Abstract
This study aims to use a machine learning (ML)-based enhanced diagnosis and survival model to predict heart disease and survival in heart failure by combining the cuckoo search (CS), flower pollination algorithm (FPA), whale optimization algorithm (WOA), and Harris hawks optimization (HHO) algorithms, which are meta-heuristic feature selection algorithms. To achieve this, experiments are conducted on the Cleveland heart disease dataset and the heart failure dataset collected from the Faisalabad Institute of Cardiology published at UCI. CS, FPA, WOA, and HHO algorithms for feature selection are applied for different population sizes and are realized based on the best fitness values. For the original dataset of heart disease, the maximum prediction F-score of 88% is obtained using K-nearest neighbour (KNN) when compared to logistic regression (LR), support vector machine (SVM), Gaussian Naive Bayes (GNB), and random forest (RF). With the proposed approach, the heart disease prediction F-score of 99.72% is obtained using KNN for population sizes 60 with FPA by selecting eight features. For the original dataset of heart failure, the maximum prediction F-score of 70% is obtained using LR and RF compared to SVM, GNB, and KNN. With the proposed approach, the heart failure prediction F-score of 97.45% is obtained using KNN for population sizes 10 with HHO by selecting five features. Experimental findings show that the applied meta-heuristic algorithms with ML algorithms significantly improve prediction performances compared to performances obtained from the original datasets. The motivation of this paper is to select the most critical and informative feature subset through meta-heuristic algorithms to improve classification accuracy.
Similar content being viewed by others
1 Introduction
Heart disease is a significant public health problem and has become the leading cause of death worldwide. Classic heart disease symptoms can be palpitations, shortness of breath, swelling in the legs and abdomen, fatigue/weakness, indigestion, hiccups and difficulty in swallowing, cough, headache, back and neck pain, fainting, and bruising [1]. Heart disease can be broken down into heart failure, coronary artery disease, vascular disease, irregular heartbeats, and many other categories [2]. Among heart diseases, heart failure has become the leading cause of death worldwide [3]. The most common symptom of this disease is the inability of the heart to pump the blood that the body needs. However, heart disease and heart failure symptoms do not play an active role in the early diagnosis of the disease to survive it. Furthermore, early precautions play a critical role in preventing life-threatening risks.
Biomarkers, defined as all kinds of biological signs, play a crucial role in patients’ early diagnosis and survival [4]. However, much data are produced daily in the healthcare industry. In the absence of modern technology, early diagnosis and survival of disease have become complex tasks. Therefore, intelligent methods are needed to make early diagnoses and disease survival from this large amount of data. Intelligent methods are in the field of ML.
ML is the core sub-field of artificial intelligence and can learn linear and nonlinear patterns from massive volumes of data. On the other hand, medical data are also complex and quite large. Considering all this, ML has become increasingly helpful and is being used in predicting disease or survival in the medical field. SVM has been trained for breast cancer [5,6,7], allergy [8, 9], COVID-19 [10,11,12] diagnosis and survival. NB has been used for the diagnosis and survival of diabetes [13, 14], and chronic kidney disease [15,16,17]. The decision tree has achieved good accuracy in diagnosing, and surviving cancer diseases [6, 18,19,20,21,22]. Neural networks have been used to classify psychological diseases [23,24,25]. In Alzheimer’s and Parkinson’s disease diagnosis, ensemble classifiers have been applied often, with success [26,27,28]. Bayesian networks have also been used to diagnose and survive many diseases [29,30,31,32,33]. LR is also one of the methods used in diagnosing and surviving diseases [34, 35]. ML methods are also used in diagnosing autism [36, 37]. There are many studies on blood disease diagnosis in the literature, namely thalassemia, blood cancer, malaria, and so on, with ML [38,39,40,41,42]. Celiac disease prediction with ML is one of the topics studied in recent years [43, 44]. KNN is one of the most used classification algorithms in many disease diagnoses, and survival [45,46,47,48,49,50].
Although ML plays an important role in diagnosing many diseases, as we mentioned above, the formation of high-dimensional datasets and the fact that these datasets may contain many irrelevant and unnecessary features are a critical disadvantage in learning algorithms. In this case, the burden of ML algorithms should be lightened. Therefore, feature selection is a significant factor in minimizing complexity, irrelevant, and redundant features. It can increase how effectively learning algorithms work. Finding the optimum feature subset in high-dimensional feature datasets is nevertheless classified as an NP-hard problem. The search space will expand exponentially as the number of features rises since a dataset with N features contains a \(2^{N-1}\) number of feature subsets. As a result, meta-heuristic algorithms have been used to identify the subset of features because exact algorithms cannot produce the desired result in a reasonable amount of time [51].
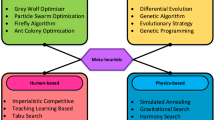
Several meta-heuristic algorithms have been developed and used in the literature to address feature selection issues: genetic algorithm (GA) [52], simulated annealing (SA) [53], ant colony optimization (ACO) [54], differential evolution (DE) [55], particle swarm optimization (PSO) [56], artificial bee colony (ABC) [57], firefly algorithm (FFA) [58], gravitational search algorithm (GSA) [59], grey wolf optimizer (GWO) [60], salp swarm algorithm (SSA) [61], bat algorithm (BA) [62], emperor penguin optimizer (EPO) [63], equilibrium optimizer (EO) [64], atom search optimization (ASO) [65], dragonfly algorithm [66], slime mould algorithm (SMA) [67], golden eagle optimizer (GEO) [68], duck travel optimization (DTO) [69], and so on.
In this study, we answer the questions can an effective feature selection be made with meta-heuristic algorithms, can successful performances be achieved with the selected features, and what is the effect of the population size and, thus, fitness value on feature selection? The experiments are conducted on the Cleveland heart disease dataset [70] and the heart failure dataset collected from the Faisalabad Institute of Cardiology [71] published at UCI.
The contribution of the study is summarized as follows:
-
To decrease the overall feature dimension and improve the overall classification accuracy of the ML algorithms, we have made a meta-heuristic-based feature selection. In other words, reducing the dimension of the feature space leads to more accurate and quicker classification.
-
To observe the impact of feature selection algorithms and the population size on the results, we have proposed a comparative study.
-
According to the classification results, although we could not comment on the population size, it has been observed that WOA tends to select fewer features than others.
-
While the most successful classification has been achieved with KNN, it has been seen that the classifications made with the reduced features by the CS algorithm are more successful on average.
-
The novelty of this study can be summarized as a comparative study examining the effect of meta-heuristic algorithms for feature selection and classification performance. Comparative studies have been conducted with CS, FPA, WOA, and HHO on two heart-related disease datasets. There are no similar comparative analyses, according to our best knowledge.
The remaining sections of the paper are structured as follows. The literature review is carried out in Sect. 2. In Sect. 3, datasets and pre-processing steps, the theoretical and mathematical background of feature selection, and classification algorithms are discussed in detail. In Sect. 4, the results of all experiments are analysed and discussed in detail. In the last section, the conclusion and future direction have been given in detail.
2 Literature review
In studies conducted for years, ML techniques have also been applied to heart diseases and heart failure for diagnosing and survival tasks. The various works that discuss the Cleveland Heart Disease dataset and the heart failure dataset are described in this section.
The studies which used the University of California Irvine’s (UCI) Cleveland Heart Disease Records are summarized here. Beulah et al. developed an ensemble-based diagnosis system for heart disease along with Brute force feature selection and obtained an accuracy of 85.48% with majority voting [72]. In Reddy et al.’s study, rough set theory-based selected features were classified with an adaptive genetic algorithm with fuzzy logic (AGAFL) and were revealed an average of 90% accuracy [73]. Kolukısa et al. applied ML algorithms with dimension reduction by using linear discriminant analysis (LDA), hybrid feature selection algorithm, and medical doctors’ recommendation-based feature selection [74]. 81.84% accuracy with SVM using medical doctors’ recommendation-based features was obtained. Li et al. developed a heart disease classification system by using ML algorithms [75]. In this system, in addition to the four classical feature selection algorithms, they used their own proposed fast conditional mutual information (FCMIM) algorithm and, with FCMIM-SVM, achieved an accuracy of 92.37%. Gupta et al. applied six classification algorithms with backward elimination and Pearson correlation coefficient for heart disease identification and achieved an accuracy of 88.16% with Naive Bayes [76]. Garate-Escamila et al. proposed a heart disease classification system and chi-square and principal component analysis (CHI-PCA) with RFs obtained the highest accuracy, with 98.7% [77]. In Tougui et al.’s study, experiments were conducted using different environments with ML algorithms [78]. The study showed that MATLAB’s ANN model was the best, with an accuracy of 85.86%. In [79], hybrid RF with a linear model (HRFLM) technique was applied to the heart disease dataset and found to have 88.7% accuracy. In [80], cluster-based decision tree learning (CDTL)-based feature selection with RF classification was applied, and 89.30% accuracy was obtained. Rani et al. developed an optimized hybrid decision support system to diagnose heart disease [81]. RandomizedSearchCV with RF gave an accuracy of 86.60%. In another study, Deepika and Balaji developed an MLP integrated with enhanced Brownian motion based on the dragonfly algorithm (MLP-EBMDA) feature selection algorithm and obtained accuracy at the rate of 94.28% [82]. In another study, Srinivas and Katarya found 94.7% accuracy by using hyOPTXg, which predicted heart disease with an optimized XGBoost classifier [83]. Lutimath et al. applied genetic algorithms for heart disease prediction tasks [84]. Gnoguem et al. combined decision tree, RF, KNN, and LR based on the maximum weighted sum of prediction with an accuracy of 92.10% [85]. Mohapatra et al. proposed an ANN, LR, and NB ensemble, and in terms of accuracy, the proposed model outperformed compared with AdaBoost and RF [86]. In Anderies et al.’s study, six different ML algorithms were compared, and SVM and LR achieved the best accuracy of 79% [87]. Shaw et al. applied maximum entropy, RF, and SVM to the dataset and obtained an accuracy level of 92.67% [88]. Goyal proposed lion optimization-based feature selection (LOFS) based on SVM, ANN, and DT. LOFS-ANN gave the best accuracy of 90.5% [89].
The studies which used the heart failure dataset collected from the Faisalabad Institute of Cardiology and at the Allied Hospital in Faisalabad are summarized here. In Chicco and Jurman’s study, the authors showed that binary classification of electronic health records of patients with cardiovascular heart problems using machine learning was done successfully [3]. It was also observed that the features selected based on machine learning and those selected based on biostatistics were compatible. Oladimeji and Oladimeji applied KNN, SVM, NB, and RF with four different feature selection algorithms [90]. Furthermore, they saw that while feature selection increased the performance of some algorithms, it caused a decrease in the performance of others. Another study used machine learning ensemble trees to construct a model for predicting heart failure survival. Over the various ensemble tree algorithms, Extreme Gradient Boosting (XGBoost) was shown to produce the most accurate results [91]. In [92], ensemble tree algorithms with feature selection were used for predicting heart failure survival. It was seen that feature selection significantly increased the classification performance of the models. Aloss et al. applied crow search algorithm-based five different ML algorithms to the dataset [93]. Swetha et al. applied several machine learning algorithms and achieved 99.1% accuracy with XGBoost [94]. In Kameswara et al.’s study, AdaBoost with Synthetic Minority Oversampling Technique (SMOTE) was applied, and an accuracy of 96.34% was obtained [95]. Karakuş and Er applied ANN, fine Gaussian SVM, fine KNN, weighted KNN, subspace KNN, boosted trees, and bagged trees to predict survival from heart failure and achieved an accuracy of 100% [96].
According to the literature review, feature selection aids in improving the classifier’s performance, and for both datasets, there are limited studies that use meta-heuristic algorithms for feature selection. Research is still needed to determine the best optimization strategy for feature subset selection.
3 Proposed methodology
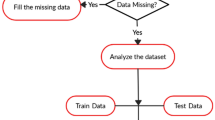
The proposed methodology to carry out experiments is given in Fig. 1. The heart disease dataset from the University of California, Irvine (UCI), and the heart failure dataset collected from the Faisalabad Institute of Cardiology and at the Allied Hospital in Faisalabad have been used for training and testing purposes. Then, CS, FPA, WOA, and HHO are used to select important features as crucial predictors of diagnosing heart disease from the original dataset. Then, the feature sets and the original dataset are given to four different classification algorithms as input.
3.1 Datasets
The datasets used in experiments are the Cleveland dataset [70] and heart failure clinical reports dataset [71] from UCI Machine Learning Repository. The first dataset consists of 303 samples; each has 14 features with two classes—healthy or patient. The detail of the feature set with the descriptions is listed in Table 1. The second dataset consists of 299 samples; each has 13 features with two classes—death event or not. The detail of the feature set with the descriptions is listed in Table 2.
The most important step after obtaining the dataset is making the data suitable for training by pre-processing. Checking for missing values is very important in the pre-processing steps. The experimental dataset is complete; there is no missing value in any feature. Then, min-max normalization is applied for numerical features to prevent domination among features due to their distance. Furthermore, all numeric features are scaled between 0 and 1. For nominal features, feature representation is applied as pre-processing. These steps are applied to both datasets.
Encoding nominal attributes with integer values only returns a ranking. However, using ranking in the similarity calculations between samples will not provide us with an accurate comparison. To cope with this situation, nominal features are transformed into binary features using one-hot encoding. Thus, each value taken by the nominal feature will be represented as a new feature. Within the scope of this study, we apply one-hot encoding to “cp”, “slope”, and “thal” features. As a result, while the “cp” feature is transformed into four new features, namely cp_1, cp_2, cp_3, and cp_4, “slope” and “thal” are transformed into three new features separately. For example, the “cp_1” feature represents the “typical angina”, and if a person has “typical angina”, the value of this attribute is 1; otherwise, 0. Therefore, the original 13 features become 20 features in the Cleveland dataset.
3.2 Meta-heuristic algorithms
3.2.1 Cuckoo search algorithm
The CS algorithm, which uses cuckoo birds’ breeding and reproduction strategies, is a meta-heuristic swarm-based approach [97]. The inspiration for the development of this algorithm is cuckoos’ brood parasitism and laying their eggs in the nests of other host birds [98]. The CS is a handy method that finds applications in many different areas, such as test functions, medical applications, data mining, machine learning and deep learning applications, image processing, path planning, and engineering problems [99].
The CS finds the nest and updates the position by realizing the steps below [100]:
-
Each cuckoo produces only one egg at a time and randomly chooses a nest of parasites to hatch,
-
The best parasite nest will be kept for the next generation,
-
There is a fixed number of available parasite nests, and the probability of detection of them is \(P_a\).
However, a biased local and global random walk is used to update the position of the cuckoo by adapting the Lévy flight signature algorithm. Lévy flight is proposed to point out the animal’s movement direction. Step length selection is drawn from the Lévy distribution, and based on step length, the algorithm moves to a new position if the new position is better than the current position. Otherwise, it stays in its current position and repeats the process. The position update process of the CS is given with (1).
In the (1), while \(X_i^{(t)}\) is the current position, \(X_i^{(t+1)}\) is the next position to be found. T represents the step size, and Levy\((\lambda )\) is the Lévy distribution-based random walk provided by \(\lambda\), which takes values between 1 and 3 [101]. \(\oplus\) realizes point-to-point multiplication between the step size and Lévy distribution.
3.2.2 Flower pollination algorithm
FPA developed by Yang is a meta-heuristic algorithm that simulates the pollination process of blossoming plants [102]. The transport of flower pollen is referred to as flower pollination. Birds, bats, insects, and other animals are the principal actors in this transfer. Some flowers and insects participate in what is known as a pollinator relationship. These blooms can attract only the birds involved in this cooperation. These insects are regarded as the primary flower pollinators.
FPA takes into account four separate rules for flower constancy, pollination behaviour, and the pollination process [103].
-
Biotic pollination is cross-pollination in which the pollinator transports pollen. This is a global pollination process, and the pollinator movement complies with the Lévy flights.
-
Abiotic or self-pollination is the process of a plant or flower reproducing itself without the aid of a pollinator. Because the pollen transfer distance is typically less than that of biotic pollination, this procedure is known as local pollination.
-
Pollinators can acquire flower stability, favouring particular blooms. The flower constant is a mathematical expression for the likelihood of reproduction. The likelihood increases in direct proportion to how similar the related flowers are.
-
To manage the sort of pollination, \(p\in [0, 1]\) has the potential to be a key. These guidelines permit the use of both local and global search strategies. The greatest solutions are discovered nearby by using local search. Additionally, global pollination effectively prevents the problem from becoming trapped in a local optimum solution.
These rules must be used to construct updated equations. For instance, pollinators like insects transfer flower pollen gametes during the global pollination stage. Pollen can travel huge distances because insects can frequently fly and cover a bigger area. (2) can therefore be used to represent Rule 1 and flower constancy numerically (Rule 3).
Here, the solution vector \(X_i\) for the pollen i or t iteration is \(X_i\), and the best solution in the current generation or iteration is \(g_*\). Here, the scaling factor \(\gamma\) is used to regulate the step size.
The Lévy flight step size parameter is \(L(\lambda )\). Insect migration can be depicted using the Lévy distribution as they travel great distances. The mathematical expression used by Lévy is presented in (3).
The usual gamma function is \(Gamma(\lambda )\) in this instance, and the step size is s. This distribution holds true for significant steps \(s>0\). Although in theory \(s_0>>0\) must exist, in practice, \(s_0\) can be as low as 0.1. Rule 2 and Rule 3 are illustrated for local pollination in (4).
In (4), the pollen type \(x_j^{(t)}\), \(x_k^{(t)}\) comes from various flowers of the same kind of plant. The Lévy distribution is used to search for several solution points throughout the search space, which is the algorithm’s most crucial property for optimization. The optimization logic of the algorithm consists of locating the solution points at a great distance using the biotic pollination model and examining the area around the solution points using the abiotic pollination model, just like in flowers.
3.2.3 Whale optimization algorithm
Mirjalili and Lewis proposed the WOA for optimization problems, drawing inspiration from humpback whales’ hunting strategies [104]. Only humpback whales have been observed bubble-net feeding as a method of foraging. When hunting, whales surround their prey by creating bubbles that travel in a circle.
Encircling Prey: Humpback whales can find and encircle their prey when hunting. Since the location of the optimal design in the search space is unknown in advance, the WOA algorithm assumes that the target prey or a state close to it is the best candidate solution that is now accessible [105]. Other search agents will attempt to move closer to the best search agent once it has been determined who the top search agent is. (5) and (6) illustrate the mathematical model of humpback whales’ prey flanking behaviour. In (5) and (6), \(\overrightarrow{X}(t)\) reflects the agent’s position, t represents the iteration and \(\overrightarrow{X^*}\) represents the optimal solution. In (7) and (8), \(\overrightarrow{A}\) and \(\overrightarrow{C}\) indicate convergence values. The random number is \(\overrightarrow{r}[0,1]\), and \(\overrightarrow{a}\) stands for the vector that decreases linearly from 2 to 0 during an iteration.
Bubble-Net Attacking Method: The bubble-net attacking method of humpback whales consists of shrinking the search environment and spiralling towards the prey while moving towards its prey. By decreasing the \(\overrightarrow{a}\) value in (8), whales reduce their search environment and exhibit prey-catching behaviours. Since the value of \(\overrightarrow{A}\) also depends on the value of \(\overrightarrow{a}\), it decreases linearly from 2 to zero. The mathematical model of the spiral shape formed by humpback whales while catching their prey is given in (9) and (10).
The distance between the whale and its preferred prey, \(D'\), is determined in (9) and (10). b is the logarithmic spiral constant, and l is a random value between \([-1,1]\). Humpback whales have a 50% chance of selecting either a spiralling or a narrowing motion pattern when travelling in the direction of their prey. In (11), the p parameter is a random number between [0, 1].
3.2.4 Harris hawk optimization algorithm
In order to solve optimization difficulties, Heidari et al. proposed the usage of HHO, which was modelled after the foraging strategy of Harris hawks [106]. Hawks complete multiple stages of cooperative foraging via tracking, flanking, and attacking [107].
Exploration Phase: Harris hawks conduct the reconnaissance phase by keeping a close eye on large trees or telegraph poles in search of their prey. The search behaviour is regarded as the global discovery phase in the HHO method. (12) provides a mathematical expression for global exploration tactics.
The Transition from Exploration to Exploitation: The Harris hawk’s present position is indicated by \(X_i^{t}\), the position vector in each iteration is indicated by \(X_i^{t+1}\), and the position vector of the prey is indicated by \(X_{rabbit}(t)\). \(r_1\), \(r_2\), \(r_3\), \(r_4\), and q are random integers and take values between 0 and 1. The population’s upper and lower bounds are denoted by ub and lb, respectively. While X meant provides the average position values of the current population of hawks, \(X_{rand}^t\) represents a randomly chosen hawk from that population. (13) is used in t iterations with N hawks to find the average location value.
Exploitation Stage: When the Harris hawks locate their prey, they surround it in a circle. Hawks base their attack strategy on the way their prey behaves. Four potential strategies are suggested to represent the attack phase, each based on the prey’s tendency to flee and Harris hawks’ pursuit tactics. Strategies depend on the prey’s energy (E) for fleeing and the random number (r). To determine if the prey may escape the encirclement ring, apply the formula r(0, 1).
When r and E are greater and equal to 0.5, a soft siege approach is used. Hawks adopt a soft siege technique because their prey has enough energy to break free of the siege ring but no possibility of doing so. (14) represents the mathematical representation of it. The vector distance between the available prey and the population is represented by the value of \(\Delta x^t\). J stands for the length of the prey’s jump during the escape, and \(r_5 (0,1)\) is an equally distributed random value.
Hard siege tactic occurs when \(r\ge 0.5\) and E\(<0.5\). The prey’s energy is also insufficient because it cannot escape. Hawks hunt with a hard siege in that situation.
When \(r<0.5\) and E \(\ge 0.5\), a soft siege tactic with quick attacks is used. In this situation, the prey can escape the siege ring since it has the requisite energy. As a result, hawks will create a more intelligent and tactful siege ring to capture the prey. There are two steps in this method. The second step updates the hawks’ position if the first step does not get them closer to their prey. In the first step, the position equation (15) found in the soft siege strategy is used. (16) models the second step, which is the update mode. It is a \(s\in \mathbb {R}^{dim}\)-dimensional random vector. (17) defines the Lévy function. In this instance, u is a random number between v(0, 1) and \(\beta\) is 1.5.
When both E and r are less than 0.5, a hard siege tactic with swift attacks are used. Because it lacks the requisite energy, the prey in this situation cannot escape the siege ring. So, hawks catch their prey in a tough siege ring and then kill it. (18) represents its mathematical modelling.
3.3 Machine learning algorithms
3.3.1 Logistic regression
LR is one of the statistical methods of supervised classification algorithms. This algorithm has recently gained importance and is being used more and more. LR is used to classify data based on a logistic function that allows multivariate analysis and models of the binary dependent variable. The dependent variable Y is drawn from a binomial distribution. By using input features X \((x_1, x_2, x_3,\ldots , x_n)\), LR calculates the conditional probability \(P(Y=1\vert X)\) or \(P(Y=0\vert X)\) to predict Y.
3.3.2 Support vector machines
SVM is an optimal classification algorithm for linear and nonlinear data. Proposed for binary and linear problems, SVM has become applicable to both nonlinear and multi-class problems. For nonlinear data, the algorithm uses kernel functions, namely linear, nonlinear, polynomial, radial basis function (RBF), and sigmoid, which provide the nonlinear mapping. Thus, original data are transformed into high-dimensional space, and in this new dimension, an optimal hyperline is found to separate data of one class from others [108]. The data that enable the hyperline to be found are called support vectors. If multiple classes are available, one-versus-one (OVO) and one-versus-rest (OVR) are used to classify these data. The graphical illustration of SVM is given in Fig. 2.
The graphical illustration of SVM [109]
3.3.3 Gaussian Naive Bayes
Bayesian classifier, a statistical classifier, uses Bayes’ theorem to make a probability-based prediction. This algorithm can predict probabilities of class membership, e.g. the probability that new unseen data belong to a predefined class. Bayesian classifiers are characterized by high accuracy and speed when applied to large databases.
Unlike Bayesian classifiers, Naive Bayes (NB) makes a simplistic assumption and asserts that the attributes are conditionally independent (i.e. there is no dependency relationship between the attributes). However, while NB works for discrete and multinomial values, it cannot classify over continuous values. The continuous values associated with each class are drawn from the Gaussian distribution when studying continuous data. Given that an X \((x_1, x_2, x_3,\ldots , x_n)\) and \(Y_i, (i=1,2,\ldots ,m)\) represent features of data sample and classes separately, \(P(X\mid Y_i)\) is calculated by using (19).
In (19), while \(\mu\) represents the mean , \(\sigma\) represents the standard deviation of the \(i^{th}\) class.
The graphical illustration of GNB is given in Fig. 3.
The graphical illustration of GNB [110]
3.3.4 Random forest
RF classification is a popular machine learning method for developing predictive models in many research areas. RFs are a collection of classification and regression trees that use binary splits on features to make predictions. Decision trees are easy to use in practice, but the decision tree’s accuracy could be higher for large datasets. In the RF, many classification and regression trees are created with randomly selected training datasets and random subsets of features to create models. The results of multiple weak decision trees are combined to make accurate predictions. Consequently, an RF often provides higher accuracy compared with decision trees.
In RF, generalization error is represented in terms of the strength of each random tree and the correlation between them [111]. For the kth tree in a random forest, a random vector \(\theta _k\) is produced. The independence rule requires that the random vector \(\theta _k\) be independent of earlier random vectors \(\theta _1,\ldots ,\theta _{k-1}\), yet with the same distribution as those earlier random vectors. To combine tree classifiers \(h_1(x), h_2(x),\ldots , h_k{x}\) the margin function is described as follows:
In (20), \(\overline{x}_k\) represents the mean value, and I is the indicator function. The generalization error based on the margin function is given with (21).
X is the training data, and Y represents the class labels. The upper bound for generalization error is given in (22).
In (22), \(\overline{p}\) is the mean correlation between classifiers, and s is the strength of the ensemble. Therefore, the strength of each classifier inside the forest and the correlation between the random trees affect generalization error. The generalization error reaches a limit as the number of trees grows.
The graphical illustration of RF is given in Fig. 4.
The graphical illustration of RF [112]
3.3.5 K-nearest neighbour
KNN is one of the most preferred prediction algorithms, which saves the training data and only constructs the model once new unseen data need to be classified. In this algorithm, data are represented as points in Euclidean space. The nearest neighbour is defined in terms of the Euclidean distance. The target function can be discrete or real-valued. For discrete values, KNN assigns the class value most common in the K learning example, closest to the new data among the training examples.
The Euclidean distance between two data, say, \(X_1 = (x_{11}, x_{12},\ldots , x_{1n})\) and \(X_2 = (x_{21}, x_{22},\ldots , x_{2n})\), is calculated based on (23).
The most important problem with this algorithm is determining K, the number of neighbours. One of the methods to be used to determine K is the elbow function. The elbow function represents the cost function arising from different facets of K. However, the improvement in error rate decreases as K increases. The value of K at which the improvement in the biased values becomes small and reaches the maximum is called the elbow, and at this value, one should stop examining the data further [113].
The graphical illustration of KNN for K=3 and K=5 is given in Fig. 5.
The graphical illustration of KNN [114]
4 Experimental study
In this section, the Cleveland dataset and heart failure are used to evaluate the effects of the CS, FPA, WOA, and HOA algorithms on the classification accuracy of machine learning algorithms.
4.1 Experimental setup and evaluation metrics
All the experiments are performed on Google Colab on a system configuration; GPU Tesla k80 with 12 GB of GDDR5 VRAM, and Intel Xeon Processor with two 2.20-GHz cores and 13 GB RAM. We use SklearnFootnote 1 library, none of the libraries on the Python platform, to develop machine learning algorithms. To realize experiments, the dataset split for each algorithm and model parameters should be determined separately at first. For KNN, the K value is only the parameter that should be determined. It is not a correct approach to randomly specify this parameter. Based on the elbow method, the number of nearest neighbours is set to K = 3 for both datasets. For LR, we employ the liblinear, which provides optimization by using the coordinate descent method. In addition, we use an L2 penalty to prevent overfitting and set the C parameter equal to 1.0. C-support vector classification (SVC), an SVM implementation in libsvm library with RBF kernel, is used. Regularization parameter C is selected as 1. The smoothing value for GNB is used as \(1\textrm{e}{-}9\). The number of estimators is selected as 100, and to measure the information gain Gini index is used for RF. CS, FPA, WOA, and HHO are used to select a subset of features.
The algorithms are compared based on F-score and validated within the dataset as 80% train set and 20% test set. Where true positive (TP) is the number of truly classified heart disease/survival patients, false positive (FP) shows the number of non-patients/survival predicted as heart disease/death patients. False negative (FN) is the incorrect classification of heart disease/death patients as non-patients/survival. True negative (TN) is the number of correctly classified non-patients/survival. The formulas of performance metrics in terms of TP, FP, FN, and TN are given with (24)–(26).
In addition to the F-score, the area under the receiver operating characteristic (ROC) curve (AUC) value is used to evaluate the performance of each model. The AUC provides an overall assessment of performance across all potential classification criteria. The AUC value is between 0 and 1. If the value is close to 1, the model has a strong capacity for classification.
4.2 Experimental results
The main motivation behind the feature selection step is to select a subset of features that best represent the dataset. In order to analyse the effect of population size, we set the population size as 10, 30, 60, and 100 and the iteration size as 1000. Due to random conditions in the mete-heuristic algorithms, the algorithms are repeated 20 times to verify the test function. Tables 3 and 4 represent the fitness values, including the minimum (min), which is the best; maximum (max), which is the worst; and standard deviation (std), average (avg), and selected features for the best for Cleveland dataset and heart failure dataset, respectively. Feature selection is realized to understand the efficiency of the meta-heuristic algorithms on classification problems.
According to Tables 3 and 4, it can be seen that the applied meta-heuristic algorithms could effectively select fewer features.
When the results of the Cleveland dataset are examined, the following conclusions are reached. CS selects 40% of the initial feature set (8 out of 20) for population sizes 10 and 60 and selects 45% of the initial feature set (9 out of 20) for population sizes 30 and 100. The best fitness value of 0.0190 is obtained with a population size of 60. The worst fitness value of 0.0357 is obtained with a population size of 10. It has been seen that from the initial feature set FPA selects 30% (6 out of 20), 35% (7 out of 20), 40% (8 out of 20), and 45% (9 out of 20) for the population sizes 10, 30, 60, and 100, respectively. The best fitness value of 0.0038 is obtained with a population size of 60. The worst fitness value of 0.0524 is obtained with a population size of 10. WOA selects 20% (4 out of 20) for the population size of 10, 25% (5 out of 20) for the population sizes of 30, and 60 and 40% (8 out of 20) for the population size of 100. The best fitness value of 0.0362 is obtained with a population size of 100. The worst fitness value of 0.0668 is obtained with a population size of 10. HHO selects 75% (15 out of 20), 25% (5 out of 20), 30% (6 out of 20), and 40% (8 out of 20) features from the initial feature set for the population sizes of 10, 30, 60, and 100, respectively, while HHO achieves the best fitness value of 0.0362 with a population size of 100, and the worst fitness value of 0.0514 with a population size of 30.
Heart failure dataset results can be summarized as follows. CS selects on average 33.33% (4 out of 12) for population size 10, selects on average 58.33% (7 out of 12) for population size 30, and for population sizes 60 and 100 algorithm selects 25% (3 out of 12) of the initial feature set. The best fitness value of 0.0245 is obtained with a population size of 60. The worst fitness value of 0.0684 is obtained with a population size of 100. FPA selects on average 41.67% (5 out of 12) for the population sizes 10, 60, and 100 and selects on average 33.33% (4 out of 12) for population size 30. The best fitness value of 0.0114 is obtained with a population size of 10. The worst fitness value of 0.0481 is obtained with a population size of 60. WOA selects on average 16.67% (2 out of 12) for the population size of 10 and 100, on average 33.33% (4 out of 12) for the population size of 30 and 25% (3 out of 12) for the population size of 60. The best fitness value of 0.1573 is obtained with a population size of 100. The worst fitness value of 0.181 is obtained with a population size of 30. HHO selects, on average, 16.67% (2 out of 12) for population sizes 10 and 60, on average 8.33% (1 out of 12) for population size 30, and the average of 41.67% (5 out of 12) 100. HHO achieves the best fitness value of 0.0236 with a population size of 10 and the worst fitness value of 0.0668 with a population size of 30.
Table 5 and Table 6 compare the F-score of LR, SVM, MNB, RF, and KNN classifiers based on the meta-heuristic algorithms under the same conditions. The KNN algorithm outperforms the LR, SVM, MNB, and RF on both datasets by providing F-score between 92.06% and 99.72%. As we can see, all meta-heuristic algorithms significantly decreased the number of features and enhanced the KNN classifier’s predictive capability. The overall classification performance remained close when comparing the original and feature-selected datasets for the remaining classifiers.
In order to measure the effect of feature selection on classification in detail, it is necessary to compare the feature selection-based results with the results obtained from the original datasets. When the results for the Cleveland dataset are examined, LR, SVM, MNB and RF show both improvement and worsening in performance. At the same time, KNN achieves an increase for each population size of each meta-heuristic algorithm. The highest increase is 11.72% obtained with KNN with FPA for population size 60. The algorithm shows an F-score of 99.72% in this case. From here, the best representative features of the Cleveland dataset are age, fbs, exang, oldpeak, ca, cp_1, thal_2, slope_0. The results for the heart failure dataset show that LR, MNB, and RF show both improvements and worsens in performance. At the same time, SVM and KNN achieve an increase for each population size of each meta-heuristic algorithm. The highest increase is 37.45% obtained with KNN with HHO for population size 10. The algorithm shows an F-score of 97.45% in this case. The best representative features of the heart failure dataset are age and time.
Performance comparisons by AUC value are given in Tables 7 and 8. When the results are evaluated according to AUC, it is clearly seen that feature selection improves the performance of algorithms. It is observed that the AUC value, which is 0.85 in the Cleveland dataset, increased up to 0.98 due to feature selection. In the heart failure dataset, it is observed that the AUC value obtained as 0.55 increased up to 0.98.
The comparison of the F-score of the applied models with alternatives in the literature is given in Tables 9 and 10 for Cleveland and heart failure datasets, respectively.
5 Conclusions
Medicine researchers regard prediction as critical for possible heart disease patients. It is challenging to choose the best representative features for medical research. The Cleveland and heart failure datasets are used to select features using CS, FPA, WOA, and HHO. Furthermore, the relevant characteristics obtained are fed into several classifiers for classification, including KNN, LR, SVM, GNB, and RF. The best features are identified using an optimization technique, improving the appropriate classifier’s accuracy. For the Cleveland dataset, KNN demonstrates superior prediction performance on FPA-selected features for the population size of 60. In the Cleveland dataset, the F-score for heart disease prediction is 99.72%. Also, for the heart failure dataset, KNN demonstrates superior prediction performance on HHO-selected features for the population size of 10. In the heart failure dataset, the F-score for survival prediction is 97.45%.
Future work is analysing various forms of heart disease datasets, such as heart sound signals and electrocardiogram (ECG) signals, to evaluate the strength of the applied algorithms.
Data availability
The datasets used in the study are public.
References
Vahini B, Sanjeev S, Narenthiran CK, Chandrasekar K (2021) A review on rheumatic heart disease. Curr Asp Pharm Res Dev 5:33–42
Javeed A, Khan SU, Ali L, Ali S, Imrana Y, Rahman A (2022) Machine learning-based automated diagnostic systems developed for heart failure prediction using different types of data modalities: a systematic review and future directions. Comput Math Methods Med 2022:9288452
Chicco D, Jurman G (2020) Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Medical Inform Decis Mak 20(1):1–16
Ekinci E (2021) Determination of biomarkers in the diagnosis of breast cancer using data mining. In: International Congress on Scientific Advances (ICONSAD’21), pp 956–958
Golagani PP, Mahalakshmi TS, Beebi SK (2021) Supervised learning breast cancer data set analysis in MATLAB using novel SVM classifier. In: Machine Intelligence and Soft Computing. Springer, pp 255–263
Shrivastava D, Sanyal S, Maji AK, Kandar D (2020) Bone cancer detection using machine learning techniques. In: Smart Healthcare for Disease Diagnosis and Prevention. Elsevier, pp 175–183
Prakash PN, Rajkumar N (2022) HSVNN: an efficient medical data classification using dimensionality reduction combined with hybrid support vector neural network. J Supercomput 78:15439–15462
Omurca Sİ, Ekinci E, Çakmak B, Özkan SG (2019) Using machine learning approaches for prediction of the types of asthmatic allergy across the Turkey. Data Sci Appl 2(2):8–12
Chan J (2021) Classifying allergic rhinitis subjects and identifying single nucleotide polymorphisms using a support vector machine approach. In: The International Young Researchers’ Conference; Virtual, pp 1–8
Dixit A, Mani A, Bansal R (2021) CoV2-detect-net: design of COVID-19 prediction model based on hybrid DE-PSO with SVM using chest X-ray images. Inf Sci 571:676–692
Hu R, Gan J, Zhu X, Liu T, Shi X (2022) Multi-task multi-modality SVM for early COVID-19 diagnosis using chest CT data. Inf Process Manag 59(1):102782
Nemati M, Ansary J, Nemati N (2020) Machine-learning approaches in COVID-19 survival analysis and discharge-time likelihood prediction using clinical data. Patterns 1(5):100074
Jackins V, Vimal S, Kaliappan M, Lee MY (2021) Ai-based smart prediction of clinical disease using random forest classifier and Naive Bayes. J Supercomput 77(5):5198–5219
Priya KL, Kypa MSCR, Reddy MMS, Reddy GRM (2020) A novel approach to predict diabetes by using Naive Bayes classifier. In: 2020 4th International Conference on Trends in Electronics and Informatics (ICOEI) (48184). IEEE, pp 603–607
Abd NS, Abdullah DA (2021) Diagnose of chronic kidney diseases by using Naive Bayes algorithm. J Al-Qadisiyah Comput Sci Math 13(2):46
Almustafa KM (2021) Prediction of chronic kidney disease using different classification algorithms. Inform Med Unlocked 24:100631
Bai Q, Chunyan S, Tang W, Li Y (2022) Machine learning to predict end stage kidney disease in chronic kidney disease. Sci Rep 12(1):1–8
Kikano EG, Tirumani SH, Suh CH, Gan JM, Bomberger TT, Bui MT, Laukamp KR, Kim KW, Dowlati A, Ramaiya NH (2021) Trends in imaging utilization for small cell lung cancer: a decision tree analysis of the NCCN guidelines. Clin Imaging 75:83–89
Musa AA, Aliyu UM (2020) Application of machine learning techniques in predicting of breast cancer metastases using decision tree algorithm. JDMGP 11(1):1–5
Das AK, Biswas SK, Mandal A (2022) An expert system for breast cancer prediction (ESBCP) using decision tree. Indian J Sci Technol 15(45):2441–2450
Sathiyanarayanan P, Pavithra S, Saranya MS, Makeswari M (2019) Identification of breast cancer using the decision tree algorithm. In: 2019 IEEE International Conference on System, Computation, Automation and Networking (ICSCAN). IEEE, pp 1–6
Huang H, Yang ZH, Gu ZW, Luo M, Xu L (2022) Decision tree model for predicting the overall survival of patients with diffused large b-cell lymphoma in the central nervous system. World Neurosurg 166:e189–e198
Niu M, Liu B, Tao J, Li Q (2021) A time-frequency channel attention and vectorization network for automatic depression level prediction. Neurocomputing 450:208–218
Sadeghi D, Shoeibi A, Ghassemi N, Moridian P, Khadem A, Alizadehsani R, Teshnehlab M, Gorriz JM, Khozeimeh F, Zhang YD et al (2022) An overview of artificial intelligence techniques for diagnosis of schizophrenia based on magnetic resonance imaging modalities: methods, challenges, and future works. Comput Biol Med 146:105554
Zhang Y, Wang L (2021) Research on classification model of BP neural network based on dl algorithm. In: 2021 International Conference on Computer Network, Electronic and Automation (ICCNEA). IEEE, pp 16–20
Khoei TT, Labuhn MC, Caleb TD, Hu WC, Kaabouch N (2021) A stacking-based ensemble learning model with genetic algorithm for detecting early stages of Alzheimer’s disease. In: 2021 IEEE International Conference on Electro Information Technology (EIT). IEEE, pp 215–222
Yang Y, Wei L, Hu Y, Wu Y, Hu L, Nie S (2021) Classification of Parkinson’s disease based on multi-modal features and stacking ensemble learning. J Neurosci Methods 350:109019
Al Sayaydeha ON, Mohammad MF (2019) Diagnosis of the Parkinson disease using enhanced fuzzy min-max neural network and OneR attribute evaluation method. In: 2019 International Conference on Advanced Science and Engineering (ICOASE). IEEE, pp 64–69
Kourou K, Rigas G, Papaloukas C, Mitsis M, Fotiadis DI (2020) Cancer classification from time series microarray data through regulatory dynamic Bayesian networks. Comput Biol Med 116:103577
Zaharchuk G (2020) Fellow in a box: combining AI and domain knowledge with Bayesian networks for differential diagnosis in neuroimaging. Radiology 295(3):638
Li Y, Chen X, Wang Y, Hu J, Shen Z, Ding X (2020) Application of group lasso regression based Bayesian networks in risk factors exploration and disease prediction for acute kidney injury in hospitalized patients with hematologic malignancies. BMC Nephrol 21(1):1–11
Ershadi MM, Seifi A (2020) An efficient Bayesian network for differential diagnosis using experts’ knowledge. Int J Intell Comput Cybern 13(1):103–126
Dekker A, Hope A, Lambin P, Lindsay P (2021) Survival prediction with Bayesian networks in more than 6000 non-small cell lung cancer patients. medRxiv
Shipe ME, Deppen SA, Farjah F, Grogan EL (2019) Developing prediction models for clinical use using logistic regression: an overview. J Thorac Dis 11(Suppl 4):S574
Lee H-A, Rau H-H, Chao LR, Hsu C-Y (2020) Establishing a survival probability prediction model for different lung cancer therapies. J Supercomput 76(8):6501–6514
Hyde KK, Novack MN, LaHaye N, Parlett-Pelleriti C, Anden R, Dixon DR, Linstead E (2019) Applications of supervised machine learning in autism spectrum disorder research: a review. Rev J Autism Dev Disord 6(2):128–146
Nogay HS, Adeli H (2020) Machine learning (ML) for the diagnosis of autism spectrum disorder (ASD) using brain imaging. Rev Neurosci 31(8):825–841
Çil B, Ayyıldız H, Tuncer T (2020) Discrimination of \(\beta\)-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system. Med Hypotheses 138:109611
Sadiq S, Khalid MU, Ullah S, Aslam W, Mehmood A, Choi GS, On B-W et al (2021) Classification of \(\beta\)-thalassemia carriers from red blood cell indices using ensemble classifier. IEEE Access 9:45528–45538
Lee YW, Choi JW, Shin E-H (2021) Machine learning model for predicting malaria using clinical information. Comput Biol Med 129:104151
Gupta A, Sharma P (2021) A review of machine learning techniques being used for blood cancer detection. Ann Romanian Soc Cell Biol 25:7796–7811
Haider RZ, Ujjan IU, Khan NA, Urrechaga E, Shamsi TS (2022) Beyond the in-practice CBC: the research CBC parameters-driven machine learning predictive modeling for early differentiation among leukemias. Diagnostics 12(1):138
Piccialli F, Calabrò F, Crisci D, Cuomo S, Prezioso E, Mandile R, Troncone R, Greco L, Auricchio R (2021) Precision medicine and machine learning towards the prediction of the outcome of potential celiac disease. Sci Rep 11(1):1–10
Koh JE, De Michele S, Sudarshan VK, Jahmunah V, Ciaccio EJ, Ooi CP, Gururajan R, Gururajan R, Oh SL, Lewis SK et al (2021) Automated interpretation of biopsy images for the detection of celiac disease using a machine learning approach. Comput Methods Programs Biomed 203:106010
Chaubey G, Bisen D, Arjaria S, Yadav V (2021) Thyroid disease prediction using machine learning approaches. Natl Acad Sci Lett 44(3):233–238
Maliha SK, Ema RR, Ghosh SK, Ahmed H, Mollick MR, Islam T (2019) Cancer disease prediction using Naive Bayes, k-nearest neighbor and J48 algorithm. In: 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT). IEEE, pp 1–7
Reza MR, Hossain G, Goyal A, Tiwari S, Tripathi A, Bhan A, Dash P et al (2021) Automatic diabetes and liver disease diagnosis and prediction through SVM and KNN algorithms. In: Emerging Technologies in Data Mining and Information Security. Springer, pp 589–599
Kaplan K, Kaya Y, Kuncan M, Ertunç HM (2020) Brain tumor classification using modified local binary patterns (LBP) feature extraction methods. Med Hypotheses 139:109696
Atallah DM, Badawy M, El-Sayed A, Ghoneim MA (2019) Predicting kidney transplantation outcome based on hybrid feature selection and KNN classifier. Multimed Tools Appl 78(14):20383–20407
Kilicarslan S, Adem K, Celik M (2020) Diagnosis and classification of cancer using hybrid model based on ReliefF and convolutional neural network. Med Hypotheses 137:109577
Beheshti Z (2022) BMPA-TVsinV: a binary marine predators algorithm using time-varying sine and v-shaped transfer functions for wrapper-based feature selection. Knowl Based Syst 252:109446
Zhou J, Hua Z (2022) A correlation guided genetic algorithm and its application to feature selection. Appl Soft Comput 123:108964
Araújo LA, e Lopes IL, Oliveira RM, Silva SHG, e Silva CSJ, Gomide LR (2022) Simulated annealing in feature selection approach for modeling aboveground carbon stock at the transition between Brazilian Savanna and Atlantic Forest biomes. Ann For Res 65(1):47–63
Hashemi A, Joodaki M, Joodaki NZ, Dowlatshahi MB (2022) Ant colony optimization equipped with an ensemble of heuristics through multi-criteria decision making: a case study in ensemble feature selection. Appl Soft Comput 124:109046
Pan J-S, Liu N, Chu S-C (2022) A competitive mechanism based multi-objective differential evolution algorithm and its application in feature selection. Knowl-Based Syst 245:108582
Shanmugam S, Preethi J (2019) Improved feature selection and classification for rheumatoid arthritis disease using weighted decision tree approach (react). J Supercomput 75(8):5507–5519
Hanbay K (2022) A new standard error based artificial bee colony algorithm and its applications in feature selection. J King Saud Univ Comput Inf Sci 34(7):4554–4567
Xie W, Wang L, Kun Yu, Shi T, Li W (2023) Improved multi-layer binary firefly algorithm for optimizing feature selection and classification of microarray data. Biomed Signal Process Control 79:104080
Ajibade SS, Oyebode OJ, Mejarito CL, Gido NG, Dayupay J, Diaz RD (2022) Feature selection for student prediction accuracy using gravitational search algorithm. J Optoelectron Laser 41(8):2022
Deep K et al (2022) A random walk grey wolf optimizer based on dispersion factor for feature selection on chronic disease prediction. Expert Syst Appl 206:117864
Balakrishnan K, Dhanalakshmi R, Khaire UM (2021) Improved salp swarm algorithm based on the levy flight for feature selection. J Supercomput 77(11):12399–12419
Eskandari S, Seifaddini M (2022) Online and offline streaming feature selection methods with bat algorithm for redundancy analysis. Pattern Recognit 133:109007
Dhiman G, Oliva D, Kaur A, Singh KK, Vimal S, Sharma A, Cengiz K (2021) BEPO: a novel binary emperor penguin optimizer for automatic feature selection. Knowl-Based Syst 211:106560
Varzaneh ZA, Hossein S, Mood SE, Javidi MM (2022) A new hybrid feature selection based on improved equilibrium optimization. Chemom Intell Lab Syst 228:104618
Too J, Rahim Abdullah A (2020) Binary atom search optimisation approaches for feature selection. Connect Sci 32(4):406–430
Too J, Mirjalili S (2021) A hyper learning binary dragonfly algorithm for feature selection: a COVID-19 case study. Knowl Based Syst 212:106553
Long W, Xu M, Jiao J, Wu T, Tang M, Cai S (2022) A velocity-based butterfly optimization algorithm for high-dimensional optimization and feature selection. Expert Syst Appl 201:117217
Eluri RK, Devarakonda N (2022) Binary golden eagle optimizer with time-varying flight length for feature selection. Knowl Based Syst 247:108771
Arumugam K, Ramasamy S, Subramani D (2022) Binary duck travel optimization algorithm for feature selection in breast cancer dataset problem. In: IOT with Smart Systems. Springer, pp 157–167
Uci (2009) Heart disease dataset, 1988. https://archive.ics.uci.edu/ml/datasets/heart+disease
Heart failure clinical records data set (2017) https://archive.ics.uci.edu/ml/datasets/Heart+failure+clinical+records
Latha CBC, Jeeva SC (2019) Improving the accuracy of prediction of heart disease risk based on ensemble classification techniques. Inform Med Unlocked 16:100203
Reddy GT, Reddy MP, Lakshmanna K, Rajput DS, Kaluri R, Srivastava G (2020) Hybrid genetic algorithm and a fuzzy logic classifier for heart disease diagnosis. Evol Intell 13(2):185–196
Kolukısa B, Hacılar H, Kuş M, Bakır-Güngör B, Aral A, Güngör V (2019) Diagnosis of coronary heart disease via classification algorithms and a new feature selection methodology. Int J Data Min Model Manag 1(1):8–15
Li JP, Haq AU, Din SU, Khan J, Khan A, Saboor A (2020) Heart disease identification method using machine learning classification in e-healthcare. IEEE Access 8:107562–107582
Gupta A, Kumar L, Jain R, Nagrath P (2020) Heart disease prediction using classification (Naive Bayes). In: Proceedings of First International Conference on Computing, Communications, and Cyber-Security (IC4S 2019). Springer, pp 561–573
Gárate-Escamila AK, El Hassani AH, Andrès E (2020) Classification models for heart disease prediction using feature selection and PCA. Inform Med Unlocked 19:100330
Tougui I, Jilbab A, El Mhamdi J (2020) Heart disease classification using data mining tools and machine learning techniques. Health Technol 10(5):1137–1144
Kondababu A, Siddhartha V, Bhagath Kumar BHK, Penumutchi B (2021) A comparative study on machine learning based heart disease prediction. Mater Today
Rani P, Kumar R, Ahmed NM, Jain A (2021) A decision support system for heart disease prediction based upon machine learning. J Reliab Intell Environ 7(3):263–275
Deepika D, Balaji N (2022) Effective heart disease prediction using novel MLP-EBMDA approach. Biomed Signal Process Control 72:103318
Srinivas P, Katarya R (2022) hyOPTXg: OPTUNA hyper-parameter optimization framework for predicting cardiovascular disease using XGBoost. Biomed Signal Process Control 73:103456
Budholiya K, Shrivastava SK, Sharma V (2020) An optimized XGBoost based diagnostic system for effective prediction of heart disease. J King Saud Univ Comput Inf Sci 34:4514–4523
Lutimath NM, Ramachandra HV, Raghav S, Sharma N (2022) Prediction of heart disease using genetic algorithm. In: Proceedings of Second Doctoral Symposium on Computational Intelligence. Springer, pp 49–58
Gnoguem C, Degila J, Bondiombouy C (2022) Predicting heart disease with multiple classifiers. In: Intelligent Vision in Healthcare. Springer, pp 59–74
Mohapatra D, Bhoi SK, Mallick C, Jena KK, Mishra S (2022) Distribution preserving train-test split directed ensemble classifier for heart disease prediction. Int J Inf Technol 14:1763–769
Anderies A, Tchin JA, Putro PH, Darmawan YP, Gunawan AA (2022) Prediction of heart disease UCI dataset using machine learning algorithms. Eng Math Comput Sci J 4(3):87–93
Shaw SK, Patidar S (2023) Heart disease diagnosis using machine learning classification techniques. In: Inventive Communication and Computational Technologies. Springer, pp 445–460
Goyal S (2023) Predicting the heart disease using machine learning techniques. In: ICT Analysis and Applications. Springer, pp 191–199
Oladimeji OO, Oladimeji O (2020) Predicting survival of heart failure patients using classification algorithms. JITCE 4(02):90–94
Moreno-Sanchez PA (2020) Development of an explainable prediction model of heart failure survival by using ensemble trees. In: 2020 IEEE International Conference on Big Data (Big Data). IEEE, pp 4902–4910
Moreno-Sanchez PA (2021) Improvement of a prediction model for heart failure survival through explainable artificial intelligence. arXiv preprint arXiv:2108.10717
Aloss A, Sahu B, Deeb H, Mishra D (2022) A crow search algorithm-based machine learning model for heart disease and cervical cancer diagnosis. In: Electronic Systems and Intelligent Computing. Springer, pp 303–311
Swetha AM, Santhi B, Brindha GR (2022) Machine learning and deep learning for medical analysis—a case study on heart disease data, chapter 8. Wiley, pp 177–209. ISBN 9781119821908. https://doi.org/10.1002/9781119821908.ch8
Kameswara Rao B, Prasan UD, Jagannadha Rao M, Pedada R, Kumar PS et al (2022) Identification of heart failure in early stages using smote-integrated adaboost framework. In: Computational Intelligence in Data Mining. Springer, pp 537–552
Özbay Karakuş M, Er O (2022) A comparative study on prediction of survival event of heart failure patients using machine learning algorithms. Neural Computing and Applications, pp 1–14
Guerrero-Luis M, Valdez F, Castillo O (2021) A review on the cuckoo search algorithm. In: Fuzzy Logic Hybrid Extensions of Neural and Optimization Algorithms: Theory and Applications, pp 113–124
Garip Z, Karayel D, Erhan Çimen M (2022) A study on path planning optimization of mobile robots based on hybrid algorithm. Concurr Comput Pract Exp 34(5):e6721
Ali W, Khan MS, Hasan M, Khan ME, Qyyum MA, Qamar MO, Lee M (2021) Introduction to cuckoo search and its paradigms: a bibliographic survey and recommendations. In: AI and Machine Learning Paradigms for Health Monitoring System. Springer, pp 79–93
Zhang Z (2021) Speech feature selection and emotion recognition based on weighted binary cuckoo search. Alex Eng J 60(1):1499–1507
Pan J-S, Song P-C, Chu S-C, Peng Y-J (2020) Improved compact cuckoo search algorithm applied to location of drone logistics hub. Mathematics 8(3):333
Mahendran G, Govindaraju C (2020) Flower pollination algorithm for distribution system phase balancing considering variable demand. Microprocess Microsyst 74:103008
Çimen ME, Garip ZB, Boz AF (2021) Chaotic flower pollination algorithm based optimal PID controller design for a buck converter. Analog Integr Circuits Signal Process 107(2):281–298
Li X, Gao L, Cao H, Sun Y, Jiang M, Zhang Y (2022) A temperature compensation method for aSix-Axis force/torque sensor utilizing ensemble hWOA-LSSVM based on improved trimmed bagging. Sensors 22(13):4809
Garip Z, Çimen ME, Karayel D, Boz AF (2019) The chaos-based whale optimization algorithms global optimization. Chaos Theory Appl 1(1):51–63
Heidari AA, Mirjalili S, Faris H, Aljarah I, Mafarja M, Chen H (2019) Harris hawks optimization: algorithm and applications. Future Gener Comput Syst 97:849–872
Garip Z, Çimen ME, Boz AF (2021) Harris şahinleri ve balina optimizasyon algoritmalarının kısıt işleme teknikleriyle uygulaması: Karşılaştırmalı bir çalışma. JISTA 4(2):76–85
Yiğit H, Köylü H, Eken S (2022) Estimation of road surface type from brake pressure pulses of abs. Expert Syst Appl 212:118726. https://doi.org/10.1016/j.eswa.2022.118726
Oyebode O, Ighravwe DE (2019) Urban water demand forecasting: a comparative evaluation of conventional and soft computing techniques. Resources 8(3):156
Dixit M, Sharma R, Shaikh S, Muley K (2019) Internet traffic detection using Naïve Bayes and k-nearest neighbors (KNN) algorithm. In: 2019 International Conference on Intelligent Computing and Control Systems (ICCS). IEEE, pp 1153–1157
Khan Z, Gul A, Perperoglou A, Miftahuddin M, Mahmoud O, Adler W, Lausen B (2020) Ensemble of optimal trees, random forest and random projection ensemble classification. Adv Data Anal Classif 14:97–116
Khan MY, Qayoom A, Nizami MS, Siddiqui MS, Wasi S, Raazi SM (2021) Automated predicti on of Good Dictionary EXamples (GDEX): A comprehensive experiment with distant supervision, machine learning, and word embedding-based deep learning techniques. Complexity 2021:1–8
Gupta G, Adarsh U, Reddy NVS, Rao BA (2022) Comparison of various machine learning approaches uses in heart ailments prediction. In: Journal of Physics: Conference Series, vol 161. IOP Publishing, p 012010
Uddin S, Haque I, Lu H, Moni MA, Gide E (2022) Comparative performance analysis of k-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci Rep 12(1):1–11
Patro SP, Padhy N, Sah RD (2022) An improved ensemble learning approach for the prediction of cardiovascular disease using majority voting prediction. Int J Model Identif Control 41(1–2):68–86
Ambrish G, Ganesh B, Ganesh A, Srinivas C, Mensinkal K et al (2022) Logistic regression technique for prediction of cardiovascular disease. In: Global Transitions Proceedings
Zhenya Q, Zhang Z (2021) A hybrid cost-sensitive ensemble for heart disease prediction. BMC Med Inform Decis Mak 21(1):1–18
El-Shafiey MG, Hagag A, El-Dahshan EA, Ismail MA (2022) A hybrid GA and PSO optimized approach for heart-disease prediction based on random forest. Multimed Tools Appl 81(13):18155–18179
Hera SY, Amjad M, Saba MK (2022) Improving heart disease prediction using multi-tier ensemble model. Netw Model Anal Health Inform Bioinform 11(1):1–13
Ishaq A, Sadiq S, Umer M, Ullah S, Mirjalili S, Rupapara V, Nappi M (2021) Improving the prediction of heart failure patients’ survival using smote and effective data mining techniques. IEEE Access 9:39707–39716
Muntasir Nishat M, Faisal F, Jahan Ratul I, Al-Monsur A, Ar-Rafi AM, Nasrullah SM, Reza MT, Khan MR (2022) A comprehensive investigation of the performances of different machine learning classifiers with SMOTE-ENN oversampling technique and hyperparameter optimization for imbalanced heart failure dataset. Sci Programm 2022:1–17
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ŞA provided software and contributed to data curation. EE was involved in supervision, conceptualization, methodology, and writing. ZG was involved in supervision, conceptualization, methodology, and reviewing and editing. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ay, Ş., Ekinci, E. & Garip, Z. A comparative analysis of meta-heuristic optimization algorithms for feature selection on ML-based classification of heart-related diseases. J Supercomput 79, 11797–11826 (2023). https://doi.org/10.1007/s11227-023-05132-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-023-05132-3