Abstract
Doubled haploids of triticale are of interest for plant breeders due to hybrid breeding programs based on cytoplasmic male sterility Tt phenomenon. However, (epi)mutations appearing during in vitro culture regeneration may lead to a phenotypic variation that makes the uniformity of plant materials questionable. Using RP-HPLC genomic DNA methylation of donor doubled haploid plants utilized as a source of tissues for the in vitro regeneration (via androgenesis and somatic embryogenesis) of triticale cv. Bogo and their consecutive generative progeny was evaluated. It was demonstrated that in vitro cultures induced a decrease of the DNA methylation of the regenerants independently of the approach used for plant regeneration. The decrease in DNA methylation of genomic DNA proceeded up to the first/second successive generations followed by the beginning of its reestablishment. Moreover, somatic embryogenesis resulted in a higher level of genomic DNA demethylation in regenerants than androgenesis and the process of methylation seems to be affected by donor plant. It is being speculated that long term changes in genomic DNA methylation may be a source of off-type individuals that may spontaneously arise during plant breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triticale is an artificial species that originated ca. 130 years ago from a cross between wheat and rye, with the first cultivars useful for breeders available in the 1960s (Mergoum et al. 2009). It exhibits high yield potential, grain quality, resistance to pathogens, favourable amino acid composition and adaptation to adverse conditions (Heger and Eggum 1991). Hybrid breeding based on cytoplasmic male sterility is a promising direction in its cultivation (Oettler et al. 2005). However, homozygous, genetically uniform and stable doubled haploids (DHs) are necessary to maximize heterosis. Such forms could be derived via androgenesis employing anther cultures or isolated microspores (Eudes and Amundsen 2005; Żur et al. 2008; Lantos et al. 2013; Würschum et al. 2013).
Tissue culture induced variation (TCIV) in regenerants (Li et al. 2007; González et al. 2013) and somaclonal variation (SV) in progeny of such regenerants (Linacero and Vazquez 1992; Ivanov et al. 1998) have been observed in crops. TCIV may be manifested at the morphological level of regenerants (Jaligot et al. 2000). Usually, off-type plants are eliminated from the breeding population. However, epigenetic changes without obvious morphological effect, are mostly not easily recognized (Bednarek et al. 2007) and may spontaneously come into being even after several generations (Brettell and Dennis 1991). Evidently, tissue culture may induce alterations in DNA methylation pattern (González et al. 2011; Shan et al. 2013) since it may impose stressful conditions (González et al. 2013).
Additionally, cell reprogramming may require resetting of the DNA methylation pattern (Baroux et al. 2011) causing epimutations. Relatively low levels of epigenetic change related to tissue culture were described in barley (Bednarek et al. 2007). Those changes occurred whether regeneration was through either androgenesis or somatic embryogenesis. However, it is not evident whether genome methylation of triticale regenerants derived via androgenesis and somatic embryogenesis and their progeny behaves similarly. Possibly, the complex genome of triticale may react differently as suggested by genomic studies (Bento et al. 2011) as well as molecular and cytological analyses (Brettell et al. 1986).
In triticale, off-type forms derived from tissue culture regeneration are not rare (Pauk et al. 2000). Cytogenetic analysis showed that triticale is genetically unstable during tissue culture manipulation (Lapitan et al. 1984). Chromosome analysis of the root tip cells of regenerated plants revealed that the rye genome was more frequently involved in chromosomal rearrangements than wheat genome (Charmet et al. 1986). Not only tissue culture regeneration, but also generative reproduction may cause genome instability (Suenaga and Nakajima 1993) of the progeny derived from regenerants.
To study the extent of changes induced by tissue culture and inherited by the progeny different methods have been used (Hossain et al. 2003; Peredo et al. 2009; Linacero et al. 2011; Díaz-Martínez et al. 2012). Most of them involved isoschizomers differing in their sensitivity towards DNA methylation sites (Ochogavía et al. 2009; Wang et al. 2013; Machczyńska et al. 2014). The limitation of the system to restriction sites recognized by the endonucleases is apparent (Schrey et al. 2013). Contrary to marker-based approaches, high performance liquid chromatography (HPLC) has been applied in studies of DNA methylation of plants regenerated via tissue culture (Renau-Morata et al. 2005; Rival et al. 2013). This technique supports general information on global DNA methylation and could be applied for the evaluation of differences among plant materials that could be related to spontaneously arising off-type plants (Johannes et al. 2009; Yi et al. 2010).
Tissue culture is a stressful environment that may lead to numerous epigenetic changes. These changes may be transmitted to the next generations. However, it is not clear how long such changes may persist in the progeny. The purpose of the study was to evaluate the level of global epigenetic changes related to genomic DNA methylation among donor triticale plants, their regenerants derived via different in vitro culture methods and consecutive generative progenies of the regenerants using RP-HPLC approach.
Materials and methods
Plant material
Four lines of DH donor plants (each line was represented by vegetatively multiplicated clones, Table 1) of winter triticale cv. Bogo (Oleszczuk et al. 2004) were used to generate regenerants via shed microspore culture (M), anther culture (A), and immature zygotic embryo culture (E). The regenerants were used to obtain the first, the second and the third generative progenies. Donor plants, their homozygous regenerants and their consecutive progenies formed a set. Each plant within the set had its complete pedigree to a specific donor from which it was derived. There were four of such sets encoded S1, S2, S3 and S4. The arrangement of the plant material is given in Table 1.
Androgenesis
Anther culture
Plants of winter triticale cv. Bogo have grown under controlled conditions in a chamber room at a photoperiod of 16 h day and 8 h night at 16 and 12 °C, respectively. The tillers were collected, when the microspores were at the mid to the late uninucleate stage and pretreated in a cool room for 4 weeks at 4 °C. Next, the spikes were sterilized (70 % ethanol then 10 % sodium hypochlorite) and rinsed three times for 1 min with sterile water. Subsequently, anthers from one half of the spikes were excised and cultured on solid induction medium 190-2 (Zhuang and Xu 1983) with Phytagel and supplemented with 90 g/l maltose, 400 mg/l glutamine, 2 mg/l 2,4-D and 0.5 mg/l kinetin at 26 °C in the dark, for a maximum of 8 weeks. Calli and embryo-like structures were transferred onto regeneration medium (190-2 with 0.5 mg/l NAA and 0.5 mg/l kinetin), when they were 2–5 cm in size. Afterwards, green plantlets were kept in an Erlenmeyer flasks, in rooting medium (190-2 with 2 mg/l IAA). After roots appeared, plants were vernalized for 6 weeks in 4 °C and transferred to soil in a greenhouse, then grew to maturity.
Shed microspore culture
The same protocol as for anther culture was used for shed microspore culture from anthers taken from the second half of the same spikes except with a liquid induction medium. Then, appearing callus pieces, as well as proembryos were transferred to solid regeneration medium.
Somatic embryogenesis
Immature zygotic embryo culture
Caryopses obtained after self-pollination of the DH mother plants were cut to remove the embryos at the coleoptile stage under aseptic conditions. Immature embryos were isolated and placed with the scutellum exposed onto MS medium (Murashige and Skoog 1962) with 30 g saccharose and 30 µM dicamba. Developing embryos were transferred onto medium supplemented with 0.9 µM 2,4-D for conversion of somatic embryos to plantlets. Cultures were maintained in controlled conditions at 26 °C under 16/8 h (day/night) photoperiod. Afterwards, the plantlets were transferred to the regeneration medium. The 5 cm long plantlets were transferred on a root formation medium (regeneration and rooting media are the same as for androgenesis). Next, small plants were kept in 4 °C for 6 weeks, then they were placed into soil in a greenhouse and grown to maturity.
Progeny
Individual DH regenerants derived via anther cultures and shed microspore cultures as well as regenerants derived from immature zygotic embryo cultures were selfed by bag isolation. Seeds of those regenerants were advanced to obtain progeny. Some plants were again bag isolated to obtain the second and third generations of progeny.
Flow cytometry
Ploidy status of regenerants after in vitro androgenesis was determined by flow cytometry (Partec; PAII; HBO lamp; Germany). Young regenerated plants were prepared according to Galbraith et al. (1983) with minor modification. Nuclei were isolated from leaves which were chopped with a razor blade in 2 ml of a lysis buffer containing DAPI, afterwards the suspension was filtered through a 30 µm pore diameter filter prior to analysis. Subsequently, histograms of the cell nuclei were generated. The ploidy level of regenerants was determined by comparing the G1 peaks of each sample to a reference (diploid plant).
DNA isolation
Total genomic DNA isolation was performed from young leaves (with flag leaf emerging) of donors, regenerants and their progeny following Plant DNeases MiniKit 250 (Qiagen) manufacturer protocol. DNA quantity was evaluated spectrophotometrically (NanoDrop 1000 Overview). The samples were tested for their integrity and purity on agarose gel with ethidium bromide (1.2 % gel, 1× TBE buffer, 160 V).
RP-HPLC analysis
The plant material used for the RP-HPLC is arranged in Table 1.
Sample preparation
DNA samples (6 µg) were dissolved in 100 µl deionised water and denatured by heating to 100 °C for 2 min following immediate placing on ice for 5 min. DNA hydrolysis was performed in a total volume of 135 µl consisting of 5 µl of 10 mM ZnSO4 and 10 µl of 1.0 U ml−1 of P1 nuclease (in 30 mM NaOAc pH 5.4), the samples were gently stirred and incubated for 17 h at 37 °C. After nuclease digestion 10 µl 0.5 M Tris pH 8.3 and 10 µl of 10 U ml−1 of alkaline phosphatase (in 2.5 M (NH4)2SO4) were added, followed by gentle mixing and incubating at 37 °C for 2 h. After digestion, the samples were centrifuged at 12,000 rpm for 5 min.
Chromatographic assessment of global DNA methylation
RP-HPLC quantification of overall DNA methylation was performed according to the procedure of Johnston et al. (2005) using Waters 625 LC System. Briefly, the 4u Max-RP C12 (250 × 4.6 mm, Phenomenex) column combined with 4u Max-RP C12 pre-column were applied using linear gradient of the eluent A comprising of 0.5 % methanol plus 10 mM KH2PO4 at a pH of 3.7 (v/v) and eluent B comprising of 10 % methanol in 10 mM KH2PO4, pH 3.7 (v/v) that changed from 100 % of A to 100 % of B within 10 min following 15 min of 100 % B eluent and ending with 100 % of the A eluent for 5 min at a flow rate equal to 1 ml min−1. The absorbance was measured spectrophotometrically (UV detector, λ = 280 nm). Chromatograph was calibrated with external standards consisting of each DNA nucleosides (5–50 µM), 15–150 µM for RNA nucleosides and 1.5–15 µM of 5-methyldeoxycytidine dissolved in deionised water. On the basis of that data the calibration curves were evaluated and used for the quantification of the nucleosides in the samples with Millennium 32v 4.0 software. The percentage of deoxycytidine methylation in relation to the total content of cytidine was calculated according to the following equation: 5mdC % = [5mdC/(5mdC + dC)] × 100, where 5mdC and dC states for 5-methyldeoxycytidine and deoxycytidine, respectively. Each RP-HPLC analysis was conducted in three repeats using DNA samples from plant material arranged in Table 1.
Statistics
One-way analysis of variance (ANOVA) with Tukey’s contrasts analysis were performed in R CRAN software (stats and multcomp packages, respectively). The regression analysis was performed in R CRAN software using stats package and polynomial regression was evaluated. For all statistical analyses significance level α = 0.01 was applied.
All RP-HPLC results (including three repeats for each sample) based on the whole plant material arranged in Table 1 were used for Tukey’s contrasts and regression analysis.
Results
Donor plants
Each donor’s genotype was represented by several vegetatively multiplicated individuals (Table 1). No morphological variation among multiplicated plants (representing the given genotype as well as all genotypes) was observed.
Plant regeneration
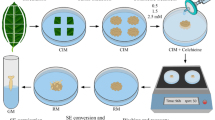
Embryos and embryo-like structures were observed after 4–8 weeks of culture in darkness in shed microspore culture (Fig. 1a). Microspore development resulted in complete embryo formation with visible cotyledons and embryo axis (Fig. 1b). Androgenesis in anther cultures resulted in lumpy embryo-like structures visible after ca. 6 weeks of culture (Fig. 1c). After transmission of such structures into the light some albino plants were detected (Fig. 1d). Somatic embryogenesis from cultured immature zygotic embryos is shown in Fig. 1e, f. Embryos appeared after 4 weeks of culture on MS medium with dicamba (Fig. 1e) and converted to plants on regeneration medium (Fig. 1f). Self-pollination of regenerants (Fig. 1g) resulted in formation of the first generative progeny (Fig. 1h).
Triticosecale Wittmack cv. Bogo regenerants derived via in vitro culture and their progeny; a androgenesis on liquid induction medium, b embryo formation from shed microspore culture, c androgenesis on solid induction medium, d albino plant derived from anther culture, e somatic embryogenesis from immature zygotic embryo, f germinating embryo, g fertile anther culture-derived plants, h progenies of regenerants
With the exception of albinos, no obvious morphological changes were observed among fertile regenerants as well as among the first, second and third generations of the progeny. However, morphological differences were visible between haploids and diploids. The haploid plants were smaller, had thinner stems and narrower leaves compared to DH plants.
Analysis of ploidy level
There were 63.3 and 71.7 % haploids among regenerants derived via shed microspore and anther culture, respectively (Table 2) with spontaneous diploidization amounting to 27.1 and 27.7 % for the two approaches, severally. Aneuploids were observed for 0.6 and 0.62 % of cases, respectively (Table 2).
Changes in global DNA methylation
The RP-HPLC approach resulted in clearly distinguishable, well-separated peaks corresponding to dC and 5mdC, with retention time equal to 5.44 and 8.43 min, respectively.
The greatest degree (25.4 %) of average global DNA methylation was observed for donors. In case of all regenerants (R) the level of DNA methylation was equal to 24.1 % and was the lowest for the first generative progeny (P1) (23.6 %). Starting from the second generative progeny, global DNA methylation was slightly greater than for P1 (Table 3).
Comparison of global DNA methylation for D, R, P1, P2 and P3 without considering sets and regeneration method
According to Tukey’s grouping (Fig. 2) donor plants differed from regenerants due to genomic DNA methylation. No difference between R and P1 was observed, however, R and P1 differed from P2 and P3 that formed separate group (Table 4). Polynomial regression (Table 5) calculated for the abovementioned data demonstrated that global DNA methylation decreased from donors to the first generative progeny and then reversed (Fig. 3).
Tukey’s grouping of donors (D), regenerants (R), the first (P 1), the second (P 2) and the third (P 3) generative progeny of the regenerants without considering sets or tissue culture regeneration method, based on global DNA methylation evaluated via RP-HPLC. Global DNA methylation is expressed as the amount of methylated cytosine to the total amount of cytosine converted to percentages. Materials forming the same group are indicated by either ‘a’, ‘b’ or ‘c’ letters
Polynomial regression of donors (D), regenerants (R), the first (P 1), the second (P 2) and the third (P 3) progenies of the regenerants (without considering sets and regeneration method). Global DNA methylation is expressed as the amount of methylated cytosine to the total amount of cytosine converted to percentages
Comparison of global DNA methylation for regenerants and their three generations of progeny by regeneration method
Comparison (ANOVA) of the global DNA methylation of the regenerants derived via shed microspore (M), anther (A) and immature zygotic embryos (E) tissue culture approaches revealed (Fig. 4) that plants obtained via androgenesis differed from those regenerated via somatic embryogenesis (Table 4). The first generative progeny of shed microspore derived regenerants differed from those obtained via anthers and the first progeny from somatic embryogenesis derived regenerants (Table 4). P 2M and P 2A did not differ from each other according to 5mdC content, although they were distinct from P 2E (Table 4). In the case of the third generative offspring, overall DNA methylation in P 3M differed from P 3E (Table 4), whereas DNA methylation level in P 3A was similar both to P 3M and P 3E .
Tukey’s grouping of the regenerants (R), the first (P 1), the second (P 2) and the third (P 3) generative progeny of the regenerants based on tissue culture method used. Grouping is indicated by small letters. The materials with the same letter (e.g. ‘a’) form the same group and are distinct from those classified as ‘b’ within the group of R, P 1, P 2 and P 3, respectively. Global DNA methylation is expressed as the amount of methylated cytosine to the total amount of cytosine converted to percentages. The regenerants derived either via M (dark grey), A (grey) or E (white) approach and their progeny are indicated by colours
Regression analysis (Fig. 5) performed for donors, RA, RM, RE and their three generations demonstrated that global genomic DNA methylation decreased in regenerants compared to the donors. The decrease in DNA methylation was observed up to the first/second generative progeny following by reversion of the process. Independently of the tissue culture approach used for plant regeneration the observed trends were significant and followed the polynomial function (Table 5). A little bit higher level of DNA demethylation for RE than for RA and RM was observed.
Polynomial regression analysis of global DNA methylation changes exhibited between D, R, P 1, P 2 and P 3 materials according to the tissue culture regeneration method. The regression line for D–RM–P 1M –P 2M –P 3M , D–RA–P 1A –P 2A –P 3A and D–RE–P 1E –P 2E –P 3E are given in black, dotted and dashed lines, respectively
Comparison of global DNA methylation for sets
Tukey’s test revealed significant differences in global DNA methylation of plant material encompassing each of the S1, S2, S3 and S4 sets (Fig. 6). In detail, in the first set D, R, P2 differed from each other, whereas differences between P1 and P3 were insignificant (Table 4). In the S2–D, P1, P2, P3 differed from each other, whereas R were similar both to P1 and P3 (Table 4). In the third set D, R, P1, P2, P3 differed from each other (Table 4) while in the fourth one D, R, P2, P3 differed from each other, whereas P1 were similar both to R and P2 (Table 4).
Tukey’s grouping of the donor plants, regenerants and consecutive generative progeny within sets without considering tissue culture approach used for plant regeneration. S 1, S 2, S 3 and S 4 state for sets comprising D, R, P 1, P 2 and P 3. For detailed description of the abbreviation see Fig 2. Materials with the same small letter (indicated on graph) of the given set belong to the same group
Global genomic DNA methylation changes followed polynomial regression in the case of all sets with local minimum at the level of the first/second generative progeny (Table 5; Fig. 7). All trends were significant. In the case of S2 and S4 local minimum was deeper compared with S1 and S3.
Discussion
There is ample evidence suggesting that tissue culture is a stressful environment that may be responsible for the induction of abiotic stress during plant regeneration (Miguel and Marum 2011). Such stress may cause the regeneration of the off-type plants that could be detected at the morphological (Li et al. 2010), cytological (Linacero and Vazquez 1992), genetic or epigenetic level (Bednarek et al. 2007; Vining et al. 2013). Some of the changes may subsequently arise among progeny originated from tissue culture-derived regenerants even after many generative cycles (Bregitzer et al. 1998; Kirikovich et al. 2003).
Thus, it is essential to have several sets of plant material encompassing homozygous donor plants, the regenerants derived from them and the progenies obtained from the regenerants to be able to identify epigenetic differences among such materials. The plant material used in our study originated from DH plants that became the source of tissues for the regeneration. The donor plants represented distinct genotypes but did not exhibit any morphological variation. Moreover, generative progeny was obtained by self-pollination under controlled conditions. Thus, any variation among our materials within sets (donor plants, regenerants and their progenies) should be attributed to the changes induced in tissue cultures and transmitted to the next generations. However, possibly due to highly homozygous plant material, as well as the limited number of plants analyzed in our study, we failed to observe at the morphological level any off-type plants among regenerants and their progeny within as well as and among sets. Nevertheless, the differences were revealed for haploids and a few aneuploids detected via flow cytometry among regenerants. The level of spontaneously derived DH plants and aneuploids was similar to prior report (Oleszczuk et al. 2004).
The lack of morphological variation or even cytological uniformity does not, however, exclude changes at the DNA level and especially those related to DNA methylation, which may take place under tissue culture conditions, as showed in barley (Bednarek et al. 2007). Our RP-HPLC study demonstrated that the triticale genome was affected by global DNA demethylation which occurred during tissue culture regeneration. The same direction of DNA methylation changes was observed in our preliminary study on triticale cv. Bogo with metAFLP approach (Machczyńska et al. 2014), where demethylation prevailed over de novo methylation. Our data are congruent with the MSAP studies of genomic DNA methylation changes performed between rye donor plants and their regenerants where the level of demethylation was greater for the regenerants (González et al. 2013). Similar results were obtained also in rye using HpaII and MspI digested DNA amplified with ISSR markers (Linacero et al. 2011). It was demonstrated that the decrease in DNA methylation at the level of regenerants may be caused by the necessity of genes activation during cell reprogramming to facilitate plant regeneration (Kaeppler and Phillips 1993).
Our RP-HPLC results showed no difference in the genomic DNA methylation level between shed microspore and anther culture approaches while androgenic regenerants exhibited greater methylation than somatic embryogenesis derived ones. It is well documented that a callus phase induces variation during plant regeneration (Bouman and Klerk 2001). In our experiment immature zygotic embryo plant regeneration encompassed an obvious callus stage whereas androgenesis, especially in the case of shed microspore cultures, largely circumvents callus. Thus, the longer callus duration of somatic embryogenesis compared to androgenesis may have been responsible for the differences in the level of DNA methylation evaluated between regenerants derived via two paths (Xu et al. 2004). Alternatively, the tissue culture medium composition could be also responsible for the difference in global DNA methylation. Plant growth regulators may enhance in vitro induced variation resulting from epigenetic modifications (Nehra et al. 1992; Bairu et al. 2006). Study on Malus xiaojinensis revealed increased DNA methylation with increasing 2,4-D (Huang et al. 2012). To induce the androgenic response in our experiment 2,4-D was supplemented to the induction medium, which may have been responsible for the greater genome-wide methylation in androgenic than in somatic embryogenesis derived regenerants. Immature zygotic embryos were cultured on a medium containing dicamba reported to cause more negative changes than 2,4-D (Rakoczy-Trojanowska 2002). It might be suggested that higher demethylation in somatic embryogenesis derived regenerants in triticale was caused by the presence of dicamba. Alternatively, the differences in global DNA methylation observed between androgenesis and somatic embryogenesis derived regenerants could have been due to cold stress used to convert the gametophytic pathway to a sporophytic one. Studies on stress-induced epigenetic changes confirm that cold treatment induces DNA demethylation (Fan et al. 2013; Shan et al. 2013). Moreover, the differences in DNA methylation observed between androgenesis and somatic embryogenesis derived regenerants could be also attributed to the ploidy level. The relationship between DNA methylation and haploids versus diploids was studied in rice (Zhang et al. 2006). The authors concluded that the higher methylation level in haploids might be a readjusting reaction to the decrease in ploidy because of survival. If DH plants inherit the increased DNA methylation e.g. via replication mechanisms then our data is congruent with the abovementioned results.
In our study significant methylation change induced in vitro and revealed by ANOVA affected not only regenerants but also their consecutive generative progeny. Reversion of the DNA demethylation in the first/second generation demonstrated by polynomial regression analysis may have reflected the very beginning of the genomic DNA methylation re-establishment. Apparently, plant regeneration via tissue cultures resulted in epigenetic changes that were not fully compensated even after several generative cycles. Based on our experiment, however, it was not possible to predict whether further generative cycles would result in DNA methylation comparable to that of donor plants or whether the process would behave differently. Although, in general, the trend of global DNA demethylation and its reversion was observed for all sets, the differences among them were obvious. Most probably this may reflect genotypic differences existing between donor plants. Obviously, revealed DNA methylation changes observed among sets suggest that triticale plant material derived via in vitro cultures and its progeny may be prone to epimutations dependent on DNA methylation that maybe exhibited among e.g. regenerants’ progeny.
Based on our HPLC analysis, we demonstrated for the first time that the level of genomic DNA methylation alterations between donor triticale plants, their regenerants derived via different in vitro culture methods and consecutive generative progenies of the regenerants was affected by in vitro culture. Moreover, genomic DNA demethylation of the regenerants depended on both regeneration method and genotype of donor plants. Evidently, re-establishment of the triticale genomic DNA methylation level after tissue culture treatment may take time and special care needs to be taken to control epimutations in regenerants.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- DAPI:
-
4′-6′ Diamidino-2-phenylindole
- DH:
-
Doubled haploid
- IAA:
-
Indole-3-acetic acid
- ISSR:
-
Inter-simple sequence repeat
- MSAP:
-
Methylation sensitive amplified polymorphism
- NAA:
-
α-Naphthaleneacetic acid
References
Bairu MW, Fennell CW, van Staden J (2006) The effect of plant growth regulators on somaclonal variation in Cavendish banana (Musa AAA cv. ‘Zelig’). Sci Hortic 108(4):347–351. doi:10.1016/j.scienta.2006.01.039
Baroux C, Raissig MT, Grossniklaus U (2011) Epigenetic regulation and reprogramming during gamete formation in plants. Curr Opin Genet Dev 21(2):124–133. doi:10.1016/j.gde.2011.01.017
Bednarek PT, Orłowska R, Koebner RMD, Zimny J (2007) Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol 7:10. doi:10.1186/1471-2229-7-10
Bento M, Gustafson JP, Viegas W, Silva M (2011) Size matters in Triticeae polyploids: larger genomes have higher remodeling. Genome 54(3):175–183. doi:10.1139/G10-107
Bouman H, Klerk G-J (2001) Measurement of the extent of somaclonal variation in begonia plants regenerated under various conditions. Comparison of three assays. Theor Appl Genet 102:111–117
Bregitzer P, Halbert SE, Lemaux PG (1998) Somaclonal variation in the progeny of transgenic barley. Theor Appl Genet 96(3–4):421–425. doi:10.1007/s001220050758
Brettell RS, Dennis E (1991) Reactivation of a silent Ac following tissue culture is associated with heritable alterations in its methylation pattern. Mol Gen Genet 229(3):365–372. doi:10.1007/BF00267457
Brettell RIS, Pallotta MA, Gustafson JP, Appels R (1986) Variation at the Nor loci in triticale derived from tissue culture. Theor Appl Genet 71(4):637–643. doi:10.1007/BF00264268
Charmet G, Bernard S, Bernard M (1986) Origin of aneuploid plants obtained by anther culture in triticale. Can J Genet Cytol 28(3):444–452. doi:10.1139/g86-067
Díaz-Martínez M, Nava-Cedillo A, Guzmán-López JA, Escobar-Guzmán R, Simpson J (2012) Polymorphism and methylation patterns in Agave tequilana Weber var. ‘Azul’ plants propagated asexually by three different methods. Plant Sci 185–186:321–330. doi:10.1016/j.plantsci.2012.01.004
Eudes F, Amundsen E (2005) Isolated microspore culture of Canadian 6× triticale cultivars. Plant Cell Tissue Organ Cult 82(3):233–241. doi:10.1007/s11240-005-0867-9
Fan H, Wei J, Li T, Li Z, Guo N, Cai Y, Lin Y (2013) DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta Physiol Plant 35(8):2445–2453. doi:10.1007/s11738-013-1278-x
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220(4601):1049–1051. doi:10.1126/science.220.4601.1049
González R, Ricardi M, Iusem N (2011) Atypical epigenetic mark in an atypical location: cytosine methylation at asymmetric (CNN) sites within the body of a non-repetitive tomato gene. BMC Plant Biol 11(1):1–11. doi:10.1186/1471-2229-11-94
González AI, Sáiz A, Acedo A, Ruiz ML, Polanco C (2013) Analysis of genomic DNA methylation patterns in regenerated and control plants of rye (Secale cereale L.). Plant Growth Regul 70(3):227–236. doi:10.1007/s10725-013-9794-7
Heger J, Eggum BO (1991) The nutritional values of some high-yielding genotypes of triticale. J Cereal Sci 14:63–71
Hossain AM, Konisho K, Mineo M, Kazuhiro N (2003) Somaclonal variation of regenerated plants in chili pepper (Capsicum annuum L.). Euphytica 130(2):233–239. doi:10.1023/A:1022856725794
Huang H, Han S, Wang Y, Zhang X, Han Z (2012) Variations in leaf morphology and DNA methylation following in vitro culture of Malus xiaojinensis. Plant Cell Tissue Organ Cult 111(2):153–161. doi:10.1007/s11240-012-0179-9
Ivanov P, Atanassov Z, Milkova V, Nikolova L (1998) Culture selected somaclonal variation in five Triticum aestivum L. genotypes. Euphytica 104(3):167–172. doi:10.1023/A:1018656903559
Jaligot E, Rival A, Beulé T, Dussert S, Verdeil JL (2000) Somaclonal variation in oil palm (Elaeis guineensis Jacq.): the DNA methylation hypothesis. Plant Cell Rep 19(7):684–690. doi:10.1007/s002999900177
Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, Bouchez D, Dillmann C, Guerche P, Hospital F, Colot V (2009) Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5(6):e1000530. doi:10.1371/journal.pgen.1000530
Johnston JW, Harding K, Bremner DH, Souch G, Green J, Lynch PT, Grout B, Benson EE (2005) HPLC analysis of plant DNA methylation: a study of critical methodological factors. Plant Physiol Biochem 43(9):844–853. doi:10.1016/j.plaphy.2005.07.015
Kaeppler SM, Phillips RL (1993) DNA methylation and tissue culture-induced variation in plants. In Vitro Cell Dev Biol Plant 29(3):125–130. doi:10.1007/BF02632283
Kirikovich SS, Svirshchevskaya AM, Levites EV (2003) Variation at isozyme loci in seed offspring of sugar beet gynogenetic lines. Sugar Tech 5(4):289–292. doi:10.1007/BF02942487
Lantos C, Bóna L, Boda K, Pauk J (2013) Comparative analysis of in vitro anther—and isolated microspore culture in hexaploid Triticale (×Triticosecale Wittmack) for androgenic parameters. Euphytica 1–11. doi:10.1007/s10681-013-1031-y
Lapitan NL, Sears RG, Gill BS (1984) Translocations and other karyotypic structural changes in wheat x rye hybrids regenerated from tissue culture. Theor Appl Genet 68(6):547–554. doi:10.1007/bf00285012
Li X, Yu X, Wang N, Feng Q, Dong Z, Liu L, Shen J, Liu B (2007) Genetic and epigenetic instabilities induced by tissue culture in wild barley (Hordeum brevisubulatum (Trin.) Link). Plant Cell Tissue Organ Cult 90(2):153–168. doi:10.1007/s11240-007-9224-5
Li R, Bruneau AH, Qu R (2010) Tissue culture-induced morphological somaclonal variation in St. Augustinegrass [Stenotaphrum secundatum (Walt.) Kuntze]. Plant Breed 129:96–99. doi:10.1111/j.1439-0523.2009.01647x
Linacero R, Vazquez AM (1992) Cytogenetic variation in rye regenerated plants and their progeny. Genome 35(3):428–430. doi:10.1139/g92-063
Linacero R, Rueda J, Esquivel E, Bellido A, Domingo A, Vázquez A (2011) Genetic and epigenetic relationship in rye, Secale cereale L., somaclonal variation within somatic embryo-derived plants. In Vitro Cell Dev Biol Plant 47(5):618–628. doi:10.1007/s11627-011-9407-y
Machczyńska J, Orłowska R, Zimny J, Bednarek P (2014) Extended metAFLP approach in studies of tissue culture induced variation (TCIV) in triticale. Mol Breed 1–10. doi:10.1007/s11032-014-0079-2
Mergoum M, Singh PK, Peña RJ, Lozano-del Río AJ, Cooper KV, Salmon DF, Gómez Macpherson H (2009) Triticale: a “new” crop with old challenges. In: Carena MJ (ed) Cereals. Handbook of plant breeding, vol 3. Springer, Berlin, pp 267–287. doi:10.1007/978-0-387-72297-9_9
Miguel C, Marum L (2011) An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot 62:3713–3725. doi:10.1093/jxb/err155
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Nehra N, Kartha K, Stushnott C, Giles K (1992) The influence of plant growth regulator concentrations and callus age on somaclonal variation in callus culture regenerants of strawberry. Plant Cell Tissue Organ Cult 29(3):257–268. doi:10.1007/BF00034361
Ochogavía A, Cervigni G, Selva J, Echenique V, Pessino S (2009) Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula. Plant Mol Biol 70(1–2):17–29. doi:10.1007/s11103-009-9454-5
Oettler G, Tams SH, Utz HF, Bauer E, Melchinger AE (2005) Prospects for hybrid breeding in winter triticale. Crop Sci 45(4):1476–1482. doi:10.2135/cropsci2004.0462
Oleszczuk S, Sowa S, Zimny J (2004) Direct embryogenesis and green plant regeneration from isolated microspores of hexaploid triticale (×Triticosecale Wittmack) cv. Bogo. Plant Cell Rep 22(12):885–893. doi:10.1007/s00299-004-0796-9
Pauk J, Puolimatka M, Lökös TK, Monostori T (2000) In vitro androgenesis of triticale in isolated microspore culture. Plant Cell Tissue Organ Cult 61(3):221–229. doi:10.1023/A:1006416116366
Peredo EL, Arroyo-García R, Revilla MÁ (2009) Epigenetic changes detected in micropropagated hop plants. J Plant Physiol 166(10):1101–1111. doi:10.1016/j.jplph.2008.12.015
Rakoczy-Trojanowska M (2002) The effect of growth regulators on somaclonal variation in rye (Secale cereale L.) and selection of somaclonal variants with increased agronomic traits. Cell Mol Biol Lett 7(4):1111–1120
Renau-Morata B, Nebauer SG, Arrillaga I, Segura J (2005) Assessments of somaclonal variation in micropropagated shoots of Cedrus: consequences of axillary bud breaking. Tree Genet Genomes 1(1):3–10. doi:10.1007/s11295-004-0001-x
Rival A, Ilbert P, Labeyrie A, Torres E, Doulbeau S, Personne A, Dussert S, Beulé T, Durand-Gasselin T, Tregear J, Jaligot E (2013) Variations in genomic DNA methylation during the long-term in vitro proliferation of oil palm embryogenic suspension cultures. Plant Cell Rep 32(3):359–368. doi:10.1007/s00299-012-1369-y
Schrey AW, Alvarez M, Foust CM, Kilvitis HJ, Lee JD, Liebl AL, Martin LB, Richards CL, Robertson M (2013) Ecological epigenetics: beyond MS-AFLP. Integr Comp Biol 53(2):340–350. doi:10.1093/icb/ict012
Shan X, Wang X, Yang G, Wu Y, Su S, Li S, Liu H, Yuan Y (2013) Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J Plant Biol 56(1):32–38. doi:10.1007/s12374-012-0251-3
Suenaga K, Nakajima K (1993) Variation in doubled haploid plants of wheat obtained through wheat (Triticum aestivum) x maize (Zea mays) crosses. Plant Breed 111(2):120–124
Vining K, Pomraning K, Wilhelm L, Ma C, Pellegrini M, Di Y, Mockler T, Freitag M, Strauss S (2013) Methylome reorganization during in vitro dedifferentiation and regeneration of Populus trichocarpa. BMC Plant Biol 13(1):92. doi:10.1186/1471-2229-13-92
Wang X, Wu R, Lin X, Bai Y, Song C, Yu X, Xu C, Zhao N, Dong Y, Liu B (2013) Tissue culture-induced genetic and epigenetic alterations in rice pure-lines, F1 hybrids and polyploids. BMC Plant Biol 13(1):77
Würschum T, Tucker M, Maurer H (2013) Stress treatments influence efficiency of microspore embryogenesis and green plant regeneration in hexaploid triticale (×Triticosecale Wittmack L.). In Vitro Cell Dev Biol Plant 1–6. doi:10.1007/s11627-013-9539-3
Xu M, Li X, Korban S (2004) DNA-methylation alterations and exchanges during in vitro cellular differentiation in rose (Rosa hybrida L.). Theor Appl Genet 109(5):899–910. doi:10.1007/s00122-004-1717-6
Yi C, Zhang S, Liu X, Bui HT, Hong Y (2010) Does epigenetic polymorphism contribute to phenotypic variances in Jatropha curcas L.? BMC Plant Biol 10:259. doi:10.1186/1471-2229-10-259
Zhang H, Peng H, Li Y, Xu P, Wang X, Wu X (2006) Patterns of DNA cytosine methylation between haploids and corresponding diploids in rice. Chin Sci Bull 51(14):1721–1728. doi:10.1007/s11434-006-2048-6
Zhuang JJ, Xu J (1983) Increasing differentiation frequencies in wheat pollen callus. In: Hu H, Vega MR (eds) Cell and tissue culture techniques for cereal crop improvement, vol 431. Science Press, Beijing
Żur I, Dubas E, Golemiec E, Szechyńska-Hebda M, Janowiak F, Wędzony M (2008) Stress-induced changes important for effective androgenic induction in isolated microspore culture of triticale (×Triticosecale Wittm.). Plant Cell Tissue Organ Cult 94(3):319–328. doi:10.1007/s11240-008-9360-6
Acknowledgments
The authors wish to thank Dr. Sylwia Oleszczuk for providing seeds of triticale cv. Bogo and for her substantial help in plant regeneration.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Machczyńska, J., Orłowska, R., Mańkowski, D.R. et al. DNA methylation changes in triticale due to in vitro culture plant regeneration and consecutive reproduction. Plant Cell Tiss Organ Cult 119, 289–299 (2014). https://doi.org/10.1007/s11240-014-0533-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-014-0533-1