Abstract
Somatic embryogenesis is a reliable and important tool, and the relevant genes controlling this process act as vital roles through the whole development of somatic embryos. However, regeneration via somatic embryogenesis in Chinese chestnut has been impeded and its molecular mechanism is not known. Therefore, firstly we described a protocol for somatic embryo initiation, development, maturation and germination. Embryogenic calli were obtained in embryo initiation medium containing 1.8 μM 2,4-D and 1.1 μM 6-BA, and then were transferred to embryo development medium without any hormones for at least 4 weeks, until cotyledonary embryos appeared. Next, the somatic embryos were transferred to embryo maturation medium containing Gamborg’s B-5 Basal Salt Mixture with 0.5 μM NAA and 0.5 μM 6-BA for 3 weeks. Finally, these mature embryos were germinated in embryo germination medium consisting of WPM with 0.5 μM NAA and 0.5 μM 6-BA, resulting in shoot regeneration with a 2.1% conversion rate. Additionally, eight embryogenesis-related genes were identified, and the expression profiles of these genes during embryogenesis were analyzed via quantitative real-time RT-PCR (qRT-PCR). The CmSERK, CmLEC1, CmWUS and CmAGL15 genes exhibited high expression in the initial embryo stages, which inferred that these genes played key roles during the initiation of embryogenesis. Studies on embryogenesis-related genes will provide an insight for further elucidating molecular mechanism during somatic embryogenesis of Chinese chestnut. Furthermore, the successful establishment of a somatic embryo regeneration system for Chinese chestnut will lay a significant foundation for a stable genetic transformation system and genetic improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Castanea plants are members of the Fagaceae family and are mainly distributed in eastern and southwestern Asia, southern Europe and North America (Li et al. 2016). Chinese chestnut (Castanea mollissima Blume) is an important economic species of the Castanea genus and is mainly cultivated in China. Chinese chestnut presents the highest yield in the context of worldwide chestnut production (Li et al. 2016). Because Chinese chestnut exhibits the benefits of broad adaptability to the environment and inherent resistance to diseases, it is widely used as a parent in crosses to breed new cultivars and germplasms with favorable characteristics (Hebard 2006; Miller et al. 2014).
Somatic embryogenesis is defined as an in vitro multiple step process involved in initiation, development, maturation and germination of somatic embryos. Due to the advantages of high totipotency and genetic stability (Verdeil et al. 2007; Ikeuchi et al. 2015), somatic embryogenesis is considered as a vital biotechnological tool that can be reliably used for a variety of plants (Sezgin and Dumanoğlu 2014; Hu et al. 2017). To date, it has been broadly utilized for early embryo rescue to solve the problem of distant hybridization incompatibility, for shortening the breeding period and speeding up the reproduction process (Giri et al. 2004), and for providing a platform for genetically modified breeding (Rugh et al. 1998; Vidal et al. 2010). Although in recent years, the studies of somatic embryogenesis have made great progress, such as in black alders, Japanese larch, strawberry tree and yam (Corredoira et al. 2013; Kim 2015; Martins et al. 2016; Manoharan et al. 2016), this effective technology in many plant species has been hindered due to explant, genotype and plant growth regulators. Within the Castanea genus, a reliable somatic embryo regeneration system and a stable genetic transformation system have been established for both American chestnut (Merkle et al. 1991; Carraway et al. 1994; Xing et al. 1999; Maynard et al. 2015) and European chestnut (Corredoira et al. 2003, 2015; Sezgin and Dumanoğlu 2014). However, in Chinese chestnut, scientists have only acquired embryogenic calli from ovules (Zhang et al. 2007) and have not been able to obtain mature somatic embryos and regenerated plantlets.
The molecular mechanism of somatic embryogenesis has been extensively investigated (Rupps et al. 2016; Orłowska et al. 2017). The current results reveal that a number of embryogenesis-related genes have been identified, and some of the genes are essential for somatic embryogenesis, particularly at the initiation stage, such as Somatic Embryogenesis Receptor-like Kinase (SERK; Hecht et al. 2001), WUSCHEL (WUS; Wang et al. 2009), Leafy Cotyledon (LECs, i.e. LEC1, LEC2 and FUS3; Gaj et al. 2005; Rupps et al. 2016) and Agamous-Like15 (AGL15; Karlova et al. 2006). The SERK gene, encoding transmembrane kinase, marks the formation of embryogenic cells in culture and may be involved in the somatic embryogenesis signaling pathway by forming a protein complex with AGL15 (Karlova et al. 2006; Cai 2011). AtSERK1 overexpression induces an enhanced capacity for somatic embryogenesis (Hecht et al. 2001). The WUS gene, belonging to WUSCHEL-related homeobox (WOX) gene family, plays a key role in early somatic embryo development (Zuo et al. 2002; Wang et al. 2009). Known WOX2, as a putative marker, is supposed to be a possible function regulating differentiation of embryos (Palovaara and Hakman 2008; Klimaszewska et al. 2011). Evidence has indicated that LEC genes appear to act as a pivotal part in the early and late stages of embryogenic development (Jia et al. 2013). LEC1 gene coding a CCAAT-binding (CBF) transcription factor, regulates embryo identity and development during embryogenesis (Kwong et al. 2003; Lee et al. 2003), and induces the expression of other members of LEC gene family, such as LEC2 and FUS3 (Lotan et al. 1998; Lee et al. 2003). However, the studies on embryogenesis-related genes have not been carried out in Castanea plants.
Taken together, the effective protocol of somatic embryogenesis in Chinese chestnut is not available, moreover, the molecular mechanism of this process has not been reported. The purpose of this study is to establish a reliable somatic embryo regeneration system and to identify embryogenesis-related genes during different stages of the somatic embryogenesis process in Chinese chestnut. In addition, molecular technologies were performed to assess somaclonal variation and genetic uniformity of somatic embryogenesis. Studies focused on somatic embryo-related genes will provide a theoretical basis for further clarifying molecular mechanism during somatic embryogenesis, and the establishment of a somatic embryo regeneration system will supply an efficient platform for genetic improvement and germplasm innovation.
Materials and methods
Plant materials
Burs of Castanea mollissima cv. ‘Yanshanhongli’ were collected from the Chestnut Experiment Station in Huairou District, Beijing, China, and immature embryos were isolated and sterilized. The tips (Fig. 1b) and ovules (Fig. 1a) of the immature embryo were used for the induction of somatic embryos. Embryogenic calli, somatic embryos of different stages, non-embryogenic calli and regenerated shoots were used for total RNA extraction. All samples subjected to RNA extraction were immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction. Somatic embryos at different stages were fixed in 3% glutaraldehyde for morphological observations under a scanning electronic microscope.
Culture procedure
Embryo initiation
For the initial explant preparations, explants were derived from immature nuts that were harvested every 3 days for 39–60 days after the first full-bloom stage. Immature nuts were separated from burs, disinfected in 75% ethanol for 1 min and 3% NaClO with continuous agitation for 6 min, and then rinsed three or four times with sterile distilled water for 30 s each time. The immature embryos were isolated from the nuts, and the tips of the immature embryos or the ovules were isolated from the immature embryos. They were then placed in embryo initiation medium (E1) in a 90 × 15-mm Petri dish (40–45 explants per dish) and cultivated for at least 8 weeks in continuous darkness at approximately 23–25 °C, until embryogenic calli were observed.
The embryo initiation medium contained 1.8 μM 2,4-dichlorophenoxyacetic acid (2,4-D) and 1.1 μM 6-benzylaminopurine (6-BA), and the other components were the same as the initiation medium used for American chestnut (Xing et al. 1999). The pH of the medium was adjusted to 5.5 before autoclaving (121 °C, 25 min).
To maintain continuous embryogenesis, a factorial test using different concentrations of 2,4-D and 6-BA in the embryo initiation medium was applied to the embryogenic calli. The test utilized four concentrations of 2,4-D (0, 0.9, 1.8, or 3.6 μM) and four concentrations of 6-BA (0, 0.55, 1.1, or 2.2 μM) for a total of 16 possible combinations. Three replicates were performed, with 40–45 embryogenic callus clumps per replication. Since Chinese chestnut browned easily in tissue culture, the browning rate, proliferation rates and callus status were assessed.
Embryo development
The embryo development medium (E2) of Chinese chestnut followed that of Xing et al. (1999). Again, a factorial test was performed using 2,4-D and 6-BA. The test utilized nine growth regulator combinations, consisting of 0, 0.45, or 0.9 μM 2,4-D and 0, 0.275 or 0.55 μM 6-BA. Five Petri dishes (50–60 embryo clumps each) were used per combination. After 4 weeks of development, the cotyledonary stage embryos of each combination were harvested for maturation. The embryos that did not reach the cotyledonary stage continued to grow in embryo development medium (E2) under continuous darkness at 23–25 °C.
Embryo maturation
After 1–2 months of development, all cotyledonary stage embryos were transferred to Petri plates containing embryo maturation medium (E3). The E3 medium was identical to the embryo maturation medium of American chestnut (Xing et al. 1999). The embryos were incubated in the dark at 23–25 °C for at least 2 weeks.
Embryo germination
The resulting embryos were cultured in embryo germination medium (E4). In reference to American chestnut embryo germination medium (Xing et al. 1999), the embryos were incubated under a 16-h photoperiod at 23–25 °C for at least 8 weeks. To promote embryo germination, the effects of different concentrations of indole-3-butyric acid (IBA), 6-BA, and α-naphthylacetic acid (NAA) on embryo germination were tested. The test utilized nine combinations of growth regulators, consisting of 0, 0.1, or 0.2 μM of IBA, 0, 0.5, or 1.0 μM of 6-BA, and 0, 0.5 or 1.0 μM of NAA, according to Orthogonal Design Assistant II V3.1.
Morphological observations of somatic embryogenesis
The morphological characteristics of typical somatic embryos, involved in globular stage, heart stage, torpedo stage and cotyledonary stage, were observed via stereomicroscopy (ZEISS SteREO Discovery.V20, Germany) and scanning electron microscopy (SEM, JEOL-5600, Japan). When performed by SEM, somatic embryos at different embryogenic stages were fixed in phosphate buffer (0.2 M, pH 6.8) and 3% (v/v) glutaraldehyde. The fixed samples were maintained at room temperature for 1 h and then kept at 4 °C for at least 24 h. Next, they were washed in the same phosphate buffer, placed on ice, and postfixed overnight in the mixture with 1% osmium tetroxide and the same phosphate buffer (v/v = 1:1) at 0 °C (Fowke et al. 1994). The fixed tissues were washed twice with phosphate buffer (0.2 M, pH 6.8) and hydrated twice in a graded series of 30, 40, 50, 60, 70, 80, 85, 90, and 95% ethanol, then three times in 100% ethanol. Subsequently, the samples were dried using a Polaron E3000 Critical Point Dryer. After drying, the samples were coated with a gold–palladium alloy and examined under a scanning electron microscope operated at 10 kV.
DNA extraction
Total genomic DNA of the parental embryogenic calli and ten germinated shoots was extracted using a modified cetyl trimethyl ammonium bromide (CTAB) method (Cheng et al. 2005). The quality of purified total DNA was observed by 1% agarose gel electrophoresis, and the concentration was detected by Nano-volume Spectrophotometry (Scan Drop, Germany). The final concentration of isolated total DNA was diluted to 50 ng/μL and stored at −20 °C for further use.
RNA isolation and cDNA synthesis
The total RNA was extracted using the Plant RNA Kit (Omega, America) based on the manufacturer’s recommendations. To remove any remaining DNA, each RNA sample was digested with DNase I (Takara, Japan). The cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen, America).
Cloning and sequence analysis of embryogenesis-related genes
Primers for the fragments of CmSERK, CmWUS, CmLEC1, CmFUS3, CmAGL15, CmABP1, CmTIR1 and CmCDC48 genes, which are related to somatic embryogenesis, were designed using the software Primer 5.0 (Premier, Canada). The primers were listed in Table 1. The cDNA for amplification was derived from embryogenic calli. Each amplification reaction was carried out in a 20 μL volume, containing 2 μL 10 × Ex Taq Buffer, 0.8 μL dNTPs (10 mM), 14 μL ddH2O, 0.2 μL Ex Taq (5 U/μL), 1 μL cDNA and 1 μL each primer (10 mM). The following program was used for polymerase chain reaction (PCR) amplification: 95 °C for 5 min followed by 36 cycles at 95 °C for 30 s, 1 min at an annealing temperature based on each pair of primers and 72 °C for 30 s, with a final step of 10 min at 72 °C. Amplification bands were separated by 1% agarose gel. Gel Extraction kit (AXYGEN, America) was used to purify the PCR products, each of which was ligated into the pMD-19 T plasmid (Takara, Japan). Then, the constructs were transformed into Escherichia coli DH5α (Takara, Japan). After identification by colony PCR, the recombinant plasmids were purified and sequenced in Shanghai Sangon Biotech Co., Ltd. The resulting nucleotide sequences of the eight genes were subsequently aligned using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the orthologs in Chinese chestnut. To define the evolutionary relationships of the eight genes, phylogenetic trees were constructed using alignments of the amino acid sequences encoded by the eight genes from Chinese chestnut and other plants using the MEGA 5.0 software with neighbor-joining algorithms. Furthermore, bootstrap analysis was inferred from 1000 replicates.
Quantitative real-time RT-PCR analysis
Quantitative real-time RT-PCR (qRT-PCR) analysis was used to assess gene expression patterns during different stages of somatic embryogenesis using a Light Cycler® 96 SW1.1 Real-Time PCR System (Roche, Germany) with SYBR Green (Takara, Japan). The CmACTIN gene was employed as the endogenous reference gene for normalization of the threshold value (Ct) of the target genes. Gene-specific primers for qRT-PCR analysis (Table 2) were designed using Beacon Designer software. These sequences were verified with the BLAST tool at NCBI, and the dissociation curves were analyzed to confirm their specificity after PCR amplification. cDNA of different embryogenic stages were used as templates. We defined embryogenic calli (EC) and globular embryos (G) as the early stages of somatic embryogenesis, heart embryos (H) and torpedo embryos (T) as the middle stages of somatic embryogenesis, cotyledonary embryos (C) as the late stage of somatic embryogenesis, and non-embryogenic calli (NEC) as the control.
Each reaction of qRT-PCR was carried out in a 10 μL volume containing 5 μL SYBR® Premix Ex Taq (2×), 3.5 μL ddH2O, 1 μL diluted template (30-fold dilution of cDNA) and 0.25 μL of each primer. The reaction program for qRT-PCR was as follows: 95 °C for 10 min, followed by 39 cycles at 95 °C for 20 s, 54 °C for 20 s, and 72 °C for 20 s, and finally melting curves were completed by increasing the temperature from 65 to 97 °C. Three technical and biological replicates were carried out by qRT-PCR and changes in gene expression among different stages of somatic embryogenesis were performed using the Light Cycler® 96 software.
Detection of somaclonal variation
Simple sequence repeats (SSR) analysis
Somaclonal variation was analyzed using 24 pairs of SSR primers (Fang et al. 2013) (Table 3). Genomic DNA derived from embryogenic calli and germinated shoots was individually amplified to detect genetic stability. Each 20-μL amplification reaction was performed, containing 2 μL 10 × LA Taq Buffer, 0.8 μL dNTPs (10 mM), 14 μL ddH2O, 0.2 μL LA Taq (5 U/μL), 1 μL DNA and 1 μL each primer (10 mM). The PCR profile was carried out by the following conditions: initial denaturation step was 5 min at 94 °C, subsequent 35 cycles at 94 °C for 30 s, 30 s at annealing temperature (Table 3), 30 s at 72 °C and final extension cycle at 72 °C for 10 min. The PCR products were separated on 8% denaturing polyacrylamide gels and observed by silver staining. For SSR analysis, a band with the same mobility was considered to be an identical band.
Gene sequencing
cDNA of embryogenic calli in embryo initiation medium and ten regenerated shoots by random selection were used as templates to amplify the fragments of CmSERK and CmWUS. Gene-specific primers were designed with Primer 5.0: CmSERK: 5′-GGTGTAGTTCCAGACAATG and 5′-AACAAGTAACCGCTCAGT, CmWUS: 5′-ATGGGAAGCATGAAGGTGCATC and 5′-TCATCTGCTTTCCGGATGTAATTG. Amplification reaction, plasmid construction and sequencing analysis were identical methods as mentioned above. The gene sequences of CmSERK and CmWUS from different individuals were aligned to detect the base variations, respectively.
Statistical analysis
The data obtained from the somatic embryo initiation, development, and germination experiments were recorded using Excel 2013. All of the trials were conducted with at least three replicates. Statistical analysis were performed using analysis of variance (ANOVA) and expressed as the mean ± standard error (SE). The significant differences were determined at 5% level with Duncan’s multiple range tests using SPSS 17.0.
Results and discussion
Establishment of a somatic embryo regeneration system for Chinese chestnut
Initiation and maintenance of embryogenic cultures
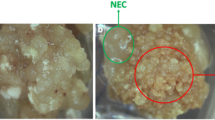
The selection of explant tissue played a key role in the successful induction of somatic embryogenesis (Lincy et al. 2009; Manoharan et al. 2016). Calli from the tips and ovules of the immature embryo used as explants in Chinese chestnut occurred after 10 days. Formation of a dense cellular clump, referred to as the proembryogenic mass, was observed after 20 and 30 days of induction. In this culturing phase, we recorded embryogenic and non-embryogenic calli, as shown in Figs. 2a, b and 3a, b. Among the 2092 tips of immature embryos that were incubated, 1952 tips were induced to calli, with 910 embryogenic callus clumps and 1042 non-embryogenic callus clumps, representing an embryogenic callus induction rate of 43.5%. However, among the 3257 cultured ovules, 1227 were induced to calli, with 919 embryogenic callus clumps and 308 non-embryogenic callus clumps, representing an embryogenic callus induction rate of 28.22% (Table 4). We found that the embryogenic callus induction rate for the tips of immature embryos was higher than that for the ovules for each replication. The embryogenic callus induction rates for the tips of immature embryos were relatively high between 45 and 54 days after the first flowering (ranged from 44.01 to 46.98%). Therefore, we selected the tips of immature embryos from 45 to 54 days after first flowering as the explants for embryogenic callus induction. Somatic embryos of European chestnut can be induced from leaf, shoot apex and zygotic embryos, while ovules were successfully used to induce somatic embryos of American chestnut (Corredoira et al. 2015; Maynard et al. 2015).
The different developmental stages of somatic embryos observed using stereomicroscopy (a–f) and regenerated shoots (g–i). a Non-embryogenic callus; b embryogenic callus; c globular stage embryos; d heart stage embryo; e torpedo stage embryo; f cotyledonary stage. Bar 500 μm; g–i regenerating shoots growing in germination medium. Bar 5 mm
In our study, we tested 16 combinations of embryo initiation medium to maintain continuous embryogenesis (Table 5). The results showed that callus morphologies observed in treatments E1-1 to E1-5 were compact, and the calli were milk white. The morphologies recorded in the E1-6 to E1-8 treatments appeared loose, with a brownish yellow coloration. Although the callus status of E1-9 and E1-10 was loose, milky white and light yellow, the browning rate was higher than in the other treatments. Not only was the callus browning rates of E1-13 to E1-16 particularly high, their callus was brownish yellow with poor status. The combinations consisting of 1.1 μM 6-BA and 1.8 or 3.6 μM 2,4-D were the two best treatments (E1-11 and E1-12). Despite that these two treatments did not show the greatest proliferation rates, their browning rates were lower, and the appearance of their calli was appropriate, with a loose morphology and light yellow coloration. It was considered that 2,4-D played a crucial role on the regulation and balance of endogenous auxin (Li et al. 2011). Many studies indicated that 2,4-D together with cytokinin was sufficient to initiate somatic embryogenesis in a number of plant species, such as Norway spruce, black alders, American chestnut and European chestnut (Hakman et al. 1985; Xing et al. 1999; Corredoira et al. 2003, 2015). Our data also showed that 2,4-D and 6-BA used together exhibited higher efficiency for the induction and maintenance of embryogenic competency in Chinese chestnut.
Development of somatic embryos
After four weeks, the embryogenic calli were transferred to embryo development medium (E2), the calli exhibited a number of somatic embryos at various developmental stages, including globular, heart, torpedo, and cotyledonary stage embryos (Fig. 2c–f). According to previous reports addressing several other plant species (Lai et al. 1998; Xing et al. 1999), increasing the sucrose concentration and reducing hormone levels in the induction medium can promote the development of somatic embryos. Thus, we tested nine growth regulator combinations of 2,4-D and 6-BA with a sucrose concentration of 60 g L−1, similar to the embryo development medium used for American chestnut (Xing et al. 1999). The results showed that the number of globular embryos in every treatment was almost the same. Heart stage, torpedo stage, and cotyledon stage embryos were observed when E2-1 medium was used, while they were rarely observed or not observed in all of the other treatments. A total of 30 globular, 2 heart, 6 torpedo, and 8 cotyledonary stage embryos per gram were obtained under these treatment conditions without 2,4-D and 6-BA (Table 6).
In general, hormone-free medium contributed to the development of somatic embryos (Li et al. 2011). This may be due to somatic embryos in high hormone concentration conditions having tissue differentiation suppressed and apical meristems that develop abnormally (Halperin and Wetherell 1964). In the present study, it was also found that the complete removal of hormones from the E2 medium was beneficial for promoting the development of somatic embryos, which was agreement with the results obtained for numerous relevant studies on somatic embryogenesis (Karami et al. 2007; Sugiura et al. 2008; Corredoira et al. 2015; Maynard et al. 2015).
Maturation of somatic embryos
Once the embryos reached the cotyledonary stage in E2 medium after 3–4 weeks, they were transferred to embryo maturation medium (E3 medium). The cotyledon embryos were matured in E3 medium for 2–4 weeks, and during this time, they became white and non-transparent (Fig. 2f), rather than light yellow and transparent. In our study, 0.5 μM NAA and 0.5 μM 6-BA were applied to the maturation of somatic embryos at the same level used for American chestnut (Xing et al. 1999). Combined with above development results of the somatic embryos, it indicated that the development and maturation of somatic embryos of Chinese chestnut had two different stages, similar to American chestnut, whereas these two stages of European chestnut somatic embryos were integrated together (Corredoira et al. 2015).
Germination of mature embryos
To germinate somatic embryos, the effects of different concentrations of IBA, 6-BA and NAA in the embryo germination medium (E4 medium) were examined. Somatic embryo germination was observed when the matured embryos were transferred to E4 medium for at least 8 weeks. The matured embryos turned from white to green after treatment under a 16-h photoperiod for 2 weeks. Germinated shoots and roots in Chinese chestnut were evaluated with references to American chestnut and European chestnut (Xing et al. 1999; Corredoira et al. 2003). Performing embryo germination in E4-2 with 0.5 μM 6-BA, and 0.5 μM NAA or in E4-5 with 0.5 μM 6-BA, 0.5 μM IBA and 1.0 μM NAA resulted in shoot regeneration. The results showed that the shoot conversion rate was higher in E4-2 (2.1%) than E4-5 (1.1%) after 8 weeks (Table 7). However, we did not obtain any regenerated shoots with the other E4 treatments. Additionally, germinated roots were also observed after 8 weeks of culture. The root conversion rate of E4-2 (13.6%) was significantly higher than that of other treatments (Table 7).
Germination difficulty of the embryos was a main limitation during somatic embryogenesis regeneration, and germination rates vary considerably in many plant species (Xing et al. 1999; Odutayo et al. 2005; Sezgin and Dumanoğlu 2014; Manoharan et al. 2016). Some species expressed relatively high germination rates (>50%), such as Pinus densiflora, Lilium ledebourii and Turkish Crocus species (Bakhshaie et al. 2010; Kim and Moon 2014; Verma et al. 2016). However the germination rates of other genotypes like American chestnut (3.3%), European chestnut (27.5%), as well as Chinese chestnuts, as seen from the result in this study (2.1%) were rather low (Xing et al. 1999; Sezgin and Dumanoğlu 2014). This may be due to different hormone ratios and genotypes resulting in the changes of regeneration rates (Verma et al. 2016). In addition, germinated shoots or roots were obtained from somatic embryos of Chinese chestnut, but whole plantlets directly conversed by somatic embryos was not observed. Possibly it was that the somatic embryos were not sufficiently mature or that the dormancy was not completely broken, leading to shoots and roots not being capable to develop synchronously (Merkle et al. 1995). Considering the relatively low regeneration rates of Chinese chestnut, the regeneration system needs to be optimized in the future.
Protocol for somatic embryo regeneration system in Chinese chestnut
According to the results of the above analysis, we developed a protocol for somatic embryo initiation, development, maturation, and germination. A summary of the procedure is shown in Fig. 4. Briefly, explants were cultured in E1 medium for approximately 2 months. During this time, both embryogenic and non-embryogenic calli were observed (Figs. 2, 3). The embryogenic calli were transferred to E2 medium, and after 7 days, globular embryos emerged (Figs. 2c, 3c). After another 1–3 weeks in development medium, the typical morphological stages of somatic embryos, such as the heart stage, the torpedo stage, and the cotyledonary stage, were also observed (Figs. 2d–f, 3d–f). Subsequently, the somatic embryos were transferred to E3 medium for 3 or 4 weeks and developed into mature embryos. Finally, the mature embryos were transferred to E4 medium for at least 8 weeks for further germination. We observed regenerated shoots (Fig. 2g–i) growing in the germination medium after 2 months.
The developmental stage of somatic embryo regeneration differs in various species. Among Castanea species, Chinese chestnut and American chestnut (Xing et al. 1999; Maynard et al. 2015) both exhibit four stages including initiation, development, maturation, and germination, while European chestnut has three stages involved in induction, maturation, and germination (Corredoira et al. 2015). For other woody plants, such as Dimocarpus longan, the somatic embryo regeneration system consisted of three stages: induction, maturation, and regeneration (Lai et al. 1998), while that of Carya cathayensis only required two stages: induction and regeneration (Zhang et al. 2011).
Morphological characteristics of somatic embryos
Embryogenic calli and non-embryogenic calli were obtained through induction of the tips of immature embryos. The non-embryogenic calli were white in color in both the induction and differentiation media (Fig. 2a). Using SEM, the non-embryogenic calli (Fig. 3a) was found to be more compact and irregular in shape than the embryogenic calli (Figs. 2b, 3b). The embryogenic calli underwent significant changes in induction and differentiation media. In the induction medium, they appeared to be yellow and granular (Fig. 2b) and contained many embryonic cells as well as embryonic cell groups and globular embryos. The SEM results showed that the surfaces of the embryogenic calli contained many tightly packed globular embryos (Fig. 3b). Embryogenic competence remained under E1 medium treatment. Various developmental stages of embryos, including the globular stage, heart stage, torpedo stage and cotyledonary stage, were also observed (Figs. 2c–f, 3c–f) in E2 and E3 media.
Assessment of somaclonal variation
SSR markers were successfully used to detect somaclonal variations in many plants (Smýkal et al. 2007; Pandey et al. 2012). In our study, SSR technology was also applied to examine the genetic variation during the germination process of somatic embryos in Chinese chestnut. The results showed that the size of amplified DNA fragments produced by 24 pairs of primers ranged from 100 bp to 500 bp. Twenty-four pairs of primers generated 45 bands with an average of 1.8 bands per primer. The amplified profiles generated among embryogenic calli and ten shoots were completely uniform, indicating a high level of genetic fidelity (additionally data are given in Online Resource 1).
The sequences of CmSERK or CmWUS gene fragments among embryogenic calli and ten individually regenerated shoots were amplified, and the results showed that these sequences were identical. It indicated that base mutations in these two genes did not occur during the germination process of somatic embryos in Chinese chestnut (additionally data are given in Online Resource 2, 3).
In the present study, somaclonal variation based on molecular analysis was not found in Chinese chestnut, which was consistent with the results reported for pea and bamboo (Smýkal et al. 2007; Singh et al. 2013). However, it did not mean that any mutation of the somatic embryo regeneration system in Chinese chestnut did not occur. Many studies indicated that somaclonal variation occurred during plant regeneration, such as coffee and sugarcane (Landey et al. 2015; Thorat et al. 2017). No variations were observed during somatic embryogenesis and regeneration in Chinese chestnut, which may be due to a very limited population detected or that the loci identified of genetic fidelity were not sufficient, as only 24 pairs of SSR primers and two gene fragment sequences were identified. However, the results based on current data showed that this regeneration system was with high genetic fidelity, and regenerated shoots had identical genetic backgrounds. We can draw a careful conclusion that this somatic embryo regeneration system in Chinese chestnut is reliable, and suitable for further genetic manipulation.
Expression analysis of somatic embryogenesis-related genes in Chinese chestnut
Identification and cluster analysis of embryogenesis-related genes
Coding fragments of eight embryogenesis-related genes related to morphogenesis, cell signalization, cell proliferation and auxins were detected, including the Somatic Embryogenesis Receptor-like Kinase gene (CmSERK), the WUSCHEL gene (CmWUS), the Leafy Cotyledon 1 gene (CmLEC1), the FUSCA3 gene (CmFUS3), the AGAMOUS-Like15 (CmAGL15), the Auxin Binding Protein 1 gene (CmABP1), the Transport Inhibitor Response 1 gene (CmTIR1), and the Cell Division Cycle 48 gene (CmCDC48).
The fragments of the eight embryogenesis-related genes of Chinese chestnut showed high similarity to genes of other species such as Citrus sinensis, Populus tomentosa, and Theobroma cacao (Table 8). For instance, the CmSERK gene shared a high degree of sequence similarity with C. sinensis (88%, NM_001288871.1) and Rosa canina (88%, HM802242.1). The CmWUS gene showed the highest similarity to P. tomentosa WOX4b (82%, KF982704.1) and T. cacao (83%, XM_007017115.2). The results of phylogenetic trees showed that CmSERK, CmLEC1, CmWUS, CmABP1, CmAGL15, CmFUS3, CmTIR1 and CmCDC48 presented the closest genetic relationships with McSERK, PtWOX4b, BoLEC1, BnABP1, CiMADS9, RcFUS3, MtTIR1 and DhCDC48, respectively (additionally data are given in Online Resource 4a-h).
Expression of embryogenesis-related genes during somatic embryogenesis process
The relative expression levels of the CmSERK, CmLEC1, CmWUS, CmABP1, CmAGL15, CmFUS3, CmTIR1, and CmCDC48 genes during embryogenesis were analyzed via quantitative real-time RT-PCR (qRT-PCR). Because expression of CmTIR1 and CmCDC48 was not detected at all stages of somatic embryogenesis, subsequent analysis did not include these two genes.
Expression profiles of CmSERK showed higher expression in all five somatic stages than in non-embryogenic stage (Fig. 5a). CmSERK expression reached a peak in EC stage and then decreased gradually, while increased expression of LdSERK in Larix decidua was observed (Rupps et al. 2016). The expression levels of CmSERK were detected in embryogenic and non-embryogenic calli, which was similar to the results reported for L. decidua, Secale cereale, as well as Medicago truncatula (Nolan et al. 2003; Gurszczyńska and; Rakoczy-Trojanowska 2011; Rupps et al. 2016). As a specific marker of embryogenic cells (Schmidt et al. 1997), combined with a high expression level at an early stage of this study, we propose that CmSERK plays a crucial role in the initiation of embryogenesis in Chinese chestnut.
Expression analysis of somatic embryogenesis-related genes during the development of Chinese chestnut. a–f The relative expression of the CmSERK (a), CmWUS (b), CmLEC1 (c), CmFUS3 (d), CmABP1 (e) and CmAGL15 (f) genes during embryogenesis was analyzed through quantitative real-time RT-PCR. Values represent the means ± SE (n = 3)
In Fig. 5b, CmWUS was found to highly express in the early stage (EC), but maintain a rather low expression level at the middle and late stages of somatic embryogenesis. The expression pattern of CmWUS during embryogenesis was in agreement with that of orthologous genes in Coffea canephora and L. decidua (Nic-Can et al. 2013; Rupps et al. 2016). The WUS gene contributed to promoting the transformation of somatic cells into embryogenic cells and the maintenance of embryogenesis, and might be considered as a putative embryo marker to predict the embryogenic potential (Zuo et al. 2002; Klimaszewska et al. 2011). Therefore, taken with the high expression during the early stage, it is believed that the role of CmWUS is to perform a function of maintaining competency of embryogenesis.
In our study, CmLEC1 expressed in all stages of embryogenesis and reached its maximum at the early stage (EC). Subsequently, CmLEC1 gene expression declined remarkably, and kept stable at the H, T and C stages (Fig. 5c). The CmLEC1 gene showed very low transcript at the NEC stage. Similar results were also obtained for D. longan and L. decidua (Cai 2011; Rupps et al. 2016) indicating that LEC1 mainly accumulated during early embryogenesis. Because the LEC1 gene was crucial for somatic embryogenic induction, LEC1 was also considered as a somatic embryogenesis marker gene (Gaj et al. 2005; Rupps et al. 2016). Based on the high expression in the early stage embryos, the role of CmLEC1 was inferred as an important regulating marker gene during Chinese chestnut embryogenesis. Previous studies demonstrated that the expression of FUS3 was activated by LEC1 transcription factor and its role was to control embryo mature genes (Braybrook and Harada 2008). In the present study, the expression of CmFUS3 was detected in all stages of embryogenesis and it achieved a higher level compared to NEC stage (Fig. 5d).
Former studies have shown that ABP1 was involved in the cell cycle and cell proliferation (Li et al. 2012). It was found that CmABP1 was expressed at a higher level at the early stage than the other stages because the cells proliferated rapidly at this stage (Fig. 5e). The results showed that CmAGL15 exhibited higher expression at the early stage, while it decreased significantly after the EC stage (Fig. 5f). The AGL15 gene was preferentially expressed during the early stage of somatic embryogenesis (Wang et al. 2002), which was similar to our data. Although some gene expression results related to embryogenesis of Chinese chestnut have been obtained, further studies are required to precisely identify their function.
In summary, CmSERK, CmLEC1, CmWUS and CmAGL15 showed a high level of expression in early embryonic stages. Moreover, these embryogenesis-related genes, except for CmWUS, presented relatively high expression levels at most of the middle and late stages during embryogenesis. Our results indicated that the CmSERK and CmLEC1 genes were marker genes of the early developmental stage associated with initiation of embryogenesis.
Abbreviations
- WPM:
-
McCown’s Woody Plant Medium salts
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- 6-BA:
-
6-Benzyladenine
- NAA:
-
α-Naphthaleneacetic acid
- IBA:
-
Indole-3-butyric acid
- BLAST:
-
Basic local alignment search tool
- SEM:
-
Scanning electron microscope
- qRT-PCR:
-
Quantitative real-time RT-PCR
- E1:
-
Embryo initiation medium
- E2:
-
Embryo development medium
- E3:
-
Embryo maturation medium
- E4:
-
Embryo germination medium
- EC:
-
Embryogenic calli
- NEC:
-
Non-embryogenic calli
- G:
-
Globular embryos
- H:
-
Heart embryos
- T:
-
Torped embryos
- C:
-
Cotyledonary embryos
- SERK :
-
Somatic embryogenesis receptor-like kinase
- WUS :
-
WUSCHEL
- LEC1 :
-
Leafy Cotyledon 1
- FUS3 :
-
FUSCA3
- AGL15 :
-
AGAMOUS-Like15
- ABP1 :
-
Auxin binding protein 1
- TIR1 :
-
Transport inhibitor response 1
- CDC48 :
-
Cell division cycle 48
References
Bakhshaie M, Babalar M, Mirmasoumi M, Khalighi A (2010) Somatic embryogenesis and plant regeneration of Lilium ledebourii (Baker) Boiss., an endangered species. Plant Cell Tissue Organ Cult 102(2):229–235
Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13(12):624
Cai Y (2011) Cloning and expression of SERK and some other somatic embryogenesis-related genes during somatic embryogenesis in Dimocarpus longan lour. Dissertation, Fujian Agriculture and Forestry University
Carraway DT, Wilde HD, Merkle SA (1994) Somatic embryogenesis and gene transfer in American chestnut. J Am Chestnut Found 8:29–33
Cheng L, Su S, Qin L, Yin W (2005) Extraction of DNA and establishment of AFLP techniques in leaves of Castanea mollissima Blume. J Beijing Univ Agric 2:5–9
Corredoira E, Ballester A, Vieitez AM (2003) Proliferation, maturation and germination of Castanea sativa Mill. Somatic embryos originated from leaf explants. Ann Bot 92(1):129–136. doi:10.1093/aob/mcg107
Corredoira E, Valladares S, Martínez MT, Vieitez AM, San José MC (2013) Somatic embryogenesis in Alnus glutinosa (L.) Gaertn. Trees 27:1597–1608
Corredoira E, Valladares S, Vieitez AM, Ballester A (2015) Chestnut, European (Castanea sativa). Methods Mol Biol 1224:163–176. doi:10.1007/978-1-4939-1658-0_14
Fang G, Blackmon BP, Staton ME, Nelson CD, Kubisiak TL, Olukolu BA, Henry D, Zhebentyayeva T, Saski CA, Cheng C, Monsanto M, Ficklin S, Atkins M, Georgi LL, Barakat A, Wheeler N, Carlson JE, Sederoff RR, Abbott AG (2013) A physical map of the Chinese chestnut (Castanea mollissima) genome and its integration with the genetic map. Tree Genet Genomes 9(2):525–537
Fowke LC, Attree SM, Rennie PJ (1994) Scanning electron microscopy of hydrated and desiccated mature somatic embryos and zygotic embryos of white spruce (Picea glauca [Moench] Voss.). Plant Cell Rep 13:612–618. doi:10.1007/BF00232931
Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222:977–988. doi:10.1007/s00425-005-0041-y
Giri CC, Shyamkumar B, Anjaneyulu C (2004) Progress in tissue culture, genetic transformation and applications of biotechnology to trees: an overview. Trees 18:115–135doi. doi:10.1007/s00468-003-0287-6
Gruszczyńska A, Rakoczy-Trojanowska M (2011) Expression analysis of somatic embryogenesis-related SERK, LEC1, VP1, and NiR, ortologues in rye (Secale cereale L.). J Appl Genet 52(1):1–8
Hakman I, Fowke LC, Arnold VS, Eriksson T (1985) The development of somatic embryos in tissue cultures initiated from immature embryos of Picea abies (Norway spruce). Plant Sci 38:53–59
Halperin W, Wetherell DF (1964) Adventive embryony in tissue cultures of the wild carrot, Daucus Carota. Am J Bot 51(3):274–283
Hebard FV (2006) The backcross breeding program of the American chestnut foundation. J Am Chestnut Found 19:55–77
Hecht V, Viellecalzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127:803–816
Hu R, Sun Y, Wu B, Duan H, Zheng H, Hu D, Zheng H, Hu D, Lin H, Tong Z, Xu J, Li Y (2017) Somatic embryogenesis of immature Cunninghamia lanceolata (lamb.) hook zygotic embryos. Sci Rep 7(1):56. doi:10.1038/s41598-017-00156-1
Ikeuchi M, Iwase A, Rymen B, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, Lucas MD, Veylder LD, Goodrich J, Brady SM, Roudier F, Sugimoto K (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1:15089. doi:10.1038/nplants.2015.89
Jia H, McCarty DR, Suzuki M (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163:1293–1305
Karami O, Kordestani GK, Mohamadi M (2007) Direct somatic embryogenesis and plant in strawberry (Fragaria ananssa). J Plant Physiol 12(4):322–332
Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18:626–638. doi:10.1105/tpc.105.039412
Kim YW (2015) Initiation of embryogenic callus from mature zygotic embryos in Japanese larch (Larix kaempferi). J Plant Biotechnol 42:223–227
Kim YW, Moon HK (2014) Enhancement of somatic embryogenesis and plant regeneration in Japanese red pine (Pinus densiflora). Plant. Biotechnol Rep 8(3):259–266
Klimaszewska K, Overton C, Stewart D, Rutledge RG (2011) Initiation of somatic embryos and regeneration of plants from primordial shoots of 10-year-old somatic white spruce and expression profiles of 11 genes followed during the tissue culture process. Planta 233:635–647. doi:10.1007/s00425-010-1325-4
Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15:5–18
Lai Z, Pan L, Chen Z (1998) Plant regeneration at high frequency from somatic embryos in longan. J Fujian Agric Univ 27:31–36. doi:10.13323/j.cnki.j.fafu(nat.sci.).1998.01.007
Landey RB, Cenci A, GuyotR, Bertrand B, Georget F, DechampE, Herrera JC, Aribi J, Lashermes P, Etienne H (2015) Assessment of genetic and epigenetic changes during cell culture ageing and relations with somaclonal variation in Coffea arabica. Plant Cell Tissue Organ Cult 122(3):517–531
Lee H, Fischer RL, Goldberg RB, Harada JJ (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. PNAS 100:2152–2156
Li M, Wang S, Feng D (2011) The advance of plant somatic embryogenesis and development. Chin Agric Sci Bull 27(03):237–241
Li H, Lai Z, Su M, Lin Y (2012) Cloning of TIR1 and ABP1 genes from embryogenic calli of longan and analysis of their expression during longan somatic embryogenesis. Acta Hortic Sin 39(2):253–264
Li Q, Shi X, Zhao Q, Cui Y, Ouyang J, Xu F (2016) Effect of cooking methods on nutritional quality and volatile compounds of Chinese chestnut (Castanea mollissima Blume). Food Chem 201:80–86. doi:10.1016/j.foodchem.2016.01.068
Lincy AK, Remashree AB, Sasikumar B (2009) Indirect and direct somatic embryogenesis from aerial stem explants of ginger (Zingiber officinale Rosc.). Acta Bot Croat 68(1):93–103
Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Manoharan R, Tripathi JN, Tripathi L (2016) Plant regeneration from axillary bud derived callus in white yam (Dioscorea rotundata). Plant Cell Tissue Organ Cult 126:481–497
Martins JF, Correia SI, Canhoto JM (2016) Somatic embryogenesis induction and plant regeneration in strawberry tree (Arbutus unedo L.). Methods Mol Biol 1359:329–339
Maynard CA, McGuigan LD, Oakes AD, Zhang B, Newhouse AE, Northern LC, Chartrand AM, Will LR, Baier KM, Powell WA (2015) Chestnut, American (Castanea dentata (Marsh.) Borkh.). Methods Mol Biol 1224:143–161
Merkle SA, Wiecko AT, Watsonpauley BA (1991) Somatic embryogenesis in American chestnut. Can J For Res 21:1698–1701
Merkle SA, Parrott WA, Flinn BS (1995) Morphogenic aspects of somatic embryogenesis. In: Thorpe TA (ed) In vitro embryogenesis in plants. Dordrecht, Kluwer Academic Publishers, pp 155–203
Miller AC, Woeste KE, Anagnostakis SL, Jacobs DF (2014) Exploration of a rare population of Chinese chestnut in North America: stand dynamics, health and genetic relationships. AoB Plants 6:11–15. doi:10.1093/aobpla/plu065
Nic-Can GI, López-Torres A, Barredo-Pool F, Wrobel K, Loyola-Vargas VM, Rojas-Herrera R, De-la-Pena C (2013) New insights into somatic embryogenesis: leafy cotyledon1, baby boom1 and WUSCHEL-related homeobox4 are epigenetically regulated in Coffea canephora. PLoS ONE 8(8):e72160. doi:10.1371/journal.pone.0072160
Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133:218–230
Odutayo O, Akinrimisi FB, Ogunbosoye I, Oso RT (2005) Multiple shoot induction from embryo derived callus cultures of cowpea (Vigna unguiculata L.) Walp. Afr J Biotechnol 4:1214–1216
Orłowska A, Igielski R, Łagowska K, pczy ska EK (2017) Identification of LEC1, L1L and Polycomb Repressive Complex2 genes and their expression during the induction phase of Medicago truncatula Gaertn. somatic embryogenesis. Plant Cell Tiss Organ Cult 129:119–132. doi:10.1007/s11240-016-1161-8
Palovaara J, Hakman I (2008) Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Picea abies somatic embryogenesis. Plant Mol Biol 66:533–549. doi:10.1007/s11103-008-9289-5
Pandey RN, Singh SP, Rastogi J, Sharma ML, Singh RK (2012) Early assessment of genetic fidelity in sugarcane (saccharum officinarum) plantlets regenerated through direct organogenesis with rapd and ssr markers. Aust J Crop Sci 6(4):618–624
Rugh CL, Senecoff JF, Meagher RB, Merkle SA (1998) Development of transgenic yellow poplar for mercury phytoremediation. Nat Biotechnol 16:925–928. doi:10.1038/nbt1098-925
Rupps A, Raschke J, Rümmler M, Linke B, Zoglauer K (2016) Identification of putative homologs of Larix decidua to BABYBOOM (BBM), LEAFY COTYLEDON1 (LEC1), WUSCHEL-related HOMEOBOX2 (WOX2) and SOMATIC EMBRYOGENESIS RECEPTOR-like KINASE (SERK) during somatic embryogenesis. Planta 243:473–488. doi:10.1007/s00425-015-2409-y
Schmidt ED, Guzzo F, Toonen MA, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124:2049–2062
Sezgin M, Dumanoğlu H (2014) Somatic embryogenesis and plant regeneration from immature cotyledons of European chestnut (Castanea sativa, Mill.). In Vitro Cell Dev Biol Plant 50:58–68
Singh SR, Dalal S, Singh R, Dhawan AK, Kalia RK (2013) Ascertaining clonal fidelity of micropropagated plants of Dendrocalamus hamiltonii, Nees et Arn. ex Munro using molecular markers. In Vitro Cell Dev Biol Plant 49(5):572–583
Smýkal P, Valledor L, Rodríguez R, Griga M (2007) Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Rep 26(11):1985–1998
Sugiura A, Matsuda-Habu Y, Gao M, Tao R (2008) Somatic embryogenesis and plant from immature persimmon (Diospyros kaki Thunb.) embryos. Hortic Sci 43(1):211–214
Thorat AS, Sonone NA, Choudhari VV, DevarumathRM, HarinathBabu K (2017) Plant regeneration from cell suspension culture in Saccharum officinarum L. and ascertaining of genetic fidelity through RAPD and ISSR markers. 3 Biotech 7(1):16. doi:10.1007/s13205-016-0579-3
Verdeil JL, Alemanno L, Niemenak N, Tranbarger TJ (2007) Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci 12:245–252. doi:10.1016/j.tplants.2007.04.002
Verma SK, Das AK, Cingoz GS, UsluE, Gurel E (2016) Influence of nutrient media on callus induction, somatic embryogenesis and plant regeneration in selected Turkish crocus species. Biotechnol Rep 10:66–74
Vidal N, Mallón R, Valladares S, Meijomín AM, Vieitez AM (2010) Regeneration of transgenic plants by Agrobacterium-mediated transformation of somatic embryos of juvenile and mature Quercus robur. Plant Cell Rep 29:1411–1422. doi:10.1007/s00299-010-0931-8
Wang H, Tang W, Zhu C, Perry SE (2002) A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J 32:831–843
Wang X, Niu QW, Teng C, Li C, Mu J, Chua NH, Zuo J (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19:224–235. doi:10.1038/cr.2008.276
Xing Z, Powell WA, Maynard CA (1999) Development and germination of American chestnut somatic embryos. Plant Cell Tissue Organ Cult 57:47–55
Zhang L, Yin W, Wang H (2007) Ovule culture in vitro of Castanea mollissima Bl. J Beijing For Univ 29:99–105
Zhang Q, Hu H, Wang Z, Yuan J, Wan J, Huang J (2011) Indirect somatic embryogenesis and plant regeneration of Carya cathayensis. Acta Hortic 38:1063–1070. doi:10.16420/j.issn.0513-353x.2011.06.010
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J Cell Mol Biol 30:349–359
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Nos. 31370679 and 31672135) and Beijing Science and Technology Plan Project (Z161100000916011).
Author information
Authors and Affiliations
Contributions
Study conception and design—LQ, QC; performing of the experiments—DL, WW, WZ, FJ, XL; analysis of data—DL, WZ; drafting of manuscript—DL, WW, QC; reading and final approval of the version to be published—LQ, QC, LDM, YX, KF, QZ.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Jose M. Segui-Simarro.
Dan Lu, Wei Wei and Wan Zhou have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lu, D., Wei, W., Zhou, W. et al. Establishment of a somatic embryo regeneration system and expression analysis of somatic embryogenesis-related genes in Chinese chestnut (Castanea mollissima Blume). Plant Cell Tiss Organ Cult 130, 601–616 (2017). https://doi.org/10.1007/s11240-017-1250-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-017-1250-3