboflow Train option to train an object model from scratch. The model PCTOC_V3 performed best in terms of the recall metric with 95.7% and mAP with 95.6% in the validation set. In an unseen test data set PCTOC_V3 outperformed the other models in mAP with 97.0% and highest recall 89.0% and was therefore selected to visualize its performance on a selection of test set images (Fig. 10) and on unseen time-series data from two culture vessels of the same experiment (SI. 8).
Object detection performance of the PCTOC_V3 model on an image selection of the test set. (A) Ground truth RGB image of the Malus ‘G214’ test set annotated with normal (Normal, green) and hyperhydric (HH, blue) explants (A1–A3). (B) Predicted objects (B1–B3) and class membership probability (0 to 1 corresponds 0 to 100%). Prediction was performed with confidence threshold and intersection of union threshold of 0.5. (Color figure online)
The PCTOC_V3 model identified multiple objects on the selection of test set images (Fig. 10) with only slightly greater predicted bounding boxes compared to ground truth. A supposedly perfect classification could be reached with the prediction settings used. However, severely hyperhydric explants (Fig. 10B1) received a lower class membership probability then explants with developing HH symptoms (Fig. 10B2). Class membership probability of normal explants was generally high on the test set (Fig. 10B3) and seemed to be stable even on the time-series RGB image data set (SI. 8, left image). For hyperhydric explants, prediction confidence increased until day 10 and decreased at day 16 in the time-series RGB image data set (SI. 8, right image).
Discussion
Time-lapse videos enable insights into early phases of HH development
HH is a serious limitation of plant tissue propagation affecting multiple phases of in vitro cultivation. The use of the novel monitoring system “Phenomenon” capturing time series image data (SI. 4 and SI. 5) identified (i) the first visual symptoms of HH to occur 5 DAT and (ii) an accelerated and higher growth of shoots of the gelrite treatment (Fig. 1A). Thereby, significant differences in the projected plant area between the two treatments were found already 5 days after transferring to the culture media in both species. As discussed previously by Kevers et al. (1984), HH may be considered as morphological response to waterlogging, which in turn induces ethylene synthesis. For A. thaliana, we observed a higher vertical growth (Fig. 1B) with hyponasty (SI. 5), which was described as ethylene-triggered strategy of ex vitro plants in waterlogging conditions to re-establish contact with air and restore successful gas exchange (Voesenek and Blom 1989). Furthermore, Vreeburg et al. (2005) described a flooding-induced petiole elongation in a two-stage process, starting with acidification of the apoplast followed by cell wall expansion. This is in agreement with our observation of a significantly higher eccentricity (deviation of the ellipse to circle) and significantly less solidity (density of the object) for explants in the gelrite treatment (SI. 1B). A more pronounced curling of the leaves was observed in Malus (SI. 4) which also resulted in epinastic leaf growth. In addition, a significant higher mean canopy height (Fig. 1B) and maximum shoot height of Malus shoots on gelrite medium (SI. 1A) indicated a more pronounced vertical orientation of growth.
Hyperhydricity induction by increased water availability
Although HH symptoms vary between different plant species and cultivars, and several factors have been described to trigger HH, a putative common underlying mechanism of apoplast flooding has been described (van den Dries et al. 2013). Several studies showed that increasing the water availability by decreasing the concentration of gelling agent, changing the type of gelling agent or the cultivation in liquid media induced HH in a large set of plant species (Dianthus sp., Casanova et al. 2008; Aloe sp., Ivanova and Van Staden 2011, Malus sp. Chakrabarty et al. 2003).
In our study, we demonstrated the HH-inducing effects of gelrite for Malus and Arabidopsis indicated by the overall increase in HH scores (Fig. 2 and SI. 3) and apoplastic liquid volume (Fig. 3 and SI. 3) over time. Gelrite differs from agar in terms of consistency and purity and resulted in a superior growth of explants at a comparable gel strength (Scherer 1987; Tsay et al. 2006; Pasqualetto et al. 1988). However, gelrite induced HH in several species (Arabidopsis sp., van den Dries et al. (2013), Malus sp. Pasqualetto et al. 1988, Prunus sp. Franck et al. 1998) limiting the use of this gelling agent. Scherer et al. (1988) could show that there is no difference in the osmotic and water potential of gelrite compared to agar. Van den Dries et al. (2013) suspected therefore a local dissolution of the culture medium due to the excretion of chelators by the explants and thus a higher water availability and water uptake. This higher water availability in gelrite-solidified media most likely explains HH-induction and accelerated growth, but other putative factors like differences in uptake of nutrients or plant hormones were also found: Higher contents of magnesium (Mg) and a higher ratio of potassium (K) to sodium (Na) were detected in the leaves of walnut explants grown on gelrite medium compared to agar, which can affect stomatal function (Barbes et al. 1993). Furthermore, Arthur et al. (2004) found a lower concentration of IAA-like compounds in gelrite than in different types of agar powder.
With the collected data and the time-lapse videos, we could narrow down crucial key points within the development of HH of the two species in time. First visual identifiable symptoms (SI. 4 and SI. 5) and significant increases in apoplastic liquid volume were observed already after 5 days of cultivation on gelrite media in both species. Time series dynamics of apoplastic liquid volume confirmed previous data for A. thaliana (van den Dries et al. 2013)—in both studies hyperhydric explants of A. thaliana had an apoplastic liquid volume of around 300 µL g−1 FM 15 days after treatment, but were carried out for the first time for Malus. Quantification of apoplastic liquid volume of A. thaliana seedlings at very early time points was limited by the very small amounts of apoplastic liquid and the distorting effect of adhering water (Fig. 3B). For Malus, the highest increase in apoplastic liquid volume for the gelrite treatment was detected within the first 4–5 days in two independent experiments (SI. 3, Fig. 3B). Furthermore, a different behavior of the HH score and the apoplastic liquid volume was found after 21 days of cultivation in Malus: While the severity of HH symptoms steadily increased over time, the apoplastic liquid volume seemed to reach saturation at later time points (SI. 3¸ Fig. 4). Therefore, we suggest the HH score to be useful to determine the symptoms of HH, whereas the quantification of apoplastic liquid volume better reflects the physiological state of the explants.
Identification of HH-specific spectral absorption features
Despite the fact that clear visible symptoms (Table 1) of HH were reported and still are the major distinguishing parameter for classification, spectroscopic analysis of HH is limited. Only Marques et al. (2021) using Fourier-transform infrared spectroscopy in attenuated total reflectance mode (FTIR-ATR), evaluated chemical properties of prepared cell walls of hyperhydric Arbutus unedo. Assuming HH as a consequence of flooding of the air-filled apoplast by water, UV–VIS–NIR–SWIR reflection spectroscopy was expected to detect these physiological changes due to higher light absorption of water compared to air. Therefore, we applied this technique to identify specific absorption features of HH essential for designing an automated detection system. However, we excluded the UV region (< 400 nm) from further analysis due to the low penetration depth of UV light in plant tissue (Qi et al. 2010), since most reflection signals can only be attributed to anatomical and biochemical properties of cuticle, trichomes and the upper epidermis.
The observed overall reduction in reflectance of hyperhydric explants (Fig. 5) compared to normal ones is consistent with the visual appearance of the observed darkening of the affected explants (SI. 4 and SI. 5). The visualization of isolated absorption features over the time course of the development of HH in Malus (Fig. 6) should give insights whether there is at least a trend in the time course of the presumed absorption characteristics. We used the continuum removal method to exclude the observed overall absolute reduction in reflection and to compare all spectra on a common base. This allowed an automated extraction of absorption peaks for the SWIR region with predominant absorptions valleys, however, produced artefacts in the VIS region. In the SWIR region, a consistent difference between absorption of normal and hyperhydric leaves was observed for the wavelengths 980 nm, 1150 nm, 1400 nm, 1520 nm, 1780 nm and 1930 nm, both over time (Fig. 6) and in the different experiments (SI. 6). Most likely, the absorption of water in the plant tissue is most responsible for the wavelengths 980 nm, 1150 nm, 1400 nm. Curran et al. (1989) described the intense absorption of liquid water at 970 nm, 1200 nm, 1400 nm and 1450 nm due to the fundamental O–H bending vibrations of the first overtone. Thus, the tendency of an increase in water absorption (970 mn, 1200 nm, 1400 nm, 1450 nm) within time is in accordance with the increase in apoplastic liquid volume over time. However, absorption bands of other compounds like proteins, lignins and sugars are located within the peak between 1300 to 1600 nm and contribute to the total absorption in this region. Curran et al. (1989) associated the absorption at 1780 nm to cellulose, sugars and starch. Since for this wavelength a higher absorption in hyperhydric leaves was observed in our study, this is in line with the detection of a higher sugar content (sucrose, glucose and fructose) in hyperhydric explants of Dianthus (Saher et al. 2005), but contradicting Kevers et al. (1987) who reported less lignin and cellulose in hyperhydric Dianthus.
Simulation of vegetation indices (Fig. 8) demonstrated traceable trends, that closely match the dynamics of the physiological reference data (Fig. 3 & SI. 3) and support the observation of time-series data (SI. 4 & SI. 5). Overall, the vegetation indices from normal explants exhibited low variance, although they were derived from different experiments. The high variance of hyperhydric explants indicated by the confidence interval can be explained by different physiological states of explants with different degrees of hyperhydricity. The simulation of a modified anthocyanin index, indicated a higher anthocyanin content in hyperhydric leaves of Malus, but not Arabidopsis, supporting our RGB image time series. The normalized difference water index (NDWI) displayed higher water contents for hyperhydric explants of both species and supported our observation that apoplastic liquid volume did not increase any more after 4 weeks of cultivation. The normalized difference lignin index (NDLI) showed in both species less lignin for hyperhydric leaves. However, the trend of the NDLI curves in both species followed inversely that of the NDWI indicating a putative dependency on plant water content. Marques et al. (2021) found no significant difference in the lignin content per dry weight of hyperhydric and normal leaves of Arbutus, whereas Kevers et al. (1987) reported a lower lignin content per fresh weight of hyperhydric tissue. It remains to be clarified, whether these divergent results are due to different species or to the fact that the fresh mass of hyperhydric explants is much higher.
Automated detection of HH by machine learning
In order to evaluate the performance of the spectral data in the classification of hyperhydric and normal leaves, we trained different ML models (Table 4), investigated the most important wavelengths of the best model (Fig. 7) and compared them against a novel vegetation index as the classical approach (Fig. 9). The ML models differed in their architecture, complexity, performance, prediction time and interpretability (Singh et al. 2016; Liakos et al. 2018; and Hesami and Jones 2020). All ML models reached a high AUCROC > 0.83 in training, however only SVM and NNET had a high accuracy > 0.80 on test data. Both ML models outperformed with an accuracy of 0.81 for NNET (balanced accuracy of 0.81) and 0.85 for SVM (balanced accuracy of 0.84) the univariate vegetation index approach with a lower accuracy of 0.63 (balanced accuracy of 0.66). Furthermore, SVM was best in classifying normal spectra indicated by highest sensitivity of 0.91 on the test set. The two-band vegetation index NDRI reached the highest specificity of 0.84 in the test data, followed by SVM with 0.76, meaning highest ratio in the identification of hyperhydric tissue, however, low sensitivity of 0.47, low accuracy of 0.63 and low F1 score of 0.59 indicated a conservative behavior of classification towards hyperhydric explants. SVM was selected due to its high performance on training and testing datasets, low training data volume requirements, performance on high-dimensional datasets, low risk of overfitting, good generalization ability, and its advantages over NNET in terms of training time, simplified structure, and interpretability (Singh et al. 2016; Liakos et al. 2018). The evaluation of the feature importance of SVM for classification (Fig. 7) supported our findings that bands (peaks with maxima at 1949 nm, 1445 nm, 1202 nm and 975 nm) associated with water absorption were crucial to distinguish between hyperhydric and normal leaves. However, the method indicated essential features importance in the VIS region with maxima at 424 nm and 676 nm. In regard of an automated HH detection system, we further evaluated two different approaches (i) HH detection based on an HSI-SWIR camera system (Fig. 9) and (ii) HH detection based on RGB camera system coupled with a deep neuronal network (DNN) to provide two putative solutions for commercial plant propagation based on our findings (Table 5, Fig. 10, SI. 8).
Following the HSI-SWIR camera system approach, we could only test the validity of our spectral classifier as a proof of concept because we only had a single HSI acquisition (SI. 7), so these results should be interpreted with caution. In addition, our analysis followed a two-class classification problem, but under the assumption that an automated HH detection system monitors explants on culture media during cultivation, culture media spectra could presumably interfere with the other classes within classification. Therefore, for further studies, we propose to include the acquisition of reflectance spectra of the culture media in the dataset. Nevertheless, we could test our ML-SVM classifier, trained on spectra from Malus ‘G214’ and A. thaliana, on the single SWIR-HSI acquisition of Malus ‘Selection 4’ segmented plant pixels, indicating the generalization ability of the classifier with respect to experimental setup and plant species/genotype. The NDRI and ML-SVM both classified most pixels correctly, however, ML-SVM segmented the borders of different classes much sharper. These preliminary results demonstrated that classification of HH is possible during in vitro cultivation and through the lid of the vessel with either an expensive SWIR-HSI system classifying with our novel ML-SVM classifier or more cost-effectively with a two-channel SWIR camera system using a novel vegetation index.
Alternatively, an RGB camera setup coupled with convolutional neural network (CNN) can be the most cost-effective solution for an automated HH-detection. Since we had identified feature importance also in the VIS region, a proof-of-concept study was conducted to demonstrate object detection via CNN. Therefore, we used the Roboflow© pipeline, which allowed an easy access to these tools and provided an interface for data annotation, pre-processing, data augmentation, training, data availability and deployment of the trained models. Comparing a self-trained YOLOv8 with the unknown object detection algorithms of Roboflow Train (Table 5), we did not reach the performance of their optimized model, which was particularly evident in the performance on test set, where PCTOC_V1 reached the highest precision with 94.4%, but with low recall of 49.5%—indicating only half of all explants could be detected. The best trained model PCTOC_V3, however had a precision of 83.8% on validation and of 97.0% on test set, indicating that prediction was mostly correct (Table 5, Fig. 10, SI. 8). In addition the explants were reliably detected (recall of 95.7% on validation set and 89.0% on test set). By using the relatively new Python library roboflow, we encountered some unsolved issues as seen in SI. 8 where non-maximum-suppression only works so far within one class, resulting in multiple predictions per object. Considering the properties of the dataset, the low amount of data (250 to 375 images), resulting from time series images (1049 explants) of only 32 individual explants, we could see already good performance on the test set and the time-series set (Fig. 10 & SI. 8).
Conclusions
To our knowledge this study is the first report of (i) identifying discriminating wavelengths in the VIS–NIR-SWIR region for the detection of HH, (ii) application of short wave infrared hyperspectral imaging to detect growth anomalies in vitro, (iii) proposing a spectral classifier for hyperhydricity. Wavelength bands (around 1940 nm, 1450 nm, 1200 nm and 970 nm) associated with absorption of water are the most distinguishable between hyperhydric and normal leaves within the analyzed spectral data set (400 nm to 2000 nm). In addition, minor important wavelengths were found in the RGB region (around 430 nm and 680 nm), whereas the NIR region seemed to be less important. Furthermore, RGB images of hyperhydric explants contain sufficient morphological and spectral features to allow a reliable detection of HH in an affordable manner via convolutional neuronal networks. However, this needs to be proven in an in-depth study. Nonetheless, these results can serve as a proof-of-concept for CNN-assisted live monitoring of plant tissue cultures and pave the way for increased use of CNN to estimate other key parameters such as multiplication rate, nutrient deficiency, and contamination.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request. RGB image dataset analysed during the current study available in the Bethge (2023) repository, [https://universe.roboflow.com/hains/hh-detection-in-vitro/dataset/8].
Abbreviations
- HH:
-
Hyperhydricity
- ML:
-
Machine learning
- UV:
-
Ultra violet
- VIS:
-
Visible radiation
- NIR:
-
Near infrared radiation
- SWIR:
-
Shortwave infrared radiation
- MWIR:
-
Mid-wave infrared radiation
- LWIR:
-
Longwave infrared radiation
- DAT:
-
Days after treatment/transfer
- CV:
-
Cross validation
- CNN:
-
Convolutional neuronal network
- HSI:
-
Hyperspectral imaging
References
Arthur GD, Stirk WA, Van Staden J, Thomas TH (2004) Screening of aqueous extracts from gelling agents (Agar and Gelrite) for root-stimulating activity. S Afr J Bot 70(4):595–601. https://doi.org/10.1016/S0254-6299(15)30197-6
Aynalem HM, Righetti TL, Reed BM (2006) Non-destructive evaluation of in vitro-stored plants: a comparison of visual and image analysis. In Vitro Cell Dev Biol-Plant 42(6):562–567. https://doi.org/10.1079/IVP2006816
Barbas E, Jay-Allemand C, Doumas P, Chaillou S, Cornu D (1993) Effects of gelling agents on growth, mineral composition and naphthoquinone content of in vitro explants of hybrid walnut tree (Juglans regia × Juglans nigra). Annales Des Sci for 50(2):177–186. https://doi.org/10.1051/forest:19930205
Bethge H (2023) HH Detection in vitro Image Dataset. https://universe.roboflow.com/hains/hh-detection-in-vitro/dataset/8. Accessed 10 Feb 2023
Bethge H, Winkelmann T, Lüdeke P (2023) Rath T (2023) Low-cost and automated phenotyping system “Phenomenon” for multi-sensor in situ monitoring in plant in vitro culture. Plant Methods 19(1):1–25. https://doi.org/10.1186/s13007-023-01018-w
Bock Biosciences GmbH (2018) RoBo®Cut. https://www.robotec-ptc.com/. Accessed 14 Feb 2023
Bradski G (2000) The openCV library. Dr. Dobb’s J Softw Tools Prof Progr 25(11):120–123
Cardoso JC, Sheng Gerald LT, Teixeira da Silva JA (2018) Micropropagation in the twenty-first century. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Plant cell culture protocols. Springer, Dordrecht, pp 17–46
Casanova E, Moysset L, Trillas MI (2008) Effects of agar concentration and vessel closure on the organogenesis and hyperhydricity of adventitious carnation shoots. Biol Plant 52:1–8. https://doi.org/10.1007/s10535-008-0001-z
Chakrabarty D, Hahn EJ, Yoon YJ, Paek KY (2003) Micropropagation of apple rootstock M. 9 EMLA using bioreactor. J Hortic Sci Biotechnol 78(5):605–609. https://doi.org/10.1080/14620316.2003.11511671
Chen C (2016) Cost analysis of plant micropropagation of Phalaenopsis. Plant Cell, Tis Organ Cult 126(1):167–175. https://doi.org/10.1007/s11240-016-0987-4
Curran PJ (1989) Remote sensing of foliar chemistry. Remote Sens Environ 30(3):271–278. https://doi.org/10.1016/0034-4257(89)90069-2
de Klerk GJ, Pramanik D (2017) Trichloroacetate, an inhibitor of wax biosynthesis, prevents the development of hyperhydricity in Arabidopsis seedlings. Plant Cell, Tiss Organ Cult 131(1):89–95. https://doi.org/10.1007/s11240-017-1264-x
Debergh P, Aitken-Christie J, Cohen D, Grout B, Von Arnold S, Zimmerman R, Ziv M (1992) Reconsideration of the term ‘vitrification’ as used in micropropagation. Plant Cell, Tissue Organ Cult 30(2):135–140. https://doi.org/10.1007/BF00034307
Dhondt S, Gonzalez N, Blomme J, De Milde L, Van Daele T, Van Akoleyen D, Storme V, Coppens F, Beemster TS, Inzé D (2014) High-resolution time-resolved imaging of in vitro Arabidopsis rosette growth. Plant J 80(1):172–184. https://doi.org/10.1111/tpj.12610
Dwyer B, Nelson J, Solawetz J (2021) Roboflow python package. https://github.com/roboflow/roboflow-python. Accessed 10 Feb 2023
Dwyer B, Nelson J, Solawetz J (2022) Roboflow (v1.0). https://roboflow.com. Accessed 14 Feb 2023
Fischler MA, Bolles RC (1981) Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun ACM 24(6):381–395. https://doi.org/10.1145/358669.358692
Franck T, Crèvecoeur M, Wuest J, Greppin H, Gaspar T (1998) Cytological comparison of leaves and stems of Prunus avium L. shoots cultured on a solid medium with agar or gelrite. Biotechnic Histochem 73(1):32–43. https://doi.org/10.3109/10520299809140504
Gamborg OL, Miller R, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50(1):151–158. https://doi.org/10.1016/0014-4827(68)90403-5
Gao BC (1996) NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens Environ 58(3):257–266. https://doi.org/10.1016/S0034-4257(96)00067-3
Gao H, Xia X, An L, Xin X, Liang Y (2017) Reversion of hyperhydricity in pink (Dianthus chinensis L.) plantlets by AgNO3 and its associated mechanism during in vitro culture. Plant Sci 254:1–1. https://doi.org/10.1016/j.plantsci.2016.10.008
Gehan MA, Fahlgren N, Abbasi A, Berry JC, Callen ST, Chavez L, Doust AN, Feldman MJ, Gilbert KB, Hodge JG, Hoyer JS (2017) PlantCV v2: image analysis software for high-throughput plant phenotyping. PeerJ 5:e4088. https://doi.org/10.7717/peerj.4088
George EF, Hall MA, De Klerk GJ (2008) Plant propagation by tissue culture. In: George EF, Hall MA, De Klerk G-J (eds) Volume I. The background. Plant propagation by tissue culture. Springer, Dordrecht
Gitelson AA, Keydan GP, Merzlyak MN (2006) Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys Res Lett. https://doi.org/10.1029/2006GL026457
Gribble K (1999) The influence of relative humidity on vitrification, growth and morphology of Gypsophila paniculata L. Plant Growth Regul 27(3):181–190. https://doi.org/10.1023/A:1006235229848
Gupta SD, Karmakar A (2017) Machine vision based evaluation of impact of light emitting diodes (LEDs) on shoot regeneration and the effect of spectral quality on phenolic content and antioxidant capacity in Swertia chirata. J Photochem Photobiol, B 174:162–172. https://doi.org/10.1016/j.jphotobiol.2017.07.029
Hesami M, Jones AM (2020) Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol 104(22):9449–9485. https://doi.org/10.1007/s00253-020-10888-2
Honda H, Takikawa N, Noguchi H, Hanai T, Kobayashi T (1997) Image analysis associated with a fuzzy neural network and estimation of shoot length of regenerated rice callus. J Ferment Bioeng 84(4):342–347. https://doi.org/10.1016/S0922-338X(97)89256-2
Huang YJ, Lee FF (2010) An automatic machine vision-guided grasping system for Phalaenopsis tissue culture plantlets. Comput Electron Agric 70(1):42–51. https://doi.org/10.1016/j.compag.2009.08.011
Ivanova M, Van Staden J (2011) Influence of gelling agent and cytokinins on the control of hyperhydricity in Aloe polyphylla. Plant Cell, Tissue Organ Cult 104(1):13–21. https://doi.org/10.1007/s11240-010-9794-5
Jocher G, Chaurasia, A, Qiu J (2023) YOLO by Ultralytics (Version 8.0.0). https://github.com/ultralytics/ultralytics. Accessed 14 Feb 2023
Kemat N (2020) Improving the quality of tissue-cultured plants by fixing the problems related to an inadequate water balance, hyperhydricity. Doctoral dissertation, Wageningen University and Research. https://doi.org/10.18174/517434
Kemat N, Visser RG, Krens FA (2021) Hypolignification: a decisive factor in the development of hyperhydricity. Plants 10(12):2625. https://doi.org/10.3390/plants10122625
Kevers C, Coumans M, Coumans-Gillès MF, Caspar TH (1984) Physiological and biochemical events leading to vitrification of plants cultured in vitro. Physiol Plant 61(1):69–74. https://doi.org/10.1111/j.1399-3054.1984.tb06102.x
Kevers C, Prat R, Gaspar T (1987) Vitrification of carnation in vitro: changes in cell wall mechanical properties, cellulose and lignin content. Plant Growth Regul 5(1):59–66. https://doi.org/10.1007/BF00035020
Kuhn M (2008) Building predictive models in R using the caret package. J Stat Softw 28:1–26. https://doi.org/10.18637/jss.v028.i05
Lee TJ, Zobayed SM, Firmani F, Park EJ (2019) A novel automated transplanting system for plant tissue culture. Biosys Eng 181:63–72. https://doi.org/10.1016/j.biosystemseng.2019.02.012
Lehnert LW, Meyer H, Obermeier WA, Silva B, Regeling B, Bendix J (2018) Hyperspectral data analysis in R: the hsdar package. arXiv Preprint. https://doi.org/10.48550/arXiv.1805.05090
Liakos KG, Busato P, Moshou D, Pearson S, Bochtis D (2018) Machine learning in agriculture: a review. Sensors 18(8):2674. https://doi.org/10.3390/s18082674
Lizárraga A, Fraga M, Ascasíbar J, González ML (2017) In vitro propagation and recovery of eight apple and two pear cultivars held in a germplasm bank. Am J Plant Sci 8(9):2238–2254. https://doi.org/10.4236/ajps.2017.89150
Mahendra PVS, Gupta SD (2004) Trichromatic sorting of in vitro regenerated plants of gladiolus using adaptive resonance theory. Curr Sci 10:348–353
Marques MP, Martins J, de Carvalho LA, Zuzarte MR, da Costa RM, Canhoto J (2021) Study of physiological and biochemical events leading to vitrification of Arbutus unedo L. cultured in vitro. Trees 35:241–253. https://doi.org/10.1007/s00468-020-02036-0
Mestre D, Fonseca JM, Mora A (2017) Monitoring of in-vitro plant cultures using digital image processing and random forests. 8th International Conference on Pattern Recognition Systems. https://doi.org/10.1049/cp.2017.0137
Mohamed SM, El-Mahrouk ME, El-Banna AN, Hafez YM, El-Ramady H, Abdalla N, Dobránszki J (2023) Optimizing medium composition and environmental culture condition enhances antioxidant enzymes, recovers Gypsophila paniculata L. hyperhydric shoots and improves rooting in vitro. Plants 12(2):306. https://doi.org/10.3390/plants12020306
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nezami-Alanagh E, Garoosi GA, Landín M, Gallego PP (2019) Computer-based tools provide new insight into the key factors that cause physiological disorders of pistachio rootstocks cultured in vitro. Sci Rep 9(1):1–5. https://doi.org/10.1038/s41598-019-46155-2
Paques M, Boxus P, Dulos M (1985) “ Vitrification”: an induceable and reversible phenomenon. In: symposium on in vitro problems related to mass propagation of horticultural plants 212. pp 253–258. https://doi.org/10.17660/ActaHortic.1987.212.38
Pasqualetto PL, Zimmerman RH, Fordham I (1988) The influence of cation and gelling agent concentrations on vitrification of apple cultivars in vitro. Plant Cell, Tissue Organ Cult 14(1):31–40. https://doi.org/10.1007/BF00029573
Patrício DI, Rieder R (2018) Computer vision and artificial intelligence in precision agriculture for grain crops: a systematic review. Comput Electron Agric 153:69–81. https://doi.org/10.1016/j.compag.2018.08.001
Peterson RA, Peterson MR (2020) Package ‘bestNormalize’. Normalizing transformation functions. R package version
Phan CT, Letouze R (1983) A comparative study of chlorophyll, phenolic and protein contents, and of hydroxycinnamate: CoA ligase activity of normal and ‘vitreous’ plants (Prunus avium L.) obtained in vitro. Plant Sci Lett 31(2–3):323–327. https://doi.org/10.1016/0304-4211(83)90071-8
Pinheiro J, Bates D, DebRoy S, Sarkar D, Heisterkamp S, Van Willigen B, Maintainer R (2017) Package ‘nlme.’ Linear Nonlinear Mixed Eff Models Vers 3(1):274
Prasad VS, Gupta SD (2008) Applications and potentials of artificial neural networks in plant tissue culture. In: Gupta SD, Ibaraki Y (eds) Plant tissue culture engineering. Springer, Dordrecht, pp 47–67. https://doi.org/10.1007/978-1-4020-3694-1_3
Qi Y, Heisler GM, Gao W, Vogelmann TC, Bai S (2010) Characteristics of UV-B radiation tolerance in broadleaf trees in southern USA. In: Gao W, Slusser JR, Schmoldt DL (eds) UV radiation in global climate change. Springer, Berlin, Heidelberg
Redmon J, Divvala S, Girshick R, Farhadi A (2016). You only look once: Unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 779–788. https://doi.org/10.1109/CVPR.2016.91
Ripley B, Venables W, Ripley MB (2016) Package ‘nnet’. R Package version. 2;7(3–12):700
Rojas-Martínez L, Visser RG, De Klerk GJ (2010) The hyperhydricity syndrome: waterlogging of plant tissues as a major cause. Propag Ornam Plants 10(4):169–175
Rstudio Team (2015) RStudio: integrated development for R. RStudio. Inc., Boston, p 879
Saher S, Fernández-García N, Piqueras A, Hellín E, Olmos E (2005) Reducing properties, energy efficiency and carbohydrate metabolism in hyperhydric and normal carnation shoots cultured in vitro: a hypoxia stress? Plant Physiol Biochem 43(6):573–582. https://doi.org/10.1016/j.plaphy.2005.05.006
Sakamoto Y, Ishiguro M, Kitagawa G (1986) Akaike information criterion statistics. D. Reidel, Dordrecht. https://doi.org/10.2307/2983028
Scherer PA (1987) Standardization of plant micropropagation by usage of a liquid medium with polyurethane foam plugs or a solidified medium with the gellan gum gelrite instead of agar. In: International Symposium on Propagation of Ornamental Plants 226. pp. 107–114. https://doi.org/10.17660/ActaHortic.1988.226.10
Scherer PA, Müller E, Lippert H, Wolff G (1988) Multielement analysis of agar and gelrite impurities investigated by inductively coupled plasma emission spectrometry as well as physical properties of tissue culture media prepared with agar or the gellan gum gelrite. In: International Symposium on Propagation of Ornamental Plants 226, pp 655–658. https://doi.org/10.17660/ActaHortic.1988.226.91
Serrano L, Penuelas J, Ustin SL (2002) Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: decomposing biochemical from structural signals. Remote Sens Environ 81(2–3):355–364. https://doi.org/10.1016/S0034-4257(02)00011-1
Shaw DR, Kelley FS (2005) Evaluating remote sensing for determining and classifying soybean anomalies. Precision Agric 6(5):421–429. https://doi.org/10.1007/s11119-005-3681-9
Singh A, Thakur N, Sharma A (2016) A review of supervised machine learning algorithms. In2016 3rd international conference on computing for sustainable global development (INDIACom). pp. 1310–1315. https://doi.org/10.35940/ijsce.E3583.1112522
Smith MA, Spomer L (1995) Vessels, gels, liquid media, and support systems. In: Aitken-Christie J, Kozai T, Smith MAL (eds) Automation and environmental control in plant tissue culture. Springer, Dordrecht, pp 371–404. https://doi.org/10.1007/978-94-015-8461-6_16
Smith MA, Spomer L, Meyer MJ, McClelland MT (1989) Non-invasive image analysis evaluation of growth during plant micropropagation. Plant Cell, Tissue Organ Cult 19(2):91–102. https://doi.org/10.1007/BF00035809
Sommer C, Straehle C, Koethe U, Hamprecht FA (2011) Ilastik: Interactive learning and segmentation toolkit. In: 2011 IEEE international symposium on biomedical imaging: From nano to macro. pp. 230–233. https://doi.org/10.1109/ISBI.2011.5872394
Sullivan C, Kaszynski A (2019) PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). J Open Sour Softw 4(37):1450. https://doi.org/10.21105/joss.01450
Terry ME, Bonner BA (1980) An examination of centrifugation as a method of extracting an extracellular solution from peas, and its use for the study of indoleacetic acid-induced growth. Plant Physiol 66(2):321–325. https://doi.org/10.1104/pp.66.2.321
Thiel M (2018) Bildgebende NIR-Hyperspektral-Technologie zur in-situ Erfassung des Blattwassergehalts. Doctoral dissertation, Leibniz Universität Hannover. https://doi.org/10.15488/3882
Tian J, Jiang F, Wu Z (2015) The apoplastic oxidative burst as a key factor of hyperhydricity in garlic plantlet in vitro. Plant Cell Tiss Organ Cult 120(2):571–584. https://doi.org/10.1007/s11240-014-0623-0
Tisserand S (2021) Vis-NIR hyperspectral cameras. Photoniques 110:58–64
Tsay HS, Lee CY, Agrawal DC, Basker S (2006) Influence of ventilation closure, gelling agent and explant type on shoot bud proliferation and hyperhydricity in Scrophularia yoshimurae—a medicinal plant. In Vitro Cell Dev Biol-Plant 42(5):445–449. https://doi.org/10.1079/IVP2006791
van Altvorst AC, Koehorst H, de Jong J, Dons HJ (1996) Transgenic carnation plants obtained by Agrobacterium tumefaciens-mediated transformation of petal explants. Plant Cell, Tissue Organ Cult 45(2):169–173. https://doi.org/10.1007/BF00048762
van den Dries N, Giannì S, Czerednik A, Krens FA, de Klerk GJ (2013) Flooding of the apoplast is a key factor in the development of hyperhydricity. J Exp Bot 64(16):5221–5230. https://doi.org/10.1093/jxb/eru497
Van Der Walt S, Colbert SC, Varoquaux G (2011) The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 13(2):22–30. https://doi.org/10.48550/arXiv.1102.1523
Van Rossum G, Drake FL (2009) Python 3 Reference manual: python documentation manual part 2. Scotts Valley, CA: CreateSpace
Vieitez AM, Ballester A, San-José MC, Vieitez E (1985) Anatomical and chemical studies of vitrified shoots of chestnut regenerated in vitro. Physiol Plant 65(2):177–184. https://doi.org/10.17660/ActaHortic.1987.212.34
Voesenek LA, Blom CW (1989) Growth responses of Rumex species in relation to submergence and ethylene. Plant Cell Environ 12(4):433–439. https://doi.org/10.1111/j.1365-3040.1989.tb01959.x
Vreeburg RA, Benschop JJ, Peeters AJ, Colmer TD, Ammerlaan AH, Staal M, Elzenga TM, Staals RH, Darley CP, McQueen-Mason SJ, Voesenek LA (2005) Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J 43(4):597–610. https://doi.org/10.1111/j.1365-313X.2005.02477.x
Zhang C, Timmis R, Hu WS (1999) A neural network based pattern recognition system for somatic embryos of Douglas fir. Plant Cell, Tissue Organ Cult 56:25–35. https://doi.org/10.1023/A:1006287917534
Zhou QY, Park J, Koltun V (2018) Open3D: a modern library for 3D data processing. arXiv Preprint. https://doi.org/10.48550/arXiv.1801.09847
Ziv M (1991) Vitrification: morphological and physiological disorders of in vitro plants. In: Debergh PC, Zimmerman RH (eds) Micropropagation. Springer, Dordrecht, pp 45–69. https://doi.org/10.1007/978-94-009-2075-0_4
Acknowledgements
We thank the technical assistants Ewa Schneider and Bärbel Ernst of the department of Woody Plant and Propagation Physiology, Institute of Horticultural Production Systems, Leibniz Universität Hannover for their excellent support in the lab. Furthermore, we thank Matthias Igelbrink and Prof. Dr. Arno Ruckelshausen at University of Applied Science Osnabrück in their support in recording the SWIR-HSI data. In addition, we are grateful for the scholarship for the completion of a dissertation of the University of Applied Science Osnabrück.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project took place within the research project “Experimentierfeld Agro-Nordwest”, which is funded by the Federal Ministry of Food and Agriculture (BMEL, Grant No.: 28DE103F18) via the Federal Agency for Agriculture and Food (BLE).
Author information
Authors and Affiliations
Contributions
HB and TW designed the experiments, HB and ZM performed the experiments and analysed the data. HB wrote the manuscript and HB, ZM, TR, TW revised the manuscript. All the authors discussed the results and collectively edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The research work was carried out in compliance with the ethical standards that do not involve the use of humans.
Additional information
Communicated by Victor M. Jimenez.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11240_2023_2528_MOESM1_ESM.png
Supplementary file 1 (PNG 3087 KB)—Maximum shoot height and shape analysis of explants of A. thaliana Col-0 and Malus ‘G214’ (Mean ±SD). A) The relative increase in mean maximum shoot height of upper 10th percentile resulted from analysis of segmented depth data collected with a scanning laser distance sensor and normalized to day 0 maximum plant height. B) Shape analysis parameter of single explants. Yellows lines indicates cultivation on standard media formulation on Gamborg-B5 (A. thaliana) and MS-Medium (Malus) solidified with 0.8% agar (w/v), while blue lines display the cultivation on induction media containing 0.25% (w/v) gelrite, inducing HH. Estimated 95% confidence interval was illustrated in light gray. C) Representative images of segmented single explants of the different plant species and treatments. Convex hull, longest path, centroid is colored in pink, while object area and perimeter are outlined in blue. Sample number (n) indicates A) individual culture containers and B) individual explants. Significance stars indicate comparisons of treatments within a time point (day) with * p < 0.05, * * p < 0.01, * * * p < 0.001. Depth data were acquired with the multisensory robot system “Phenomenon” (Bethge et al. 2023)
11240_2023_2528_MOESM2_ESM.png
Supplementary file 2 (PNG 1478 KB)—Visual scoring and apoplastic liquid volume of Malus ‘G214’ Experiment IV & V. Samples from 0 days after transfer (DAT 0) represent the starting plant material cultured on control media. Yellows bars indicate cultivation on standard media formulation MS-Medium with 0.8% (w/v) agar, while gray bars display the cultivation on induction media containing 0.25% (w/v) gelrite. Dashed lines represent the medians of each histogram. Sample number (n) indicates the individual explants. Different letters resulting from Kruskal-Wallis-Test followed by Fisher´s LSD (p <0.05) indicate significant differences between histograms. Different letters resulted from Tukey’s HSD test at p < 0.05 and show significantly different time points within one treatment, while asterisks indicate comparisons of treatments within a time point with * = p < 0.05, ** = p < 0.01, *** = p < 0.001
11240_2023_2528_MOESM3_ESM.png
Supplementary file 3 (PNG 10898 KB)—Sample holder for reflectance measurement with Perkin-Elmer Lambda 900 UV-VIS-NIR-SWIR spectrometer. Leaf samples were placed with adaxial orientation inside the 3D-printed sample holder
Supplementary file 4 (AVI 235807 KB)—Hyperhydricity induction time-lapse video in Malus ‘G214’. Left side represents control MS-medium solidified with 0.8% agar (w/v), while right image displays the cultivation on induction media containing 0.25% (w/v) gelrite. Both media were supplemented with 1 g L-1 titanium dioxide. RGB Images were acquired with the multisensory robot system “Phenomenon” (Bethge et al. 2023)
Supplementary file 5 (AVI 151178 KB)—Hyperhydricity induction time-lapse video in A. thaliana . Left side represents control B5-medium solidified with 0.8% agar (w/v), while right image displays the cultivation on induction media containing 0.25% (w/v) gelrite. Both media were supplemented with 1 g L-1 titanium dioxide. RGB images were acquired with the multisensory robot system “Phenomenon” (Bethge et al. 2023)
11240_2023_2528_MOESM6_ESM.png
Supplementary file 6 (PNG 1656 KB)—Similarities in spectral reflectance of Malus sp. 'G214' and A. thaliana 'Col-0'. Upper row represents extracted absorption features after segmented convex-hull removal of raw spectra and bottom row demonstrates difference spectrum of the absorption peaks. Normal (NN, colorized in green) and hyperhydric (HH, colorized in blue) spectra membership were based on visual scoring of HH (NN, 0-1; HH 2-4). Colorized arrows indicating potential major biochemical leave compounds absorbing in the given wavelength region, according to Curran(1989). Reflection spectra were measured with UV/VIS/NIR Spectrometer (PerkinElmer Lambda 950) in wavelength range of 200 – 2000 and a resolution of 1 nm
Supplementary file 7 (AVI 21151 KB)—SWIR-hyperspectral image stack/video of Malus ‘Selection 4’. Image data set were acquired with the imaging system developed and described by Thiel (2018). This system consisted of an EVK Helios Core NIR Line-scan camera (240 px × 1 px and 252 spectral channels in the wavelength region of 900 nm to 1700 nm), two 65 W halogen spot lights and a conveyer-belt system to move the sample. Spectral reflectance was recorded top-down through the lids of the culture containers
Supplementary file 8 (AVI 273432 KB)—Automated detection of hyperhydricity via CNN, while the induction of HH in Malus ‘G214’ in time-lapse video. Left side represents control MS-medium solidified with 0.8% agar (w/v), while right image displays the cultivation on induction media containing 0.25% (w/v) gelrite. RGB images were acquired with the multisensory robot system “Phenomenon” (Bethge et al. 2023). Class membership predictions resulted from the trained PCTOC_V3 object detection model, available under Bethge (2023)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bethge, H., Mohammadi Nakhjiri, Z., Rath, T. et al. Towards automated detection of hyperhydricity in plant in vitro culture. Plant Cell Tiss Organ Cult 154, 551–573 (2023). https://doi.org/10.1007/s11240-023-02528-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-023-02528-0