Abstract
Right and timely expression of the stress regulatory genes is required for plants to compete against abiotic stresses; it necessitates the isolation and characterization of stress-responsive promoters for improving crops' tolerance to abiotic stresses. Dehydration Responsive Element Binding (DREB) regulates the expression of numerous stress-responsive genes in plants and leads an inevitable role in the adaptation of plants to abiotic stresses. In this study, the promoter region of Phoenix dactylifera (Date palm, a major fruit crop of the arid region) PdDREB1G gene was isolated and characterized for the first time. A comparison of the activity of two promoter fragments, 880 bp (DS) and 1.6 kb (DF) of PdDREB1G to AtRD29A was performed. Histochemical assay displayed remarkable GUS staining and RT-qPCR analysis confirmed the induction of GUS expression in T3 plants of transformed tobacco subjected to different abiotic stresses. Furthermore, compared with the widely used AtRD29A promoter, the relative expression of GUS in leaves by DS and DF was three and twofold higher under salt stress, respectively, while it was twofold in polyethylene glycol (PEG) and abscisic acid (ABA) for DS. Under SA stress, DF and DS displayed 1.5 and onefold expression in leaves, respectively. In the root, DS showed a fourfold increased expression in salt, threefold in PEG and ABA, and twofold in SA. Hence, the DS promoter characterized in the present study becomes a choice over RD29A for abiotic stress responses and is useful to develop stress-tolerant transgenic plants by inducing the expression of stress-inducible genes on stress.
Key message
Novel inducible promoter DREB1G cloned from the Date palm, a major fruit crop of the arid region, exhibiting high fold expression over AtRD29A to drought and salinity stress is useful to develop stress-tolerant transgenic plants by inducing the expression of stress-inducible genes on stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are constantly challenged by various environmental stresses especially drought, salinity, and temperature. These abiotic stresses hinder plant growth by affecting biochemical, physiological, and molecular activities from germination onwards causing severe loss to crop productivity. Nevertheless, some plant species are adapted to compete against the stress through an integrated network of various mechanisms like the activation of genes by inducible promoters.
Promoters are crucial regulators of gene expression due to the presence of important cis-acting elements. Promoters are upstream gene regulatory sequences recognized by transcription factors (TFs) involved in controlling transcription initiation and progression. In modern crop improvement strategies, the design of efficient gene constructs relies on promoter efficiency, tissue specificity, and other characteristics that allow the introgression of agronomically relevant traits to overcome biotic and abiotic stresses (de Melo et al. 2021). The widely used promoter for the development of transgenic plants, pCaMV35S is reported to have other effects like gene silencing and metabolic penalties impacting plant fitness (de Melo et al. 2021). The constitutive promoters cause unnecessary high gene expression all the time which leads to interference with other cellular pathways of plant development (Zhang et al. 2016; Jiang et al. 2018). Variability in the expression pattern of genes under CaMV 35S promoter across different plant species, among and within tissues, and under different environmental conditions has been exemplified (Schnurr et al. 2000). Post-transcriptional and translation silencing by overexpression of genes under 35S promoter has been documented well (Rajeevkumar et al. 2015). The pros and cons of 35S promoter with its limitation to use it as a preferred promoter are also reviewed (Amack and Antunes 2020). To circumvent the negative impacts of constitutive promoters, the identification and functional characterization of plant-derived stress-inducible promoters be on focus as an alternative to drive the expression of the genes on stress with no negative impact on plant growth. The inducible promoters that cause temporal expression of a gene based on certain stimuli are enable to eradicate the negative effects of the constitutive promoters. They are used to express stress-tolerant genes under certain stressful conditions such as drought, heat, cold, dehydration, and oxidative environment (Rai et al. 2009; van Essen et al. 2010). Overexpression of AtDREB1A under stress-inducible AtRD29A promoter in transgenic Arabidopsis and rice showed improved tolerance to abiotic stress with no abnormalities in plant growth (Kasuga et al. 2004; Kong et al. 2016). Besides, the right expression of the stress regulatory genes in a spatiotemporal fashion is required to target diverse abiotic stresses (Kasuga et al. 1999), and it mandates a well-characterized inducible promoter. The review on the impact of inducible promoters in transgenic plant production and crop improvement (Misra and Ganesan 2021) signifies the importance to look for act-on stress promoters. High-level expression by inducible promoters has been reported under different stresses: salinity and osmotic stress-inducible GAPP promoters from maize (Hou et al. 2016), heat-inducible Apx, Dhn and Hsc70 from pearl millet (Divya et al. 2019), osmotic and cold stress-inducible BBX24 promoter from chrysanthemum (Imtiaz et al. 2015), drought stress-inducible promoters RD29A and RD29B from Arabidopsis (Bihmidine et al. 2013), and osmotic stress-inducible DREB2, DREB6, and Wdreb2 from wheat (Wang et al. 2021). Among various abiotic stress promoters characterized, the AtRD29A promoter is the prime one with stronger activity under drought stress (Yamaguchi-Shinozaki and Shinozaki 1993, 1994), therefore, it has been successfully used to drive the expression of drought-tolerant genes in different plant species (Kasuga et al. 2004; Bihmidine et al. 2013).
Dehydration Responsive Element Binding (DREB) gene has been widely gaining attention as these are important plant transcription factors involved in pathways for enhancing abiotic stress tolerance by regulating the expression of many stress-inducible genes. However, the functionally characterized promoters of DREB transcription factors are relatively limited: DREB3 from soybean (Xiao et al. 2008), DREB1B from rice (Gutha and Reddy 2008), DREB2C from Arabidopsis (Chen et al. 2012), DREB1 from buckwheat (Fang et al. 2015) and DREB2, DREB6 and Wdreb2 from wheat (Wang et al. 2021). The studies on DREB1G are limited to the expression of the OsDREB1G gene of rice which is documented as a cold stress-responsive DREB gene (Moon et al. 2019).
Date palm (Phoenix dactylifera L.), an important fruit crop of the Arecaceae family is a perennial and dioecious monocot widely cultivated in arid and semi-arid regions of North Africa and the Middle East. Its adaptability to arid and semi-arid regions, especially to withstand temperature fluctuations, ranging from 56 to 60˚C to a few degrees below zero (Safronov et al. 2017) makes it a choice to explore the genome to design strategies like cloning of stress-inducible promoters as well as genes imparting tolerance and thereby the development of resilient crops.
We have investigated the promoter potential of the upstream region of the date palm DREB1G gene for the first time. The activity of the promoter fragments cloned from Date palm was evaluated in transgenic tobacco plants under different stress conditions viz., salinity, drought, cold, abscisic acid (ABA), and salicylic acid (SA). Further, in the present study, the promoter activity was also compared to the widely used stress-inducible promoter AtRD29A.
Materials and methods
Plant material and cloning of the promoter fragments
Leaf tissues of Date palm (Phoenix dactylifera L.) cv. Khalas were collected from the Date Palm Research Center, UAE University, Al Ain. DNA was extracted from the leaf tissues ground in liquid nitrogen using the Bioline DNA extraction kit (Meridian Biosciences, TN, USA) following the kit protocol. The DNA was quantified using Nanodrop 2000 (ThermoScientific, USA). The primers for the DREB1G promoter were designed from the upstream sequence of the translational start codon of the PdDREB1G gene sequence retrieved from the date palm genome database (NW_008246748.1:c253756-251297 Phoenix dactylifera cultivar Khalas unplaced genomic scaffold, DPV01 pdS000242) using Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012; https://bioinfo.ut.ee/primer3-0.4.0/). The two promoter fragments (1.6 kb and 880 bp) were amplified using the primers DREB-F1: CGGAATTCCCGTGCTATGGCATGATTA, DREB-R: TGCCTAGGGTTTCTCGGGGACTGATTGG, and DREB-F2: CGGAATTCAATGGTGCCA TGAATTGGAT (DREB-F1 and R for 1.6 kb and F2 and R for 880 bp (the bases in italics represents the restriction sites of EcoR1 and Avrll for the forward and reverse primers respectively). The PCR was performed with the primers using Phusion High-Fidelity Taq polymerase (NEB, USA) following the program: initial denaturation at 98 °C for 30 s, and 30 cycles of denaturation (at 98 °C for 10 s), annealing (at 55 °C for 20 s) and extension (at 72 °C for 40 s) with a final extension at 72 °C for 10 min. The RD29A promoter (730 bp, AB019226.1) from Arabidopsis thaliana DNA (extracted using DNeasy Plant Mini kit, Qiagen, Hilden, Germany) using the primers RD29A-F (CGGAATTCGTGAATTAAGAGGAGAGAGGAGG) and RD29A-R (TGCCTAGGTTTCCAAAGATTTTTTTCTTTCC) by following the above PCR program. The amplified PCR products were electrophoresed on 1% (w/v) Agarose gel and the fragments were purified using a QIAquick PCR gel extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The purified amplicons were cloned into the pCR®-Blunt II-TOPO® vector using the Zero Blunt TOPO PCR Cloning Kit (ThermoFisher, MA, USA) following the kit protocol. The cloned fragments were confirmed by sequencing (Macrogen, Seoul, South Korea).
Analysis and comparison of Cis-elements
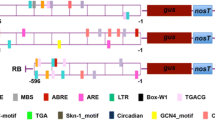
The sequence of PdDREB1G promoter fragments was scanned for the presence of cis-acting elements using the program PlantCARE (Lescot et al. 2002; http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The regulatory elements were analyzed using the PLACE (Higo et al. 1999; http://www.dna.affrc.go.jp/PLACE/) database. The upstream region 1.6 kb and 880 bp from the start codon of the PdDREB1G gene were analyzed and compared to AtRD29A promoter.
Construction of the promoter-GUS reporter vectors
The promoter fragments were cloned upstream of the gusA reporter gene of the plant transformation plasmid vector pCAMBIA1391Z. The gusA gene has a 5’-extension with a catalase intron to ensure the expression in plants but not in bacteria. The plasmid has genes for selecting kanamycin (nptI) resistance in bacteria and hygromycin (enhanced 35S:hpt) resistance in plants. The promoter fragments digested from the pCR®-Blunt II-TOPO® vector with EcoR1 and Avrll restriction enzymes were ligated into the pCAMBIA1391Z vector containing gusA gene digested with the same enzymes using T4 DNA ligase (NEB, USA) to construct the plant transformation vector pCAMBIA1391Z-PDREB1G-1.6::GUS named as DF and pCAMBIA1391Z-PDREB1G-880::GUS named as DS (Fig. 1). The plasmid vector pCAMBIA1391Z-PRD29A::GUS (as RD29A hereafter) was developed by ligating the digested fragment of the RD29A promoter as above. The ligated plasmids were transformed into DH5α by the chemical transformation method. The plasmids of the positive clones confirmed by PCR using the promoter-specific primers as described previously were extracted using QIAprep spin Miniprep kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quantity of the plasmids was measured by Nanodrop 2000 and the sequence was confirmed by sequencing (Macrogen, Seoul, South Korea). The recombinant plasmids of DS, DF, RD29A, and also pCAMBIA1391Z-35S::GUS were mobilized into disarmed hypervirulent A. tumefaciens strain EHA105 (Hood et al. 1993) through electrotransformation. The positive colony (confirmed by PCR with specific primers) selected was used for tobacco transformation.
Agrobacterium-mediated transformation of tobacco
The binary vector constructs in A. tumefaciens strain EHA105 streaked on solid LB medium (10 g l–1 Tryptone; 5 g l–1 Yeast extract; 5 g l–1 NaCl; 15 g l–1 bacteriological grade agar) plates containing 25 mg l–1 rifampicin (Phytotechnology Laboratory, KS, USA) and 50 mg l–1 kanamycin (Phytotechnology Laboratory, KS, USA). Single colonies of the bacteria were transferred into 25 ml liquid LB medium containing antibiotics in a 50 ml sterile Falcon tube and incubated overnight horizontally on a shaker (250 rpm) at 28 °C. The cells were pelleted by centrifugation at 8000 xg for 7 min after reaching the culture at 0.5–0.6 OD (OD600). The LB medium was replaced with an equal volume of liquid MS (Murashige and Skoog 1962; Phytotechnology Laboratory, KS, USA) medium (3% w/v sucrose) containing 200 μM acetosyringone (Acros Organics, Geel, Belgium). After dispersing the pellet, the bacterial culture was incubated at 50 rpm for 1–2 h at room temperature (22–23 °C).
Nicotiana tabacum cv. SR1 was transformed for promoter characterization. Leaf segments of tobacco (1.5 to 2.5 cm segments excised from in vitro grown tobacco plants on half-strength MS medium with 2% (w/v) sucrose were immersed in the Agrobacterial suspension for 20 min and was followed by vacuum infiltration of 10 min. The infected leaf segments were blot-dried on sterile filter paper and co-cultivated on MS medium (with 3% sucrose) supplemented with 9.4 μM Kinetin (Sigma, USA), 5.7 μM Indole-3-acetic acid (IAA; Sigma, USA), and 200 μM acetosyringone. Co-cultivated plates were incubated in dark at 25 ± 2 °C for 2 days. The co-cultivated leaf segments were washed with 300 mg l–1 timentin (Phytotechnology Laboratory, KS, USA) solution, blot dried, and cultured on a regeneration medium with the selection agent i.e., MS medium with 9.4 μM Kinetin, 5.7 μM IAA, 300 mg l–1 timentin, and 25 mg l–1 hygromycin (Phytotechnology Laboratory, KS, USA). The shoots grown were transferred on MS basal medium supplemented with 100 mg l–1 timentin, and 25 mg l–1 hygromycin in 3–4 weeks. The well-grown shoots were rooted on half-strength MS medium (with 2% sucrose) containing 100 mg l–1 timentin, and 25 mg l–1 hygromycin. The pH of all plant tissue culture media was adjusted to 5.8 before the addition of 0.8% (w/v) agar (PlantMedia, Ohio, USA) in the case of solid media and was autoclaved at 121 °C (15 lb) for 20 min. Antibiotics and filter-sterilized plant growth regulators dissolved in respective solvents were added to the media after autoclaving. All the plant cultures were incubated in a growth room at 25 ± 1 °C (60–80% humidity) in 16 h light: 8 h dark photoperiod under white fluorescent tubes (40 μmol m−2 s−1) unless otherwise mentioned.
Molecular confirmation of transgenic plants
Genomic DNA extracted from leaves of rooted shoots growing on half-strength MS medium (with 2% sucrose) containing 100 mg l–1 timentin, and 25 mg l–1 hygromycin using the cetyltrimethylammonium bromide (CTAB; Sigma, St. Louis, USA) method as described by Dutta et al. (2013). Putatively transformed plants (9 independent lines) were confirmed by the presence of DS and DF with respective primers as described earlier and the gusA gene using PCR. PCR reactions were performed in a volume of 20 μl containing 1 × reaction buffer, 100 ng of DNA template, 1.5 mM MgCl2, 0.2 mM of each of the dNTPs, 0.2 μM of each primer, and 2.5 unit of HotStar Taq DNA polymerase (HotStar Taq DNA polymerase kit, Qiagen, Hilden, Germany). The PCR conditions of gusA gene-specific primers (GUS-F: ATGAACATGGCATCGTGGTGATTG; GUS-R: GAGATCGCTGATGGTATCGGTGTG) consist of 95 °C for 15 min, 25 cycles of 94 °C for 20 s, 60 °C for 20 s, 72 °C for 30 s and a final extension of 72 °C for 5 min. The plasmid DNA (pCAMBIA1391Z) was used as a positive control, whereas DNA isolated from untransformed plants (WT) was used as the negative control.
Acclimatization of transgenic (T0) plants
PCR confirmed plantlets after washing were planted in a soil mix (sand and peat; 1:1) in small pots (8 cm w × 8.5 cm ht). The pots were initially covered by a polyethylene bag for 10 days to retain moisture during the acclimatization in the plant room (25 ± 2 °C, 70% humidity, 16 h light: 8 h dark photo-period, 400 μmol/m2/s—Heliospectra LED Lights). The well-growing plantlets were transplanted into 5 L pots (21 cm w × 19 cm ht) and grown till seed harvest (5–6 months).
Generation of T2 and T3 plants
The transgenic T1 tobacco seeds with promoters and gusA gene collected from different T0 plant lines were surface sterilized separately by treating with 10% (v/v) Clorox solution in 2 ml Eppendorf tubes for 10 min with inversion. After washing three times with sterile water, the blot-dried seeds were cultured on half-strength MS (2% sucrose) containing 25 mg l–1 hygromycin for germination. The healthy plants derived from T1 seeds after 20 days were selected based on segregation and were transplanted into the soil as described previously. T2 transgenic tobacco seeds germinated on hygromycin containing half-strength MS were subsequently transplanted into soil and collected the seeds from the dried pods. The T3 plantlets were used for the promoter analysis studies.
Abiotic stress treatments
For characterization and elucidating the response of PdDREB1G promoter to different abiotic stresses, 25-day-old transgenic T3 plantlets (three lines each) of DS, DF, RD29A, 35S, and untransformed plantlets (WT) were subjected to 150 mM NaCl, 10% (w/v) PEG6000, 20 µM ABA, 10 µM SA, 4 °C for 24 h. After 24 h some of the treated plantlets were collected for GUS staining; the remaining plantlets (leaves and roots) were frozen in liquid nitrogen and stored at -80 °C for the real-time qPCR analysis. All experiments were repeated three times.
Histochemical GUS assay
Seedlings (15- and 25-days) of WT and transgenic plants of DS, DF, RD29A, and 35S were collected for GUS expression analysis in response to various abiotic stresses. Histochemical staining was performed following the procedure described by Jefferson et al. (1987). The samples collected from transgenic plantlets and WT with and without stress were transferred into the GUS assay buffer (1 mg m l–1 X-Gluc, 100 mM sodium phosphate buffer pH 6.8, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 100 μM Na2EDTA) and after applying vacuum (10 min) were incubated overnight at 37 °C. The chlorophyll was removed by repeated washing with absolute alcohol: acetic acid (3:1, v/v) solution. After destaining, the samples were stored in 5% (v/v) acetic acid and photographed (Nikon D5300). For each construct, samples were collected from at least three different transgenic lines.
RNA isolation and RT‐qPCR analysis
Total RNA from the leaves and roots of the transformed tobacco seedlings and control was extracted using Maxwell® RSC Plant RNA Kit (Promega, Madison, USA). The quantity and quality of RNA were checked by Nanodrop 2000 and gel electrophoresis, respectively. cDNA was synthesized from 1 μg total RNA using a QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The qRT-PCR reactions were carried out in a total of 10 μl volume containing 2 μl of diluted cDNA (1:5), 200 nM of gusA gene primers (GUS-qF: GAATACGGCGTGGATACGTTAG; GUS-qR: GATCAAAGACGCGGTGATACA) and 5 μl of 2 × Fast SYBR Green PCR Master Mix (Applied Biosystems, CA, USA) in an optical 96 well plate using a StepOnePlusTM Real-Time PCR System (Applied Biosystems, CA, USA). qRT-PCR temperature cycle was set up as 95 °C for 10 min, 40 cycles of 95 °C for 10 s, 58 °C for 15 s, and 72 °C for 20 s. The transcript level of the gusA gene under the control of different promoters between samples was compared by the 2−ΔΔCt method (Livak and Schnittgen 2001). Actin (LOC_107821481) tobacco (NtACT-qF: AGGCTGTCCTTTCCTTGTATG; NtACT-qR: CAAAGCATGACCCTCGTAGAT) was used as an endogenous reference gene for normalization and calculating the fold change of transgene. Three technical replicates were analyzed for each biological replicate.
Statistical analysis
For all the stress experiments, the relative expression of the genes and standard error values were analyzed using the Microsoft Excel program. The data values represent means ± SE from three independent experiments. Student t-test was performed to assess the significant differences between control and treatment conditions. Differences of P < 0.05 were considered significant.
Results
Bioinformatic analysis of the PdDREB1G promoter sequence
A 1.6 kb fragment (DF) and 880 bp fragment (DS) located upstream of the start codon (ATG) of PdDREB1G were isolated from the genomic DNA of the date palm cv. Khalas. The analysis of the promoter sequences using PLACE and PlantCARE showed many abiotic stress-responsive cis-elements. The frequency of cis-elements related to abiotic stress in the DF, DS fragments, and RD29A was compared (Table 1). The results revealed that the promoter sequences contain several water-deficit responsive cis-acting elements such as EBOXBNNAPA (CANNTG), DRE1COREZMRAB17 (ACCGAGA), ABRELATERD1 (ACGTG), ACGTATERD1 (ACGT), MYB2AT (TAACTG), MYBCORE (CNGTTR), MYB1AT (WAACCA), MYB2CONSENSUSAT (YAACKG). G-box (CACGTG) and TC-rich repeats (GTTTTCTTAC) are responsible for dehydration and salinity tolerance. The LTRECOREATCOR15 (CCGAC) represented low-temperature tolerance. The DF fragment contained one SA-responsive TCA (CCATCTTTTT) element. The numbers of ABRE elements accountable for the activation of genes under early dehydration were 11 and 10 in DF and DS fragments, respectively. EBOXBNNAPA was intended for dehydration, and the ABA-responsive elements were nine in the DF region and three in the DS fragment. Two G-box regions in DF and one in DS were responsible for dehydration, high salinity, and ABA response. There were two DRE1COREZMRAB17 and TC-rich repeats in the DF fragment and one each in the DS fragment which is significant for drought and salinity tolerance. The promoter sequence showed two cold-responsive LTR elements which were on the DS region. There were two MYBCORE and one MYB2AT region in DF and one each MYB1AT region in DF and DS fragments which are important for dehydration stress response (Fig. 2 & 3).
PCR confirmation, GUS assay, and characteristics of the PdDREB1G promoter in response to various abiotic stresses
Putatively transformed plants (9 independent lines) were confirmed by the presence of DS and DF (with respective primers) with an amplicon of 880 bp and 1.6 kb, respectively (Fig. 4a). All transgenic plants were also confirmed by the gusA gene (an amplicon of 353 bp) using PCR (Fig. 4b). The GUS assay of the transgenic seedlings with the gusA gene under the promoter fragments of the present study (DF and DS), under AtRD29A and 35S, showed GUS activity. The GUS staining of the transgenic plants with the gusA gene under the control of DF and DS exposed to salt and PEG treatment showed intense blue staining (Fig. 5). Treatment with ABA showed comparatively less staining (Fig. 5). The staining after exposure to SA and cold was more intense in DS compared to DF (Fig. 5). The positive controls with 35S promoter displayed staining in all the stresses, but was less intense to non-stressed of the same (Fig. 5). AtRD29A deep blue staining in all stress conditions except cold. No GUS activity was detected in non-transgenic control (WT) plants (Fig. 5).
Quantitative gusA gene expression levels
Quantitative gusA gene expression levels of the DS and DF fragments measured via real-time PCR analysis after exposure of T3 tobacco seedlings to 150 mM NaCl, 10% polyethylene glycol 6000 (PEG 6000), 20 µM ABA, and 10 µM SA revealed high transcript levels of gusA compared to control (WT) indicated high promoter activity (Fig. 6a-e). The transcript levels were high in leaves compared to the root (Fig. 6a-e). Among these stresses, treatment with NaCl caused the most substantial changes in gusA gene expression, followed by ABA, PEG, and SA (Fig. 6a-e). The expression level was lowest under cold treatment (Fig. 6a-e). The DS showed an increased expression of gusA gene in all the given stresses in the leaf and root compared to DF (Fig. 6a-e). The only exception was in cold stress where there was no difference between DF and DS even though there was a significant increase in gusA gene expression level compared to the control (Fig. 6e).
Transcriptional expression levels of gusA in leaves and roots of transgenic tobacco plants under different promoters (DS, DF, and RD29A) and WT seedlings after 24 h of exposure to abiotic stresses. a. Salt, b. PEG, c. ABA, d. SA, e. Cold. The actin gene was used as an endogenous control for normalization. The relative expression was calculated with respect to WT plants. The relative gene expression was calculated by the 2^−ΔΔCt method. In all cases, data values represent means ± SE from three independent experiments. Asterisks indicate statistical significance (∗∗P < 0.01, Student’s t-test) of differences between transgenic lines and WT seedlings. (WT-Untransformed; NaCl-sodium chloride; PEG-polyethylene glycol; ABA-abscisic acid; SA-salicylic acid)
When compared with the RD29A promoter, in the case of leaves the expression by DS was threefold higher and DF was twofold under salt stress (Fig. 7a). The DS was two times higher in PEG and ABA. The DS, DF, and RD29A showed no significant difference under SA and cold stress (Fig. 7a). In the case of root, expression of the gusA gene in DS was fourfold higher in salt, threefold higher in PEG and ABA, twofold higher in SA, and no significant difference under cold stress (Fig. 7b). Expression of the gusA gene by DF displayed no significant difference compared to the RD29A promoter (Fig. 7b).
Transcriptional expression levels of gusA in leaves and roots of 25-day-old transgenic tobacco plants under different promoters (DF, DS, and RD29A) after 24 h of exposure to abiotic stresses (Salt, PEG, ABA, SA, and Cold). a. Leaves. b. Roots. The actin gene was used as an endogenous control for normalization. Relative expression of DS and DF were calculated with respect to RD29A plants. The relative gene expression was calculated by the 2^-ΔΔCt method. In all cases, data values represent means ± SE from three independent experiments. Asterisks indicate statistical significance (∗P < 0.05 and.∗∗P < 0.01, Student’s t-test) of differences of DS and DF with respect to the RD29A line. (WT-Untransformed; NaCl-sodium chloride; PEG-polyethylene glycol; ABA-abscisic acid; SA-salicylic acid)
Spatiotemporal expression of the PdDREB1G promoter
Since GUS staining with DS promoter was the highest under salt stress and more prominent than DF, DS transgenic plants were used to determine whether promoter fragment DS is developmentally regulated. The patterns of the GUS histochemical staining were monitored during different plant developmental stages (15-, 25-day-old and mature plants), and in tissues such as roots, leaves, stems, flowers, pods, and seeds under salt stress (Fig. 8a-h). In 2 week-old tobacco seedlings, the intensity of GUS staining was weak (Fig. 8a). In a 25-day-old seedling, there was GUS staining in almost all tissues, but intense in the vascular region (Fig. 8b). In well-grown plants, GUS activity was prominent in leaves, roots, and stems (Fig. 8c-h). GUS staining was higher in leaves than in roots (Fig. 8c). In the case of the root, GUS staining was intense in the elongation zone of the root and root tips (Fig. 8d). The stem hairs and vascular tissues exhibited intense blue staining (Fig. 8e). There was considerable staining on petals and pods (Fig. 8f, g), but was negligible in seeds (Fig. 8h). The results showed that the inducible promoter potential of the DS fragment in driving gene expression was in a spatiotemporal manner under stress.
Discussion
Stress tolerance of plants is materialized by inducing the expression of genes by the promoters which control the binding of RNA polymerase to DNA. The selection of the promoter i.e., a stretch of DNA comprising the core-promoter region and multiple repeats or combinations of heterologous upstream regulatory elements (cis-motifs or TF-binding sites) is significant in designing a transformation cassette that would enable the precise control of transgene activity (Ali and Kim 2019). The selection of a promoter, to confer constitutive, spatial, and/or temporal transgene expression, is one of the vital components for the development of genetically modified plants. Of the promoters, constitutive promoters are of wide use but are reported to have undesirable effects such as gene silencing due to methylation, etc. on the expression of transgenes (Okumura et al. 2016; Amack and Antunes 2020). Validation of promoters for precise spatial and temporal control of transgene expression contributes to the improvement of crop productivity and sustainable agriculture. The process of transcription is essential for gene regulation and is accomplished through sequence-specific binding of transcription factors to their target promoters (Ali and Kim 2019). The pivotal role in controlling processes is not played by encoding sequences, but by regulatory elements which dynamically enhance or restrict gene expression levels within an organism (Venter 2007). Targeted activation of promoters by environmental stresses inducing the expression of genetic information leading to the final product has great significance, and the review by Misra and Ganesan (2021) exemplifies its importance in transgenic plant production and crop improvement. The understanding of the regulation of plant gene expression at the cis-acting elements level of a promoter cloned from a native plant gene of date palm, an arid fruit crop tailored to gusA gene and its functional validation at the plant level is an add-on to the development of stress competent crops.
Date palm is one of the widely cultivated fruit crops in arid regions, the genes and the mechanism involved in its tolerance to various abiotic stress are relatively unknown. The functional analysis of the promoter fragments of the DREB1G gene from the Date palm in the present study revealed multiple cis-acting elements that modulate transcription in a stress-inducible and tissue-specific manner. DREBs are important transcription factors belonging to the family of AP2/ERF transcription factors containing conserved AP2/ERF domain and are further subdivided into six subgroups of A-1 to A-6. These DREBs bind to CRT/DRE cis-elements (A/GCCGAC) in the promoters which regulate genes playing a pivotal role in plants' tolerance to biotic and abiotic stresses (Zhou et al. 2010). It is proven now that most DREB genes are regulated by abiotic stresses, and this induction may be ABA-dependent or ABA-independent. (Yoshida et al. 2014). Further, the DREB1/CBF-type TFs are activated by four or fewer major abiotic stresses (cold, heat, drought, and salinity), although the pattern of expression of orthologous genes in different species varied (Yang et al. 2020; Li et al. 2021).
The efficacy of the date palm promoters of the present study and the Arabidopsis-derived stress-inducible promoter RD29A in driving the expression of the gusA reporter gene in transgenic tobacco was analyzed. In this study, we have compared long (1.6 kb) named DF and short (880 bp) named DS of the PdDREB1G promoter fragments to find out the optimum promoter length for higher activity under abiotic stress. The well-known drought AtRD29A promoter was kept for comparison as the positive control. The screening of cis-elements showed that the PdDREB1G promoter fragments were enriched in stress-related cis-elements related to salinity, dehydration, ABA, and temperature (Fig. 3). This result directly correlated with an excellent performance of the promoter under salinity, dehydration and ABA treatment (Fig. 5 and Fig. 6a-e). The DF and DS showed similar or greater GUS activity than the AtRD29 promoter. Compared to the DF promoter and AtRD29, the performance of DS was significantly high in salinity, PEG, and ABA treatment (Fig. 7a, b). The expression level was comparatively less in SA and cold treatment (Fig. 7a, b). Most of the cis-elements related to drought, salinity, and cold like ACGTATERD1(ACGT), ABRELATERD1(ACGTG), EBOXBNNAPA (CANNTG), GBOX (CACGTG), LTR (CCGAC) are concentrated in the DS (880 bp) fragment (Fig. 2). For example, the occurrence of 10 ACGTATERD1 regions and 6 ABRELATERD1 and 3 EBOXBNNAPA regions in the DS fragment could be the reason for the high GUS expression under salinity, PEG and ABA (Guiltinan et al. 1990). ABA signaling is also known to play an important role in salinity and drought stress tolerance. ABRE functioned in response to low-temperature, high-salinity, and dehydration treatments, but not to ABA. Studies showed that the G-box family core (ACGT) is the most conserved element among different plant species and responds to abiotic stresses especially water-deficit and salt stresses (Mehrotra et al. 2013). Other cis-elements are also present in DF and DS such as MYBs, which are known to respond to water deficit and ABA (Hussain et al. 2021). Functional characterization of the TkSRPP promoter in response to hormones and wounding stress in transgenic tobacco has also been reported (Dong et al. 2023). Increased expression of GUS in DS compared to positive control AtRD29A could be because of fewer number cis-elements in its promoter region e.g., five ACGTATERD1 regions, one ABRELATERD1, and two EBOXBNNAPA regions (Table 1). Responsiveness to SA indicates that PdDREB1G has a role in oxidative stress and disease response in plants as had been documented in OsDREB1B (Gutha and Reddy 2008). The cis-acting element (TACCGACAT), namely the dehydration-responsive element (DRE), is absent in the DF and DS promoters of the present study but has been present in the promoter regions of many dehydration and low-temperature stress-inducible genes (Shinozaki and Yamaguchi-Shinozaki 2000). Fang et al. (2015) notified high GUS activity under drought stress conditions by the FeDREB1 promoter of common buckwheat. Further, in coffee plants, three different promoter haplotypes of CcDREB1D consist of different cis-elements that are involved in the tissue-specific expression, ABA and light regulation has been reported (Alves et al. 2018).
The differences in GUS expression between DS and DF under different types of abiotic stresses are probably related to the distribution pattern of specific cis-regulatory sequences and the co-localization and aggregation of motifs nearby as documented in rice and sorghum (Srivastav et al. 2010). The activation of genes depends not only on the cis-elements in the promoter but also on their positions and the presence of enhancers, regulatory sequences, repressors, and other synergistic cis-elements (Sawant et al. 2005). In spite of the presence of a high number of stress-responsive cis-elements in DF, the expression of GUS under salinity, PEG and ABA are significantly less in DF compared to DS. A recent study showed the presence of repressor region 5′-AATGATA-3′ region in the promoter could negatively affect the expression of genes under salt stress or hypoxia (Seok et al. 2022). The DF fragment, which is not a part of the DS fragment contains one repressor region (Fig. 3). The reduced activity of the DF promoter compared to DS may be due to the presence of the repressor elements in the distal region of the promoter i.e., specific to the DF fragment. Though there are more cis-elements in the DF, their relative distance of them from the transcription start site may also be a reason behind the reduced activity over DS. The upstream region of several gene promoters was found to contain positive or negative regulatory elements, some of which were characterized as enhancers or silencers (Timko et al. 1985; Tyagi 2001). The DS region may also contain an uncharacterized new element crucial for salinity stress response because of its ability to respond to salt stress in high profoundness.
GUS staining revealed that the DS promoter was active in different tissues and organs of the tobacco plants, such as roots, stems, leaves, flowers, and pods except in seeds (Fig. 8a-h). This result was similar to the PsDREB2 promoter in Paeonia suffruticosa (Liu et al. 2019), and the GmPRP2 promoter in soybean (Chen et al. 2014). PdDREB1G showed stress-specific and organ-specific expression as reported by the OsDREB1B promoter (Gutha and Reddy 2008). The results proved that the promoter PdDREB1G is highly activated under salinity than cold stress which is different from the result obtained with overexpression of OsDREB1G which was highly activated only with cold stress and not with drought and salinity or ABA (Moon et al. 2019). In line with it, the expression in the present study was high in the leaf. High low-temperature activation has also been demonstrated by OsDREB1B (Gutha and Reddy 2008). The differences in the result with respect to our study are due to the promoter modules enriched in cis-acting elements which drive more consistent gene expression and it reinforces the idea of a synergistic effect of cis-elements in gene promoter sequences.
Very specific expression patterns of transgenes enable elucidation of the cellular regulation mechanisms; in such cases, inducible promoters are an open choice as they switch on or off the gene of interest under certain conditions or at certain developmental stages. The comparison of the DS promoter to well-studied stress inducive promoter RD29A in the present study establishes that DS is a strong inducible promoter over RD29A except in cold stress and will be a choice in designing transformation-cassettes in the development of genetically modified crops for stress tolerance.
Conclusions
Validation of promoters with respect to the reflexes of agronomically important plants like Date palms growing well in arid regions is of great significance in the development of genetically modified plants with improved stress competence. The present study for the first time places a strong stress-inducible promoter cloned from the arid fruit crop, Date palm is an open choice for the development of stress-tolerant crops.
Data availability
The data are available from the corresponding author’ on request.
Abbreviations
- ABA:
-
Abscisic acid
- DF:
-
DREB full (1.6 kb)
- DS:
-
DREB short (880 bp)
- GUS:
-
β-Glucuronidase
- IAA:
-
Indole-3-acetic acid
- PEG:
-
Polyethylene glycol
- SA:
-
Salicylic acid
References
Ali S, Kim W-C (2019) A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front Plant Sci 10:1433. https://doi.org/10.3389/fpls.2019.01433
Alves GSC, Torres LF, de Aquino SO et al (2018) Nucleotide diversity of the coding and promoter regions of DREB1D, a candidate gene for drought tolerance in Coffea species. Tropical Plant Biol 11:31–48. https://doi.org/10.1007/s12042-018-9199-x
Amack SC, Antunes MS (2020) CaMV35S promoter – A plant biology and biotechnology workhorse in the era of synthetic biology. Curr Plant Biol 24:100179
Bihmidine S, Lin J, Stone JM et al (2013) Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 237:55–64. https://doi.org/10.1007/s00425-012-1740-9
Chen H, Je J, Song C, Hwang JE, Lim CO (2012) A proximal promoter region of Arabidopsis DREB2C confers tissue-specific expression under heat stress. J Integr Plant Biol 54:640–651. https://doi.org/10.1111/j.1744-7909.2012.01137.x
Chen L, Jiang B, Wu C et al (2014) GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant Biol 14:1471–2229. https://doi.org/10.1186/s12870-014-0245-z
de Melo BP, de Moura SM, Morgante CV et al (2021) Regulated promoters applied to plant engineering: an insight over promising soybean promoters under biotic stress and their cis-elements. Biotechnol Res Innov 5:e2021005. https://doi.org/10.4322/biori.202105
Divya K, Kavikishor PB, Bhatnagar-Mathur P et al (2019) Isolation and functional characterization of three abiotic stress-inducible (Apx, Dhn, and Hsc70) promoters from pearl millet (Pennisetum glaucum L.). Mol Biol Rep 46:6039–6052. https://doi.org/10.1007/s11033-019-05039-4
Dong G, Fan M, Wang H et al (2023) Functional characterization of TkSRPP promoter in response to hormones and wounding stress in transgenic tobacco. Plants 12:252
Dutta I, Kottackal M, Tumimbang E, Tajima H, Zaid A, Blumwald E (2013) Sonication-assisted efficient Agrobacterium-mediated genetic transformation of the multipurpose woody desert shrub Leptadenia pyrotechnica. Plant Cell Tiss Organ Cult 112:289–301. https://doi.org/10.1007/s11240-012-0236-4
Fang ZW, Xu XY, Gao JF et al (2015) Characterization of FeDREB1 promoter involved in cold- and drought-inducible expression from common buckwheat (Fagopyrum esculentum). Genet Mol Res 14:7990–8000. https://doi.org/10.4238/2015.July.17.7
Guiltinan MJ, William R, Marcotte J, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250:267–271. https://doi.org/10.1126/science.2145628
Gutha LR, Reddy AR (2008) Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Bio 68:533–555. https://doi.org/10.1007/s11103-008-9391-8
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database 1999. Nucleic Acids Res 27:297–300
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218. https://doi.org/10.1007/BF01977351
Hou J, Jiang P, Qi S et al (2016) Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-11 H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PlosOne 11:e0154041. https://doi.org/10.1371/journal.pone.0154041
Hussain Q, Asim M, Zhang R et al (2021) Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity Stress. Biomol 11:1159. https://doi.org/10.3390/biom11081159
Imtiaz M, Yang Y, Liu R et al (2015) Identification and functional characterization of the BBX24 promoter and gene from chrysanthemum in Arabidopsis. Plant Mol Biol 89:1–19. https://doi.org/10.1007/s11103-015-0347-5
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO 6:3901–3908. https://doi.org/10.1002/j.1460-2075.1987.tb02730.x
Jiang P, Zhang K, Ding Z et al (2018) Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotechnol 18:59. https://doi.org/10.1186/s12896-018-0470-x
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291. https://doi.org/10.1038/7036
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible promoter improved drought and low temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350. https://doi.org/10.1093/pcp/pch037
Kong X, Zhou S, Yin S et al (2016) Stress-inducible expression of an F-box gene TaFBA1 from wheat enhanced the drought tolerance in transgenic tobacco plants without impacting growth and development. Front Plant Sci 7:1295. https://doi.org/10.3389/fpls.2016.01295
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291
Lescot M, Déhais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li Z, Wang G, Liu X et al (2021) Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genom 22:456. https://doi.org/10.1186/s12864-021-07799-5
Liu H, Zhu K, Tan C et al (2019) Identification and characterization of PsDREB2 promoter involved in tissue-specific expression and abiotic stress response from Paeonia suffruticosa. Peer J 7:7052. https://doi.org/10.7717/peerj.7052
Livak KJ, Schmittgen TD (2001) Analysis of the relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mehrotra R, Sethi S, Zutshi I, Bhalothia P, Mehrotra S (2013) Patterns and evolution of ACGT repeat cis-element landscape across four plant genomes. BMC Genom 14:1471–2164. https://doi.org/10.1186/1471-2164-14-203
Misra S, Ganesan M (2021) The impact of inducible promoters in transgenic plant production and crop improvement. Plant Gene 27:100300
Moon S-J, Min MK, Kim J-A et al (2019) Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front Plant Sci 10:297. https://doi.org/10.3389/fpls.2019.00297
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 5:473–497
Okumura A, Shimada A, Yamasaki S et al (2016) CaMV-35S promoter sequence-specific DNA methylation in lettuce. Plant Cell Rep 35:43–51. https://doi.org/10.1007/s00299-015-1865-y
Rai M, He C, Wu R (2009) Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res 18:787–799. https://doi.org/10.1007/s11248-009-9263-2
Rajeevkumar S, Anunanthini P, Sathishkumar R (2015) Epigenetic silencing in transgenic plants. Front Plant Sci 6:693. https://doi.org/10.3389/fpls.2015.00693
Safronov O, Kreuzwieser J, Haberer G et al (2017) Detecting early signs of heat and drought stress in Phoenix dactylifera (Date palm). PLoS ONE 12:e0177883. https://doi.org/10.1371/journal.pone.0177883
Sawant SV, Kiran K, Mehrotra R et al (2005) A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J Exp Bot 56:2345–2353. https://doi.org/10.1093/jxb/eri227
Schnurr JA, Guerra GA (2000) The CaMV-35S promoter is sensitive to shortened photoperiod in transgenic tobacco. Plant Cell Rep 19:279–282
Seok H-Y, Tran HT, Lee S-Y, Moon Y-H (2022) AtERF71/HRE2, an Arabidopsis AP2/ERF transcription factor gene, contains both positive and negative cis-regulatory elements in its promoter region involved in hypoxia and salt stress responses. Int J Mol Sci 23:5310. https://doi.org/10.3390/ijms23105310
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223. https://doi.org/10.1016/s1369-5266(00)80068-0
Srivastav A, Mehta S, Lindlof A, Bhargava S (2010) Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal Behav 5:775–784. https://doi.org/10.4161/psb.5.7.11769
Timko MP, Kausch AP, Castresana C et al (1985) Light regulation of plant gene expression by an upstream enhancer-like element. Nature 318:579–582. https://doi.org/10.1038/318579a0
Tyagi AK (2001) Plant genes and their expression. Transgenic rice: a valuable monocot system for crop improvement and gene research. Curr Sci 80:161–169
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3 - new capabilities and interfaces. Nucleic Acids Res 40:e115
van Essen D, Zhu Y, Saccani S (2010) A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol Cell 39:750–760. https://doi.org/10.1016/j.molcel.2010.08.010
Venter M (2007) Synthetic promoters: genetic control through cis engineering. Trends Plant Sci 12:118–124. https://doi.org/10.1016/j.tplants.2007.01.002
Wang H, Zhu Y, Yuan P et al (2021) Response of wheat DREB transcription factor to osmotic stress based on DNA methylation. Int J Mol Sci 22:7670. https://doi.org/10.3390/ijms22147670
Xiao S, Jian-Hu D, Ming C et al (2008) Isolation and regulative region analysis of promoter of stress-related gene GmDREB3 from soybean. Acta Agrono Sin 34:1475–1479. https://doi.org/10.3724/SP.J.1006.2008.01475
Yamaguchi-Shinozaki K, Shinozaki K (1993) Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol 101:1119–1120. https://doi.org/10.1104/pp.101.3.1119
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6:251–264. https://doi.org/10.1105/tpc.6.2.251
Yang Y, Al-Baidhani HHJ, Harris J et al (2020) DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: impact on plant development, stress tolerance and yield. Plant Biotechnol J 18:829–844. https://doi.org/10.1111/pbi.13252
Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21:133–139. https://doi.org/10.1016/j.pbi.2014.07.009
Zhang H, Hou J, Jiang P et al (2016) Identification of a 467 bp promoter of maize phosphatidylinositol synthase gene (ZmPIS) which confers high-level gene expression and salinity or osmotic stress inducibility in transgenic tobacco. Front Plant Sci 7:42. https://doi.org/10.3389/fpls.2016.00042
Zhou M-L, Ma J-T, Pang J-F et al (2010) Regulation of plant stress response by dehydration responsive element binding (DREB) transcription factors. Afr J Biotechnol 9(54):9255–9279
Acknowledgements
The authors acknowledge the financial support of the Ministry of Presidential Affairs, United Arab Emirates. We thank Dr. Ouwerkerk PBF, Institute of Biology, Leiden University, Leiden, The Netherlands, for providing the plasmid pCAMBIA1391Z.
Author information
Authors and Affiliations
Contributions
MK Conceived the project, PK designed and carried out the experiment mostly, drafted the Manuscript, KW carried out initial experiments, SS, GL, SK, and SRHA assisted with the experiments, and KA overlook the experiment. All authors reviewed and corrected the Manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest, financial or otherwise.
Ethical approval
The results in the Manuscript are the author's original work. The authors confirm that this manuscript has not been previously published and is not currently under consideration by any other journal. Additionally, all of the authors have approved the contents of this paper and have agreed to the journal’s submission policies.
Additional information
Communicated by Amita Bhattacharya.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kodackattumannil, P., Whitley, K., Sasi, S. et al. Novel inducible promoter DREB1G cloned from date palm exhibits high fold expression over AtRD29 to drought and salinity stress. Plant Cell Tiss Organ Cult 154, 367–380 (2023). https://doi.org/10.1007/s11240-023-02460-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-023-02460-3