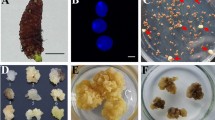
Abstract
The ability to produce doubled haploid (DH) plants has found broad application in research and breeding. For major crop species such as maize (Zea mays L.) and barley (Hordeum vulgare L.), routine large-scale production of DHs has enabled the acceleration of breeding processes, for example through efficient generation of homozygous lines. However, in forage crops such as perennial ryegrass (Lolium perenne L.), low and genotype-specific responses to in vitro anther culture (AC) still limit wide-spread use of DHs. Here, we report the responses of nine bi-parental populations, segregating for microspore embryogenesis and plant regeneration capacity, to an effective AC protocol. Genotypes of exceptionally high androgenic ability, producing over 200 green plants per 100 anthers cultured, could be selected. Continuous and distinctly shaped distributions for the evaluated traits were indicative of quantitative polygenic control and the presence of different alleles in each population. An insignificant association of embryo production with plant regeneration, as well as a low correlation between green and albino plant yield (ρ = 0.20), suggested that different genes influence these traits. The populations evaluated here provide a rich source of alleles needed for the introgression of high levels of androgenic capacity into recalcitrant material. Moreover, this germplasm is ideally suited for use in future genotyping and mapping studies so that the genetic control of androgenic capacity in perennial ryegrass can be elucidated. Ultimately, our results will help to realize the potential of DH induction in one of the world’s most important forage crop species.
Similar content being viewed by others
Introduction
The ability to produce doubled haploid (DH) plants significantly increases the efficiency and efficacy of basic and applied research as well as plant breeding programs. Access to completely homozygous material has been invaluable for the creation of mapping populations for genetic and genomic studies (Forster et al. 2007) and the simplification of genome sequence assembly (Dunwell 2010). Through self-pollination, DH populations can be propagated indefinitely and stored as seed, thus allowing for multi-year and multi-location phenotyping for QTL discovery (Guo et al. 2013), which has been particularly useful in cereal species (Cabral et al. 2014; Sannemann et al. 2015; Obsa et al. 2016). In breeding programs, DH production replaces inbred line development through repeated self-pollination, so that major reductions in the time to cultivar release are achieved (Dwivedi et al. 2015). Many of the modern barley (Hordeum vulgare L.), rice (Oryza sativa L.) and wheat (Triticum aestivum L.) cultivars are DHs (Devaux and Kasha 2009; Niu et al. 2014; Mishra and Rao 2016; Niak et al. 2016). When used as parents in hybrid breeding, DHs allow for effective exploitation of hybrid vigour (heterosis) (Birchler et al. 2010). Doubled haploid techniques have, for example, been essential in bringing about last century’s major increases in maize (Zea mays L.) yields (Geiger and Gordillo 2009).
Immature microspores are the starting material of choice for in vitro DH induction, since they are usually abundantly available and, by applying a stress treatment, can be induced to develop into embryos and then plants (Seguí-Simarro and Nuez 2008; Ferrie and Caswell 2011). Even though a plethora of anther and microspore culture protocols are available (Maluszynski et al. 2003; Dwivedi et al. 2015), their success is highly species- and genotype-specific (Seguí-Simarro 2010). In practice, access to DH production and its associated benefits is therefore restricted to the few crops in which methods are sufficiently developed to allow routine application, such as barley, tobacco (Nicotiana tabacum L.) and rapeseed (Brassica napus L.) (Seguí-Simarro 2015). Despite the relatively advanced state of DH techniques in perennial ryegrass (Lolium perenne L.), the world’s most economically valuable forage crop, they are not yet part of the standard repertoire of researchers and breeders (Arias Aguirre et al. 2011; Pembleton et al. 2015). A self-incompatibility system prevents efficient development of inbred lines in this species (Cornish et al. 1979), so that DH induction through androgenesis may well be the most practical way to obtain homozygous germplasm and thus enable, for instance, hybrid seed production (Begheyn et al. 2016a). Since DH research in perennial ryegrass began in the late 1970s, anther culture (AC) protocols have been successfully adapted from wheat (Olesen et al. 1988) and barley (Boppenmeier et al. 1989), improved by optimizing the sugar source (Bante et al. 1990) and cold pre-treatment duration (Opsahl-Ferstad et al. 1994a), and even converted to isolated microspore culture (IMC) (Andersen et al. 1997).
Nevertheless, it was soon realized that high androgenic capacity in perennial ryegrass is not only strongly genotype-dependent, but, similar to wheat, also a rare trait in breeding germplasm (Olesen et al. 1988; Bante et al. 1990; Madsen et al. 1995; Nielsen et al. 2015). In addition to improved induction protocols, introgression of androgenic capacity was therefore deemed a prerequisite for the implementation of DH production in perennial ryegrass breeding programs (Halberg et al. 1990). Investigations into the DH induction responses of genotypes derived from parents with known performances revealed that distinct genetic factors control embryo formation, plant regeneration frequency, and the percentage of green plants (Andersen et al. 1997). The presence of a few recessive genes with large effects, controlling embryo formation in an additive manner and regeneration and green plant frequencies in a dominant manner (Opsahl-Ferstad et al. 1994b), explained the observed genetic interactions. Genotypes exhibiting a high production of embryos and green plants were intercrossed to combine their beneficial alleles and resulted in the selection of genotypes with up to 53.5 fold increases in green plant production (Halberg et al. 1990). These ‘inducers’ were then used to introgress androgenic capacity into unresponsive material, which resulted in a 7.3 fold increase in green plant production (Madsen et al. 1995).
By the middle of the 1990s, it was supposed that AC would soon find wide application in breeding programs because an in vitro protocol was available and the ‘inducer’ approach was demonstrated to be effective (Opsahl-Ferstad et al. 1994a). Remarkably, the two decades that followed saw neither scientific communications on DHs in perennial ryegrass nor their use in commercial breeding, possibly due to the time consuming technical and empirical efforts required to achieve methodological improvements (Forster et al. 2007). Significant advances in fields such as genetics, genomics and biotechnology made during that time, resulted in unprecedentedly effective tools to potentially unravel the genetic mechanisms behind DH induction capacity. Moreover, the synergy between many of these technologies and DH techniques, microspore transformation for direct transgene fixation for example (Guo et al. 2009; Schedel et al. 2016), are sufficient reason to believe that there have never been more advantageous and impactful applications of DHs possible in perennial ryegrass research and breeding than there are now (Begheyn et al. 2016a, b).
By using appropriate germplasm in concert with current genotyping and mapping approaches, discovery of the genetic loci involved in androgenic capacity became feasible. For example, a recent genome-wide association study (GWAS) in wheat found two QTL on chromosomes 1B and 7B that explained 53% of the variation in green plant production (Nielsen et al. 2015). Similarly, genes on chromosome 4A seem to be responsible for total and green plant production in triticale (×Triticosecale Wittm.) (Krzewska et al. 2012, 2015). Molecular markers have been developed through such studies, and now enable introgression of androgenic capacity into recalcitrant breeding germplasm. In perennial ryegrass however, the genetic resources to perform this type of investigation have yet to be established, so that a modern reboot of research on the production of DHs can be initiated (Andersen et al. 1997; Begheyn et al. 2016a).
The main objective of this research was to create such a resource by evaluating the response to in vitro AC of different perennial ryegrass populations segregating for androgenic capacity. Specifically, we aimed to investigate whether (1) our AC method was effective in this germplasm, (2) the genetic control of the component traits of androgenic capacity could be elucidated, (3) highly androgenic genotypes could be selected for use in introgression breeding, (4) the regenerated green plantlets were homozygous, diploid and had an acceptable performance in the field, and (5) the studied populations would be suitable for future genotyping and mapping studies.
Materials and methods
Plant material
The plant material used in this study was developed at the research and breeding station of DLF A/S in Store Heddinge, Denmark, and originated from a DH induction program. Paircrosses were made between genotypes with differing responses to in vitro AC. Harvested seeds were sown in autumn 2014, vernalized under natural conditions in an unheated greenhouse over winter. Fifty randomly selected progenies of each paircross were grown in an unheated greenhouse in soil filled 13 cm diameter (1 L pots) during 2015 and 2016 at the experimental research station of ETH Zurich in Eschikon, Lindau, Switzerland. Paircross parents and their offspring populations (anther donors) are numbered according to Fig. 1. The paircross parents capable of producing green plants will hereafter be referred to as being androgenic (P2, P102 and P169) while all others will be referred to as being non-androgenic (Supplementary Table S1).
Haploid and doubled haploid induction
In order to select and prepare donor spikes for in vitro AC, the microspore developmental stage was assessed by harvesting anthers from the middle of a spike for a squash preparation in a drop of water on a glass slide, and observing the cells using a stereomicroscope. Spikes with anthers in which the majority of the microspores were in the late-uninucleate stage were harvested and kept at 4 °C in the dark for 24–72 h before being used for AC.
After the cold treatment, spikes were surface sterilized for 1 min in 70% ethanol with a drop of Tween-20, followed by stirring for 10 min in 2.5% sodium hypochlorite with a drop of Tween-20, and four rinses in sterile demineralized water. Adapted versions of the 190-2 medium described by Wang and Hu (1984) were used for all culture steps. In all cases, the Ca(NO3)2·4H2O concentration was increased to 140 mg L−1, the MnSO4·H2O concentration was decreased to 4.9 mg L−1, the FeSO4·7H2O concentration to 13.9 mg L−1, and the Na2EDTA·2H2O concentration to 18.6 mg L−1. For the induction medium, 1-naphthaleneacetic acid (NAA) was replaced by 1.5 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D), and sucrose by 90 g L−1 maltose. Hormone concentrations were reduced to 0.25 mg L−1 in the regeneration medium and omitted altogether in the resting medium. A concentration of 0.35% Gelrite (Duchefa Biochemie, Haarlem, The Netherlands) was used for solidification and the pH was set to 6. All culture steps took place at 26 °C, with a 16 h photoperiod.
Aseptically excised anthers were placed in a 90 mm Petri dish containing induction medium. A density of 126 anthers per dish, derived from the 7 spikelets in the middle of each spike, was used. Depending on flower availability, between 126 and 252 anthers were cultured for the majority of anther donor genotypes. After 6–8 weeks, ELS were moved onto the regeneration medium for shoot and root induction. Green regenerants were recovered after 3–4 weeks and grown on resting medium in ‘De Wit’ culture tubes (Duchefa Biochemie, Haarlem, The Netherlands) for another 2–3 weeks before transfer to the greenhouse. Anthers of some genotypes produced so many embryos and/or green plants that representative subsamples had to be taken for further culture and analysis.
Phenotypic evaluation of the response to haploid and doubled haploid induction
The following observations were recorded for each anther donor genotype: (1) the percentage of embryogenic anthers, which were those producing macroscopic embryo-like structures (ELS), (2) embryo formation, expressed as the number of ELS per 100 anthers cultured (AC), (3) plant yield, expressed as plant production (number of plants per 100 AC) or plant regeneration (per 100 ELS cultured (EC)), and (4) green plant yield, expressed as green plant production (number of green plants per 100 AC) or green plant regeneration (per 100 EC). The percentages of green and albino regenerants per genotype were also calculated.
Statistical analysis
All analyses were performed and all figures were generated in RStudio (RStudio Team 2015) running R version 3.3.1 (R Core Team 2016) and using the packages ggplot2 (Wickham 2009) and cowplot (Wilke 2016). Rank-based, non-parametric tests (Kruskal–Wallis with Bonferroni corrected Dunn’s tests post hoc (P ≤ 0.05), Wilcoxon–Mann–Whitney tests and Spearman’s rank-order correlation tests) were used for the statistical analyses, since the data violated the necessary assumptions for parametric testing and transformation did not offer significant improvement (Manninen 2000).
Genetic characterization of the plant material
DNA was extracted from anther donor plants and green regenerants using a 96-well plate KingFisher Flex Purification System and KingFisher Pure DNA Plant Kits (Thermo Fisher Scientific, Waltham, MA, USA). Simple sequence repeat (SSR) markers G05_065 and G05_134 (Studer et al. 2008) and high resolution melting curve analysis (HRM) marker 05_02833 (Manzanares et al. 2016), selected because of their heterozygosity in a large number of anther donor genotypes, were used to characterize the green regenerants. A detailed primer description and the PCR amplification protocol used for the SSR markers can be found in Studer et al. (2008). Amplification products were separated, visualised and scored using an ABI 3130 16 capillary electrophoresis system (Applied Biosystems, Foster City, CA, USA) and the GeneMarker software version 1.5 (SoftGenetics, PA, USA). HRM analysis was performed as described in Studer et al. (2009) and Manzanares et al. (2016). Briefly, PCR amplicons were melted between 60 and 98 °C using a 96-well LightScanner and genotyping was performed with the LightScanner software package (BioFire Diagnostics, UT, USA).
Leaf tissue samples of the green regenerants were prepared and analysed as described in Doležel et al. (2007), following steps 1C and 2B. Samples were analysed using a CyFlow Space flow cytometer (Sysmex Partec GmbH., Görlitz, Germany) equipped with a UV led diode array. At least 2500 events were acquired per sample and only measurements with a coefficient of variation for the G0/G1 peaks of <2.5% were accepted.
Results
Phenotypic responses to anther culture
A total of 351 genotypes, derived from nine distinct paircross populations, were subjected to in vitro AC during two flowering seasons. Depending on flower availability, between 17 and 50 genotypes could be evaluated per population. A total of 75,726 anthers were cultured, 17.1% of which were embryogenic. On average, the embryo induction rate was 86.7 ELS per 100 AC, the regeneration frequency 76.2 plants per 100 AC (or 53.4 plants per 100 EC) and 38.2 green and 38.0 albino plants were obtained per 100 AC (or 20.1 green and 33.3 albino plants per 100 EC).
In 2015, 74.6% of the 295 evaluated genotypes produced ELS, 56.6% produced plants, 30.1% produced green plants (3.1% did so exclusively) and 26.8% produced only albinos. The genotypes responded differently for the evaluated traits and a wide segregation within each population was observed (Fig. 2; Supplementary Fig. S1). The averages of most populations were close to zero for plants per 100 AC and green plants per 100 AC or EC, with population 11 having the lowest overall performance. Nevertheless, for each evaluated trait, at least several responsive genotypes could be identified within each population. Remarkably embryogenic genotypes were observed in populations 2, 3 and 4 (Fig. 2b), while populations 3, 4 and 7 included genotypes producing high numbers of plants (Fig. 2c). Population 6 included genotypes with a relatively high plant regeneration frequency (Fig. 2d) and green plant regeneration per 100 EC (Fig. 2f), as well as the anther donor from which the most green plants per 100 AC were obtained (Fig. 2e). Several genotypes producing over 100 green regenerants per 100 AC could be identified in all populations, except population 11.
Box plots of the responses of the evaluated populations to anther culture. The letters above each plot denote significant differences between populations (P ≤ 0.05). a Percentage of embryogenic anthers; b number of embryo-like structures (ELS) per 100 anthers cultured (AC); c number of plants regenerated per 100 AC; d number of plants regenerated per 100 ELS cultured (EC); e number of green plants (GP) regenerated per 100 AC; f number of GP regenerated per 100 EC. For graphical reasons, observation 1530 from population 6 has been omitted from graph e
Genotypic differences were observed for the percentages of green plants that were regenerated, both within and between the evaluated populations (Fig. 3). Within populations 1, 7 and 8, no genotypes produced more than 65% green plants for instance, whereas anther donors of population 11 produced either green or albino plantlets exclusively.
Highly androgenic genotypes, those producing embryos in large numbers as well as having a very high plant regeneration frequency, high green plant production and green plant percentage, could be selected from the evaluated populations (Table 1). For example, anther donor 47 from population 3 had 71% embryogenic anthers and produced 646 ELS per 100 anthers, from which 1002 plants including 858 green plants (86%) per 100 AC could be regenerated.
Influence of paircross parents’ androgenic capacity
Different combinations of pair cross parents influenced the performances of the populations for the evaluated traits. For the percentage of embryogenic anthers for example, progeny from a cross between non-androgenic P133 and androgenic P102 or P169 performed significantly better (P ≤ 0.05) compared to progeny from a cross between P133 and androgenic P2 (Fig. 4a-II). Similarly, anthers of genotypes with androgenic P169 as one of their parents were significantly more embryogenic (P ≤ 0.01) if the non-androgenic parent was P133 instead of P10 (Fig. 4b-I). The same effect was observed for plants and albino plants per 100 AC (P ≤ 0.05; Supplementary Table S2). Progeny of the cross between androgenic P2 and non-androgenic P175 had a significantly lower performance for percentage of embryogenic anthers (P ≤ 0.001), ELS per 100 AC (P ≤ 0.01), plants per 100 AC (P ≤ 0.01) and green plants per 100 AC (P ≤ 0.05) than progeny of the cross between androgenic P2 and most other non-androgenic parents (Fig. 4b-III; Supplementary Table S1).
Box plots with attached density plots of the percentage of embryogenic anthers. a Grouped by common non-androgenic paircross parent (P10, P133, P144, P175 and P48); b grouped by common androgenic paircross parent (P169, P102 and P2). Combination P133 and P2 performed significantly worse (P ≤ 0.05) than the other combinations in section a-II. The cross between P169 and P133 performed significantly better (P ≤ 0.01) than the cross P169 and P10 (b-I). Combination P2 and P175 performed significantly worse than all other combinations (P ≤ 0.001; b-III)
Correlations between traits and years
Ranked correlations (Spearman’s ρ) were calculated between the evaluated traits, using observations from all populations evaluated in 2015, but excluding genotypes that produced <20 plants per AC (n = 102) in order to detect inter-trait relationships relevant in more androgenic material (Table 2). Many positive and highly significant (P ≤ 0.001) correlations were found, such as ρ = 0.80 between plants per 100 AC and green plants per 100 AC. Embryo production had a high positive correlation with plant production (ρ = 0.83; P ≤ 0.001) but was not significantly correlated with plant regeneration. Green plant production had a relatively low, positive correlation with albino production (ρ = 0.20; P ≤ 0.05) and was not significantly correlated with albino regeneration. Green plant regeneration was not significantly correlated with albino plant production and regeneration.
Populations 1, 4, 6 and 7, selected because of their widely segregating and high responses to in vitro AC, were evaluated again in 2016. Due to flower availability, the number of genotypes evaluated in both 2015 and 2016 (n = 78) is rather low in all but population 1 (n = 45). A clear effect of the year was observed and the anther donors performed significantly better in 2015 for almost all traits (Fig. 5). Ranked positive correlations between the years ranged from ρ = 0.45 for plants or green plants per 100 EC (P ≤ 0.001) to ρ = 0.54 for the two embryo related traits (P ≤ 0.001).
Box plots of the responses of populations 1, 4, 6 and 7 to anther culture in both 2015 (left) and 2016 (right). Spearman’s ρ for the overall correlation between years per trait is given within the plots. Significance levels of the differences between the years for each trait and population are given above the plots. a Percentage of embryogenic anthers; b number of embryo-like structures (ELS) per 100 anthers cultured (AC); c number of plants regenerated per 100 AC; d number of plants regenerated per 100 ELS cultured (EC); e number of green plants (GP) regenerated per 100 AC; f number of GP regenerated per 100 EC. For graphical reasons, observation 1530 from population 6 (2015) has been omitted from graph e. ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns not significant
Green regenerants
Loss of green plantlets during and after transplantation to the greenhouse was negligible and 1111 green putative DHs were raised. The selected markers were informative for 529 plants, 99.6% of which were homozygous at loci where the anther donor was heterozygous, confirming their microspore origin. The ploidy levels of all 1111 green regenerants were analysed with flow cytometry and 472 haploid (n), 572 diploid (2n), 48 tetraploid (4n), and even a few triploid (3n), heptaploid (7n), and mixoploid (e.g. n and 2n) plants were found. In the summer of 2016, 539 diploid and 40 tetraploid green regenerants were transplanted into the field, where the conditions were significantly hotter and dryer than most years. Both vigorously growing and extremely weak phenotypes were observed. On a scale from 0 to 9 for overall vigour, 3.7% of the diploid plants scored a 7 or higher, while all of the tetraploid plants scored a 6 or lower. Spikes were produced by 69 diploid and 1 tetraploid plants, out of which 11 diploid plants set seed.
Discussion
Androgenic capacity
In this study, we applied an effective in vitro AC protocol to a large number of perennial ryegrass genotypes derived from crosses between distinct androgenic and non-androgenic plants, and generated thousands of microspore-derived embryos and green DH plants. Interestingly, even though it is difficult to compare between tissue culture studies (Oleszczuk et al. 2014; Seldimirova and Kruglova 2015), quite a few genotypes outperformed the most androgenic plants reported to date, except for our earlier pilot study (Begheyn et al. 2016b). For example, ten anther donors produced more than 354 green plants per 100 AC, which is three times the highest number reported before (Opsahl-Ferstad et al. 1994b). One highly androgenic genotype from population 6 generated over 1500 green plants per 100 AC, which is comparable to yields of the responsive model barley cultivar ‘Igri’ (Jacquard et al. 2006) but far better than ‘Svilena’, the model used in wheat (Lantos et al. 2013). These high yields are probably due to the superior genetics of the studied plant material rather than the relatively minor differences between our method and previous practices (Olesen et al. 1988; Opsahl-Ferstad et al. 1994a, b). Indeed, an unprecedentedly large number of beneficial alleles should be present within the paircross parents, since they originate from a recurrent selection programme to improve androgenicity (Niels Roulund, personal communication; Andersen et al. 1997). Further increases in production efficiency and yield may be achieved by utilizing recent novel understanding of microspore embryogenesis processes (Żur et al. 2014; Fábián et al. 2015; Sinha and Eudes 2015) and progress in optimization of (pre-)culture conditions in other cereal species such as barley, wheat or triticale (Castillo et al. 2014; Würschum et al. 2015; Echávarri and Cistué 2016).
Genotypes with a uniquely high androgenic capacity could be selected in the present study (Table 1) and can now be used for the introgression of this trait into recalcitrant germplasm (Halberg et al. 1990; Madsen et al. 1995). In fact, population 11, derived from the cross between androgenic P2 and non-androgenic P175, may be considered as a test case of this ‘inducer’ approach (Halberg et al. 1990). Even though one of its parents is incapable of microspore embryogenesis or plant regeneration, population 11 included genotypes regenerating as many as 64 green plants per 100 AC. Recalcitrant genotypes can thus be ‘induced’ to exhibit acceptable productivity levels (Murigneux et al. 1994; Madsen et al. 1995).
Genetic control of androgenic capacity
The evaluated populations showed a continuous distribution for each component trait of androgenic capacity, which confirms their previously reported quantitative polygenic control (Halberg et al. 1990; Opsahl-Ferstad et al. 1994b; Madsen et al. 1995). Additionally, variation in the inheritance patterns between the populations indicates the presence of different numbers of distinct alleles in their paircross parents (Madsen et al. 1995; Torp et al. 2001; Nielsen et al. 2015). For example, non-androgenic parent P133 may harbour superior alleles for microspore embryogenesis and plant regeneration compared to P10.
Because all non-androgenic parents in this investigation (except those of populations 10 and 11) are capable of microspore embryogenesis and albino plant regeneration, the remarkably high levels of embryo and plant production observed in their offspring could be due to additive allelic effects or the presence of similar or identical alleles in both parents (Fig. 2). Alleles from androgenic parent P2 must be responsible for the plant regeneration, and possibly also the green plant regeneration, observed in population 10, since its non-androgenic parent (P144) lacks the ability to regenerate plants. This would suggest a certain level of dominance of the genetic control of these two traits (Opsahl-Ferstad et al. 1994b). Although populations 10 and 11 share androgenic parent P2, the non-androgenic parent of the latter (P175) is unable to produce embryos. This resulted in a significantly lower embryo and therefore plant production in population 11 compared to population 10.
Embryo and plant production capacity were highly correlated (ρ = 0.83; P ≤ 0.001), suggesting their control by related, linked or even identical genes. This scenario was also proposed in maize, where one QTL was found to be correlated with both traits (Murigneux et al. 1994). Indeed, all traits expressed as production per 100 AC were highly and significantly correlated, which is to be expected since the reverse would be biologically impossible. Drawing conclusions on genetic control from these relationships is, therefore, likely to be of limited value. More informative is, for example, the insignificant correlation observed between embryo production and plant regeneration, which supports the hypothesized distinct genetic control of the two traits in perennial ryegrass (Stanis and Butenko 1984; Olesen et al. 1988; Boppenmeier et al. 1989). In both wheat and barley, no genetic correlation between these two traits was found either (Tuvesson et al. 1989; Larsen et al. 1991).
Conversely, the high correlation between plant and green plant regeneration (ρ = 0.72; P ≤ 0.001), again supported by findings in wheat (Tuvesson et al. 1989), is unlikely to be due to similar genetic control mechanisms, since many of the non-androgenic parents are capable of plant but not green plant production. Additionally, the absence of a correlation between QTL explaining variation in green plant production with embryo formation or plant regeneration was reported in wheat (Torp et al. 2001; Nielsen et al. 2015). Several other possibilities may be considered instead; for example, dominant control of green plant production by several genes, alleles for which are contributed solely by the androgenic parents (Tuvesson et al. 1989; Opsahl-Ferstad et al. 1994b), or complementation of recessive beneficial alleles present in a heterozygous state within the non-androgenic parents (Madsen et al. 1995). Alternatively, environmental conditions before or during AC could have been more conducive to green plant production than when the androgenic capacity of the paircross parents was examined. External influences at that time may have caused potential green regenerants to be afflicted by albinism, thus masking the presence of the capacity for green plant production of those genotypes now classified as non-androgenic (see also below) (Opsahl-Ferstad et al. 1994b).
Albinism
The evaluated populations and genotypes differed in the number and percentage of regenerated albinos. Similar to reports in triticale, a low or insignificant correlation was found between green and albino plant production as well as regeneration (González et al. 2005; Krzewska et al. 2015). Opsahl-Ferstad et al. (1994b) found an insignificant correlation of r = 0.29, similar to the ρ = 0.20 (P ≤ 0.05) found here, and hypothesized that different genes must be responsible for green and albino plant production. Also in accordance with their results, a highly negative correlation (ρ = −0.83; P ≤ 0.001) between the number of green plants produced and the percentage of albinos was found, which is expected in the presence of both green and albino plant production promoting alleles in a population (Opsahl-Ferstad et al. 1994b). Additionally, the low negative correlation (ρ = −0.29; P ≤ 0.01) between the number of albino plants produced and green plant percentage, suggests that the environment is of greater influence on albino production than genetics (Kumari et al. 2009; Makowska and Oleszczuk 2014), even though QTL for albinism have been reported in several cereal species (He et al. 1998; Chen et al. 2007; Krzewska et al. 2015). The presence of alleles required for green plant production can thus be ‘phenotypically invisible’, if potential green regenerants suffer from environmentally induced albinism.
Robustness of androgenic capacity
A significant effect of the year was observed in the response to AC and a better overall performance of the genotypes was recorded in 2015, possibly due to late and erratic flowering caused by incomplete vernalization in 2016 (Fig. 5). Influence of the growing conditions of the anther donor has been well documented in perennial ryegrass, and responses may be better at the start of the growing season (Olesen et al. 1988; Boppenmeier et al. 1989; Bante et al. 1990). Similarly, anther response dropped around 50% when barley spikes were harvested in the second half of the year (Jacquard et al. 2006), and seasonal variations in AC response levels of the same genotype was reported in oat (Kiviharju et al. 2017). Higher correlations than the observed ρ = 0.45–0.54 (P ≤ 0.001) would probably have been found between the years, if 2016 had contributed less zeros to the dataset. Nevertheless, genotypes with a high embryo and/or plant production in 2015 were frequently highly androgenic in 2016 as well. One genotype from population 4 produced the most total plants and green plants in both years for example. Indeed, the three best green plant producers within population 1 in 2016 were in the top five in 2015.
To investigate the stability of androgenic responses over time and environments, several highly androgenic genotypes were artificially vernalized and induced to flower in a climate chamber during the winter of 2016. Many embryos and green plants were produced under such conditions as well (data not shown). Based on these findings, it seems that highly androgenic genotypes do exhibit a robust DH production and, as has been reported elsewhere, the heritability of this ability is relatively high (Lazar et al. 1984; Opsahl-Ferstad et al. 1994b; Moieni et al. 1997).
Characteristics of the green regenerants
Andersen et al. (1997) reported that 99.7% of the 913 green regenerants they screened were homozygous, a nearly identical finding to the 99.6% homozygosity observed in this study. Perennial ryegrass AC does not seem to be sensitive to undesirable regeneration of heterozygous plantlets from sporophytic tissue or unreduced gametes. The observed spontaneous chromosome doubling rate of 52% of this study is at the lower end of the 50–80% range reported in literature (Olesen et al. 1988; Bante et al. 1990; Halberg et al. 1990; Begheyn et al. 2016b). Besides spontaneous polyploidization, chromosome doubling may also be induced by in vitro or in vivo application of chromosome doubling agents such as colchicine (Melchinger et al. 2016; Ślusarkiewicz-Jarzina et al. 2017). In contrast to triticale (Lantos et al. 2014) and wheat (Rubtsova et al. 2013) however, this is not considered necessary in perennial ryegrass due to a high level of natural chromosome doubling (Andersen et al. 1997).
Our field evaluation of almost 600 regenerated DHs confirmed that, even though most were weak and showed low fertility, it was possible to select vigorous and fertile genotypes, similar to findings reported by Bante et al. (1990), Opsahl-Ferstad (1993) and Andersen et al. (1997). Perennial ryegrass DHs perform similar to those produced in rye, another obligate outcrossing member of the grass family in which AC is being studied. One investigation, performed on fewer genotypes than used here, found that only 10–36% of rye DHs were suitable for research or breeding purposes due to low vigour, fertility, survival and abnormal growth (Tenhola-Roininen et al. 2006). On the other hand, the maintenance and multiplication of DHs of tall fescue (Lolium arundinaceum (Schreb.) Darbysh. [syn. Festuca arundinacea Schreb.]) through seed obtained by self-fertilization was not found to pose any problems, even though this species is known to be self-incompatible (Kindiger 2016). More investigations into the vigour and fertility of perennial ryegrass DHs are needed in order to better characterize to what extend inbreeding depression affects their agronomic performance.
Concluding remarks
Here, we have demonstrated the effectivity of the present in vitro AC method for DH production in perennial ryegrass, as well as the value of the used plant material as a resource for future inquiries into the genetic control of androgenic capacity. The variation in embryo production, plant regeneration and green plant production observed between and within the populations was large, and its pattern indicates the presence of different genes and distinct alleles involved in the control of androgenic capacity. Through approaches like GWAS or QTL mapping, genomic regions involved in the control of microspore embryogenesis, plant regeneration, green and albino plant production may now be identified within this germplasm (Wędzony et al. 2015). Apart from their academic interest, these types of studies facilitate the development of molecular markers for androgenic ability, which, in concert with highly responsive ‘inducer’ genotypes such as the ones selected here, could rapidly and dramatically increase the perennial ryegrass gene pool from which DHs can efficiently be produced. This expansion of the applicability of DH techniques has the potential to accelerate the progress of basic and applied research as well as breeding programs in the forage grasses.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- AC:
-
Anther(s) culture(d)
- EC:
-
ELS cultured
- ELS:
-
Embryo-like structures
- DH:
-
Doubled haploid
- GWAS:
-
Genome-wide association study/studies
- HRM:
-
High resolution melting
- NAA:
-
1-Naphthaleneacetic acid
- QTL:
-
Quantitative trait locus/loci
- SSR:
-
Simple sequence repeat
References
Andersen SB, Madsen S, Roulund N, Halberg N, Olesen A (1997) Haploidy in ryegrass. In: Jain SM, Sopory SK, Veilleux RE (eds) In vitro haploid production in higher plants. Springer, Dordrecht, pp 133–147
Arias Aguirre A, Studer B, Frei U, Lübberstedt T (2011) Prospects for hybrid breeding in bioenergy grasses. Bioenergy Res 5:10–19
Bante I, Sonke T, Tandler RF, van den Bruel AMR, Meyer EM (1990) Anther culture of Lolium perenne and Lolium multiflorum. In: Sangwan RS, Sangwan-Norreel BS (eds) The impact of biotechnology in agriculture. Springer, Dordrecht, pp 105–127
Begheyn RF, Lübberstedt T, Studer B (2016a) Haploid and doubled haploid techniques in perennial ryegrass (Lolium perenne L.) to advance research and breeding. Agronomy 6:60
Begheyn RF, Vangsgaard K, Roulund N, Studer B (2016b) Efficient doubled haploid production in perennial ryegrass (Lolium perenne L.). In: Roldán-Ruiz I, Baert J, Reheul D (eds) Breeding in a world of scarcity: proceedings of the 2015 meeting of the section “Forage Crops and Amenity Grasses” of Eucarpia, Ghent, Belgium. Springer, Cham, pp 151–155
Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA (2010) Heterosis. Plant Cell 22:2105–2112
Boppenmeier J, Zuchner S, Foroughi-Wehr B (1989) Haploid production from barley yellow dwarf virus resistant clones of Lolium. Plant Breed 103:216–220
Cabral AL, Jordan MC, McCartney CA, You RM, Humphreys DG, MacLachlan R, Pozniak CJ (2014) Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.). BMC Plant Biol 14:340
Castillo AM, Nielsen NH, Jensen A, Vallés MP (2014) Effects of n-butanol on barley microspore embryogenesis. Plant Cell Tissue Organ Cult 117:411–418
Chen X-W, Cistué L, Muñoz-Amatriaín M, Sanz M, Romagosa I, Castillo A-M, Vallés M-P (2007) Genetic markers for doubled haploid response in barley. Euphytica 158:287–294
Cornish MA, Hayward MD, Lawrence MJ (1979) Self-incompatibility in ryegrass. I. Genetic control in diploid Lolium perenne L. Heredity 43:95–106
Devaux P, Kasha KJ (2009) Overview of barley doubled haploid production. In: Touraev A, Forster BP, Jain SM (eds) Advances in haploid production in higher plants. Springer, Dordrecht, pp 47–64
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2:2233–2244
Dunwell JM (2010) Haploids in flowering plants: origins and exploitation. Plant Biotechnol J 8:377–424
Dwivedi SL, Britt AB, Tripathi L, Sharma S, Upadhyaya HD, Ortiz R (2015) Haploids: constraints and opportunities in plant breeding. Biotechnol Adv 33:812–829
Echávarri B, Cistué L (2016) Enhancement in androgenesis efficiency in barley (Hordeum vulgare L.) and bread wheat (Triticum aestivum L.) by the addition of dimethyl sulfoxide to the mannitol pretreatment medium. Plant Cell Tissue Organ Cult 125:11–22
Fábián A, Földesiné Füredi PK, Ambrus H, Jäger K, SzabóL, Barnabás B (2015) Effect of n-butanol and cold pretreatment on the cytoskeleton and the ultrastructure of maize microspores when cultured in vitro. Plant Cell Tissue Organ Cult 123:257–271
Ferrie AMR, Caswell KL (2011) Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell Tissue Organ Cult 104:301–309
Forster BP, Heberle-Bors E, Kasha KJ, Touraev A (2007) The resurgence of haploids in higher plants. Trends Plant Sci 12:368–375
Geiger HH, Gordillo GA (2009) Doubled haploids in hybrid maize breeding. Maydica 54:485–499
González JM, Muñiz LM, Jouve N (2005) Mapping of QTLs for androgenetic response based on a molecular genetic map of × Triticosecale Wittmack. Genome 48:999–1009
Guo Y-D, Hisano H, Shimamoto Y, Yamada T (2009) Transformation of androgenic-derived Festulolium plants (Lolium perenne L. × Festuca pratensis Huds.) by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 96:219–227
Guo B, Wang D, Guo Z, Beavis WD (2013) Family-based association mapping in crop species. Theor Appl Genet 126:1419–1430
Halberg N, Olesen A, Tuvesson IKD, Andersen SB (1990) Genotypes of perennial ryegrass (Lolium perenne L.) with high anther-culture response through hybridization. Plant Breed 105:89–94
He P, Shen L, Lu C, Chen Y, Zhu L (1998) Analysis of quantitative trait loci which contribute to anther culturability in rice (Oryza sativa L.). Mol Breed 4:165–172
Jacquard C, Asakaviciute R, Hamalian AM, Sangwan RS, Devaux P, Clément C (2006) Barley anther culture: effects of annual cycle and spike position on microspore embryogenesis and albinism. Plant Cell Rep 25:375–381
Kindiger B (2016) Generation of paternal dihaploids in tall fescue. Grassl Sci 62:1–5
Kiviharju E, Moisander S, Tanhuanpää P (2017) Oat anther culture and use of DH-lines for genetic mapping. In: Gasparis S (ed) Oat: methods in molecular biology, vol 1536. Springer, New York, pp 71–93
Krzewska M, Czyczyło-Mysza I, Dubas E, Gołebiowska-Pikania G, Golemiec E, Stojałowski S, Chrupek M, Żur I (2012) Quantitative trait loci associated with androgenic responsiveness in triticale (× Triticosecale Wittm.) anther culture. Plant Cell Rep 31:2099–2108
Krzewska M, Czyczyło-Mysza I, Dubas E, Gołebiowska-Pikania G, Żur I (2015) Identification of QTLs associated with albino plant formation and some new facts concerning green versus albino ratio determinants in triticale (× Triticosecale Wittm.) anther culture. Euphytica 206:263–278
Kumari M, Clarke HJ, Small I, Siddique KHM (2009) Albinism in plants: a major bottleneck in wide hybridization, androgenesis and doubled haploid culture. Crit Rev Plant Sci 28:393–409
Lantos C, Weyen J, Orsini JM, Gnad H, Schlieter B, Lein V, Kontowski S, Jacobi A, Mihály R, Broughton S, Pauk J (2013) Efficient application of in vitro anther culture for different European winter wheat (Triticum aestivum L.) breeding programmes. Plant Breed 132:149–154
Lantos C, Bóna L, Boda K, Pauk J (2014) Comparative analysis of in vitro anther- and isolated microspore culture in hexaploid triticale (× Triticosecale Wittmack) for androgenic parameters. Euphytica 197:27–37
Larsen ET, Tuvesson IK, Andersen SB (1991) Nuclear genes affecting percentage of green plants in barley (Hordeum vulgare L.) anther culture. Theor Appl Genet 82:417–420
Lazar MD, Baenziger PS, Schaeffer GW (1984) Combining abilities and heritability of callus formation and plantlet regeneration in wheat (Triticum aestivum L.) anther cultures. Theor Appl Genet 68:131–134
Madsen S, Olesen A, Dennis B, Andersen SB (1995) Inheritance of anther-culture response in perennial ryegrass (Lolium perenne L.). Plant Breed 114:165–168
Makowska K, Oleszczuk S (2014) Albinism in barley androgenesis. Plant Cell Rep 33:385–392
Maluszynski M, Kasha KJ, Forster BP, Szarejko I (2003) Doubled haploid production in crop plants: a manual. Kluwer Academic Publishers, Dordrecht
Manninen OM (2000) Associations between anther-culture response and molecular markers on chromosomes 2 H, 3 H and 4 H of barley (Hordeum vulgare L.). Theor Appl Genet 100:57–62
Manzanares C, Barth S, Thorogood D, Byrne SL, Yates S, Czaban A, Asp T, Yang B, Studer B (2016) A gene encoding a DUF247 domain protein cosegregates with the S self-incompatibility locus in perennial ryegrass. Mol Biol Evol 33:870–884
Melchinger AE, Molenaar WS, Mirdita V, Schipprack W (2016) Colchicine alternatives for chromosome doubling in maize haploids for doubled-haploid production. Crop Sci 56:559–569
Mishra R, Rao GJN (2016) In-vitro androgenesis in rice: advantages, constraints and future prospects. Rice Sci 23:57–68
Moieni A, Lokos-Toth K, Sarrafi A (1997) Evidence for genetic control and media effect on hapioid regeneration in the anther culture of hexaploid wheat (Triticum aestivum L.). Plant Breed 116:502–505
Murigneux A, Bentolila S, Hardy T, Baud S, Guitton C, Jullien H, Ben Tahar S, Freyssinet G, Beckert M (1994) Genotypic variation of quantitative trait loci controlling in vitro androgenesis in maize. Genome 37:970–976
Niak N, Rout P, Umakanta N, Verma RL, Katara JL, Sahoo KK, Singh ON, Samantaray S (2016) Development of doubled haploids from an elite indica rice hybrid (BS6444G) using anther culture. Plant Cell Tissue Organ Cult 128:679–689
Nielsen NH, Andersen SU, Stougaard J, Jensen A, Backes G, Jahoor A (2015) Chromosomal regions associated with the in vitro culture response of wheat (Triticum aestivum L.) microspores. Plant Breed 134:255–263
Niu Z, Jiang A, Abu Hammad W, Oladzababbasabadi A, Xu SS, Mergoum M, Elias EM (2014) Review of doubled haploid production in durum and common wheat through wheat × maize hybridization. Plant Breed 133:313–320
Obsa BT, Eglinton J, Coventry S, March T, Langridge P, Fleury D (2016) Genetic analysis of developmental and adaptive traits in three doubled haploid populations of barley (Hordeum vulgare L.). Theor Appl Genet 129:1139–1151
Olesen A, Andersen SB, Due IK (1988) Anther culture response in perennial ryegrass (Lolium perenne L.). Plant Breed 101:60–65
Oleszczuk S, Tyrka M, Zimny J (2014) The origin of clones among androgenic regenerants of hexaploid triticale. Euphytica 198:325–336
Opsahl-Ferstad H-G (1993) Androgenetic response in grasses. I. Anther culture in perennial ryegrass (Lolium perenne L.). II. Molecular studies of embryogenesis in barley (Hordeum vulgare L.). Dissertation, Agricultural University of Norway
Opsahl-Ferstad H-G, Bjørnstad Å, Rognli OA (1994a) Influence of medium and cold pretreatment on androgenetic response in Lolium perenne L. Plant Cell Rep 13:594–600
Opsahl-Ferstad H-G, Bjørnstad Å, Rognli OA (1994b) Genetic control of androgenetic response in Lolium perenne L. Theor Appl Genet 89:133–138
Pembleton LW, Shinozuka H, Wang J, Spangenberg GC, Forster JW, Cogan NOI (2015) Design of an F1 hybrid breeding strategy for ryegrasses based on selection of self-incompatibility locus-specific alleles. Front Plant Sci 6:764
R Core Team (2016) R: a language and environment for statistical computing. https://www.r-project.org/. Accessed 1 Feb 2017
RStudio Team (2015) RStudio: integrated development for R. http://www.rstudio.com/. Accessed 1 Feb 2017
Rubtsova M, Gnad H, Melzer M, Weyen J, Gils M (2013) The auxins centrophenoxine and 2,4-D differ in their effects on non-directly induced chromosome doubling in anther culture of wheat (T. aestivum L.). Plant Biotechnol Rep 7:247–255
Sannemann W, Huang BE, Mathew B, Léon J (2015) Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol Breed 35:86
Schedel S, Pencs S, Hensel G et al (2016) RNA-guided Cas9-induced mutagenesis in tobacco followed by efficient genetic fixation in doubled haploid plants. Front Plant Sci 7:1995
Seguí-Simarro JM (2010) Androgenesis revisited. Bot Rev 76:377–404
Seguí-Simarro JM (2015) Editorial: doubled haploidy in model and recalcitrant species. Front Plant Sci. doi:10.3389/fpls.2015.01175
Seguí-Simarro JM, Nuez F (2008) How microspores transform into haploid embryos: changes associated with embryogenesis induction and microspore-derived embryogenesis. Physiol Plant 134:1–12
Seldimirova OA, Kruglova NN (2015) Androclinic embryoidogenesis in vitro in cereals. Biol Bull Rev 5:156–165
Sinha R, Eudes F (2015) Dimethyl tyrosine conjugated peptide prevents oxidative damage and death of triticale and wheat microspores. Plant Cell Tissue Organ Cult 122:227–237
Ślusarkiewicz-Jarzina A, Pudelska H, Woźna J, Pniewski T (2017) Improved production of doubled haploids of winter and spring triticale hybrids via combination of colchicine treatments on anthers and regenerated plants. J Appl Genet. doi:10.1007/s13353-016-0387-9
Stanis VA, Butenko RG (1984) Developing viable haploid plants in anther culture of ryegrass. Dokl Biol Sci 275:249–251
Studer B, Asp T, Frei U et al (2008) Expressed sequence tag-derived microsatellite markers of perennial ryegrass (Lolium perenne L.). Mol Breed 21:533–548
Studer B, Jensen LB, Fiil A, Asp T (2009) “Blind” mapping of genic DNA sequence polymorphisms in Lolium perenne L. by high resolution melting curve analysis. Mol Breed 24:191–199
Tenhola-Roininen T, Immonen S, Tanhuanpää P (2006) Rye doubled haploids as a research and breeding tool—a practical point of view. Plant Breed 125:584–590
Torp AM, Hansen ALA, Andersen SB (2001) Chromosomal regions associated with green plant regeneration in wheat (Triticum aestivum L.) anther culture. Euphytica 119:377–387
Tuvesson IK, Pedersen S, Andersen SB (1989) Nuclear genes affecting albinism in wheat (Triticum aestivum L.) anther culture. Theor Appl Genet 78:879–883
Wang X, Hu H (1984) The effect of Potato II medium for triticale anther culture. Plant Sci Lett 36(3):237–239
Wędzony M, Żur I, Krzewska M, Dubas E, Szechyńska-Hebda M, Wąsek I (2015) Doubled haploids in triticale. In: Eudes F (ed) Triticale. Springer, Cham, pp 111–128
Wickham H (2009) ggplot2: elegant graphics for data analysis. https://cran.r-project.org/package=ggplot2. Accessed 1 Feb 2017
Wilke CO (2016) cowplot: streamlined plot theme and plot annotations for ‘ggplot2’. https://cran.r-project.org/package=cowplot. Accessed 1 Feb 2017
Würschum T, Tucker MR, Maurer HP, Leiser W (2015) Ethylene inhibitors improve efficiency of microspore embryogenesis in hexaploid triticale. Plant Cell Tissue Organ Cult 122:751–757
Żur I, Dubas E, Krzewska M, Janowiak F, Hura K, Pociecha E, Bączek-Kwinta R, Płażek A (2014) Antioxidant activity and ROS tolerance in triticale (× Triticosecale Wittm.) anthers affect the efficiency of microspore embryogenesis. Plant Cell Tissue Organ Cult 119:79–94
Acknowledgements
This work was supported by ETH Research Grant ETH-34 14-1 and the Swiss National Science Foundation (SNSF Professorship grant No.: PP00P2 138988). We thank Jana Čížková, Štěpán Stočes, Sonja Reinhard, Diana Zwahlen, Verena Knorst and Daniel Grogg for their assistance with diverse technical aspects of this work. We are grateful to Dr. Simon Bull, Dr. Steven Yates and Dr. Roland Kölliker for fruitful discussions and helpful suggestions. We are indebted to Prof. Dr. Achim Walter and the Crop Science group for having hosted the Molecular Plant Breeding group at ETH Zurich during most of this project.
Author information
Authors and Affiliations
Contributions
BS, NR and KV conceived the idea, designed the experiments and edited the manuscript. NR and KV developed the methodology and the in vitro protocol. RB conducted the experiments, analysed the data and wrote the manuscript. DK facilitated and helped conduct the flow cytometry experiments and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Alison M. R. Ferrie.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Begheyn, R.F., Roulund, N., Vangsgaard, K. et al. Inheritance patterns of the response to in vitro doubled haploid induction in perennial ryegrass (Lolium perenne L.). Plant Cell Tiss Organ Cult 130, 667–679 (2017). https://doi.org/10.1007/s11240-017-1255-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-017-1255-y