Abstract
Neuroendocrine neoplasms (NENs) comprise a highly heterogeneous group of tumors arising from the diffuse neuroendocrine system. NENs mainly originate in gastrointestinal, pancreatic, and pulmonary tissues, and despite being rare, show rising incidence. The molecular mechanisms underlying NEN development are still poorly understood, although recent studies are unveiling their genomic, epigenomic and transcriptomic landscapes. RNA was originally considered as an intermediary between DNA and protein. Today, compelling evidence underscores the regulatory relevance of RNA processing, while new RNA molecules emerge with key functional roles in core cell processes. Indeed, correct functioning of the interrelated complementary processes comprising RNA biology, its processing, transport, and surveillance, is essential to ensure adequate cell homeostasis, and its misfunction is related to cancer at multiple levels. This review is focused on the dysregulation of RNA biology in NENs. In particular, we survey alterations in the splicing process and available information implicating the main RNA species and processes in NENs pathology, including their role as biomarkers, and their functionality and targetability. Understanding how NENs precisely (mis)behave requires a profound knowledge at every layer of their heterogeneity, to help improve NEN management. RNA biology provides a wide spectrum of previously unexplored processes and molecules that open new avenues for NEN detection, classification and treatment. The current molecular biology era is rapidly evolving to facilitate a detailed comprehension of cancer biology and is enabling the arrival of personalized, predictive and precision medicine to rare tumors like NENs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Neuroendocrine neoplasms (NENs) are rare tumors that arise from cells of the diffuse neuroendocrine system, and their incidence is rising in the last decades [1, 2]. In the USA, the incidence rate went from 1.09 per 100,000 people in 1973 to 6.98 per 100,000 in 2012, probably due to improved diagnostic methods, which enable a detection in earlier stages [3]. Also, the new attempts to classify NENs according to their grade and location have facilitated clinicians to improve their diagnoses. The most recent of these classifications was proposed by the International Agency for Research on Cancer (IARC) and World Health Organization (WHO), classifying NENs in different grades (G1 to G3) based on the Ki-67 index and the mitotic rate [4]. NENs may also be classified, according to their histological differentiation, into well differentiated neuroendocrine tumors (NETs) and poorly-differentiated neuroendocrine carcinomas (NECs) [4].
NENs may be detected earlier if they are accompanied by any hormone overproduction due to their neuroendocrine nature, which can give rise to hormonal or metabolic syndromes or clinical conditions. This is the case for the carcinoid syndrome, caused by serotonin and histamine hypersecretion in gastrointestinal and lung NENs (GI-NENs and LungNENs, respectively); or different conditions caused by pancreatic hormones, like insulin, somatostatin, or gastrin in pancreatic NENs (PanNENs) [5, 6]. However, NENs are often difficult to diagnose due to the lack of precise biomarkers, chromogranin A and synaptophysin being the most common neuroendocrine biomarkers, but with limited sensitivity, specificity, and prognosis capacity [7]. The only curative treatment for NENs is surgery, whenever possible. Current medical treatments include somatostatin analogs, mTOR inhibitors, antiangiogenic/tyrosine kinase inhibitors and the novel peptide receptor radionuclide therapy [8]. However, NENs treatment and clinical management are commonly complicated by their high heterogeneity, which is present at different layers (genetic, cellular, histopathological, anatomical, etc. [6]), and more diagnostic and therapeutic biomarkers must be explored to better understand these pathologies [9]. Among the molecular layers of NEN heterogeneity, the best studied is the genomic landscape [10, 11]. Indeed, along with the development of next generation sequencing, whole genomic studies have been deployed in different NEN types, which have enabled to refine the classification of various NENs according to their mutations and other genetic alterations. Specifically, mutations in certain genes have been observed with high frequency in NENs, like MEN1 or ATRX/DAXX in PanNENs, SMAD genes in SI-NENs, chromatin remodeling genes in pulmonary carcinoids or TP53 and RB1 for SCLC [12,13,14,15,16]. Nevertheless, an increasing number of publications have been recently focusing on transcriptomics in NENs, exploring not only the patterns of mRNA levels but exploring also different RNA molecules, which are commonly found to be dysregulated in tumors and cancer, in a reciprocal pathological interplay linked with genomic alterations [17].
In this review, we have focused on the dysregulations of RNA biology in the most common NEN types: PanNENs, GI-NENs, and small intestine NENs (SI-NENs), and the LungNENs, which comprise the well-differentiated NETs, i.e., typical (TC) and atypical carcinoids (AC), and the poorly-differentiated NECs, i.e., large-cell neuroendocrine carcinoma (LCNEC) and small cell lung cancer (SCLC). Other types of endocrine/neuroendocrine neoplasms like pheochromocytoma, paraganglioma, pituitary tumors, thyroid cancer, or Merkel cell carcinoma, have been typically considered separate entities from classic NENs and are not included in this review. Nevertheless, this topic has been insufficiently studied in those tumors, and thus, the knowledge generated about their molecular landscape remain vastly unexplored, and therefore, deserve further attention.
2 RNA biology and surveillance
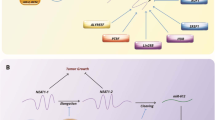
RNA was originally viewed as a passive intermediate between the coding information encased into the DNA, and its ultimate cellular effectors, proteins [18]. However, incessant research in the last decades fully overturned this simplistic concept, as many other RNA molecules have been discovered, which have their own function without necessarily been translated into proteins, while multiple regulatory roles were deciphered for known and new RNA-related molecules and processes. Indeed, in recent decades many different RNA molecules and their functions have been described, besides classic coding messenger RNAs (mRNAs), and non-coding ribosomal RNA (rRNA) and transfer RNA (tRNA). These novel RNA types are also non-coding RNAs (ncRNA), which have important roles in gene expression regulation [19], modifying gene expression and cell signaling through the interaction with mRNAs, and thereby regulating transcription, RNA processing or translation. Moreover, they are able to form fully functional complexes with proteins [20], and can be involved in many complex processes, such as splicing and processing other RNAs or even the maintenance of genome stability [21] (Fig. 1).
Schematic illustration summarizing the main players comprising the biology of RNA. The four major fields involved in the regulation of RNA function and metabolism are shown: Protein Synthesis; RNA Catalysis; Gene expression regulation; and RNA processing. mRNA: messenger RNA, tRNA: transfer RNA, piRNA: PIWI-Interacting RNA, rRNA: ribosomal RNA, TERC: telomerase RNA component, snRNA: small nuclear RNA, siRNA: small interference RNA, miRNA: microRNA, aRNA: antisense RNA, lncRNA: long non coding RNA, circRNA: circular RNA, snoRNA: small nucleolar RNA
The term RNA surveillance refers to a growing set of processes and mechanisms involved in ensuring the fidelity of RNA production and quality to RNA molecules. These mechanisms act at various steps of the RNA biogenesis pathway to detect and degrade transcripts that have not been properly processed [22]. RNA molecules are finely surveyed and refined at various steps and through different procedures, which include capping in 5’, splicing, 3’ end formation and nuclear export, while being in a kinetic competition with decay targeting machineries, like nonsense, nonstop or no-go mediated decays or RNA exosome [23]. In these macromolecular-guided processes, correct RNA-protein and protein-protein interactions need to be ensured and meticulous regulation is necessarily required [24]. In this sense, machineries like the exon-junction complex (EJC) interacts with RNA processing or decays complexes to ensure correct RNA maturation [25].
In this scenario, the discovery of many “new” RNA molecules, which could not be detected before, arose with the arrival of deep sequencing technologies [26]. This RNA biology field was soon directly connected with the study of disease development, and particularly with cancer. Indeed, an emerging number and variety of RNA molecules have been demonstrated to be dysregulated in tumor cells, conferring malignant properties related to core cell functions: proliferation, migration, metastasis, apoptosis, etc. [27, 28]. Transcriptomics studies uncovered the first dysregulations in RNA biology in tumoral pathologies and remain critically important and informative on RNA expression changes. In NENs, numerous transcriptomic studies have identified differentially expressed genes potentially related to tumorigenesis [29,30,31], while allowing to stratify NENs in new molecular subgroups, which may help to better understand their heterogeneity [32]. Moreover, efforts have been made to apply these discoveries to improve NEN diagnosis and prediction prognosis through specific tests such as the NETest [33], a gene expression assay developed to diagnose NENs based on PCR of blood-isolated RNA samples, which has evolved and is being tested to improve diagnosis and detect residual disease in NENs [34, 35]. However, much remains to be known about the different types of RNA species and RNA-related biological processes in NENs. Thus, we will summarize the available evidence and reflect on the future of RNA biology in these heterogeneous tumors.
2.1 Alternative splicing and NENs
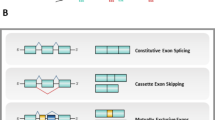
In eukaryotes, the generation of mature RNA molecules from their DNA templates requires a pivotal step known as splicing (Fig. 2), whereby non-required segments contained in the sequence of the pre-mature RNA, or introns, are removed, while the remaining segments, or introns, are bound together. This process of maturation enables RNA to perform its function and is carried out by the spliceosome, a macromolecular ribozyme complex comprised by a set of small nuclear RNAs (snRNAs) bound to their proteins forming a discrete core of ribonucleoproteins, in association to a numerous group of ancillary proteins which are known to interact with the splicing machinery during the process: the splicing factors. As previously reviewed, these splicing factors can also exert other biological functions related with RNA biology [36]. In fact, splicing is a highly complex process, as 95% of human genes undergo alternative splicing, which, from a single gene, can give rise to various, distinct mature RNAs and subsequently, in the case of mRNA, different proteins. As a result, alternative splicing increases the complexity and enhances the versatility of the transcriptome and the proteome of each cell without increasing the size of the genome, as described in detail in a recent review, which highlights the importance of RNA alternative splicing in the physiology of animal organisms, and how this may offer multiple opportunities when this process is altered in different diseases [37]. However, this complicated process requires a fine regulation, and its failures have been associated to several diseases, including cancer [38].
Schematic representation of the splicing process. On top, the spliceosome assembly is represented, showing all the intermediate macrocomplexes that are formed during the splicing process since the spliceosome joins the RNA molecule until it is spliced. Below, the two steps catalyzed by the spliceosome are represented. At the bottom, a scheme with the different patterns of alternative splicing is shown
In NENs, alternative splicing and its dysregulation are still scarcely explored but new evidence is arising. Failures of this system may be due to mutations or dysregulations in some of the components of the splicing machinery. Recently, a targeted transcriptomic analysis of the core splicing machinery (i.e., 17 spliceosome elements and 27 selected splicing factors) in PanNENs demonstrated a profound dysregulation of the main components of this complex system. Specifically, half of the components studied were upregulated in tumor tissue as compared to non-tumoral adjacent tissue, being some key spliceosome elements and splicing factors (i.e., NOVA1, PRPF8, RAVER1, SRSF5 and SNW1) associated with clinical parameters [39]. Moreover, in vitro and in vivo studies in PanNENs cell lines unveiled the ability of NOVA1 to modulate proliferation and senescence by altering critical signaling pathways and splicing mechanisms. Most importantly, NOVA1 could also influence the response to current pharmacological treatments (e.g., everolimus), suggesting that it may provide a promising candidate to develop novel biomarkers and therapeutic targets in PanNENs [39].
In SCLC cells, knockdown of the splicing factor SRRM4 led to cell death in vitro and suppressed tumor growth in SCLC mouse models, decreasing serum miR-4516 levels, which is upregulated in SCLC patients [40]. Also in SCLC, the splicing factor RBM10 has been related with major processes associated to cell proliferation and transformation, through a mechanism involving yet another splicing factor, RBM5, known to possess tumor suppressor properties, which seems to regulate RBM10 expression postranscriptionally, interacting with RBM10 splice variants [41]. In chemoresistant SCLC cells, the splicing factor ESRP1 is downregulated, which changes alternative splicing pattern of the arginine methyltransferase CARM1, and, in turn, activates TGF-β/Smad pathway and induces epithelial to mesenchymal transition [42]. Likewise, NUDT21 is also downregulated in SCLC cells, where it may alter the expression of two isoforms of GLS1, giving rise to an altered glutamine metabolism [43].
Defects in alternative splicing commonly lead to the genesis of splicing variants that can exert oncogenic functions. This is the case of the aberrant splicing variants of the G protein coupled, 7-transmembrane domain (TMD) somatostatin receptor subtype 5, SST5, which encodes two truncated receptors that only conserve 4 and 5 TMD and are consequently termed SST5TMD4 and SST5TMD5, respectively [44]. These two receptors, particularly SST5TMD4, have been reported to enhance oncogenic and aggressiveness features in various endocrine and neuroendocrine tumors and hormone-related cancers [45,46,47,48,49,50,51], and are also overexpressed in PanNENs tissue, where SST5TMD4 expression is associated with malignant characteristics and confers oncogenic properties to PanNEN cell models [52]. In line with this, a splice variant of CCK2R gene, which diminishes CCK2R membrane density and activity, is expressed in insulinomas [53]. Also, a splice variant of the fusion transcript INS-IGF2 is known to be expressed in insulinomas, while it is not expressed in normal pancreatic tissue [54]. Actinin-4 splice variant (Actn-4sv) is also expressed in PanNENs, but not in normal islets cells. This splice variant is more expressed in low-grade tumors and positively correlated with survival of patients [55]. Splicing variant of the Receptor for Hyaluronic Acid-Mediated Motility (RHAMMB), which is highly expressed in PanNENs, has been used to specifically deliver a small interfering RNA against another splicing variant related with apoptosis inhibition: Bcl-xL, which may be used as a novel therapy in PanNENs [56]. In SCLC tissue, when the glycoprotein NCAM, which has several splicing variants, expresses its exon-18-variant, it could have certain diagnostic potential for this disease [57]. Interestingly, the full phosphoprotein DARPP-32 and its truncated isoform, t-DARPP, are expressed in SCLC tissue (but not in normal lung), where they promote tumor growth by increasing Akt/Erk and decreasing anti-apoptotic signaling [58].
These results illustrate the emerging evidence supporting the dysregulation of alternative splicing, its machinery and its resulting products, in PanNENs and SCLC, as well as their pathological implications. Thus, further research is warranted to elucidate in detail how alternative splicing is altered in NENs, especially in GI-NENs and other LungNENs, how it contributes to the pathology, and to ascertain its potential as a new pharmacological target. On the other hand, other processes and machineries regulating RNA biology, from nonsense mediated decay to the RNA exosome complex [38] have not been explored in NENs to date as they have been studied in other types of cancer and deserve growing attention.
3 RNA species and NENs
Advances in RNA sequencing in the last decades have enabled the discovery of novel RNA molecules, which have been subsequently linked to various diseases, particularly cancer. These RNA species include microRNAs (miRNA), long non-coding (lncRNAs), circular (circRNA), PIWI-interacting (piRNA), small nuclear (snRNA), and other less known or abundant RNAs. The importance of different aspects of RNA biology and these RNA species in cancer has been previously reviewed in detail [59,60,61]. However, the relatively low incidence, diversity and heterogeneity of NENs often preclude their inclusion in general reviews on cancer and its related molecular mechanisms. Nevertheless, a growing body of evidence indicate that some of these RNA species can be implicated in NENs malignancy and that they could represent useful biomarkers in the management of these tumors. In this section, we will review the most relevant publications in this emerging field (Table 1).
3.1 MicroRNAs and NENs
MicroRNAs or miRNAs are short non-coding RNA molecules that target other RNAs, mainly in their 3’ region. These miRNAs regulate RNA expression through complementary binding and causing destabilization, degradation or even impeding translation. They are involved in many important cell functions and are frequently dysregulated under pathological conditions [122]. In fact, some studies have attempted to classify different types of NENs according to their miRNA expression profiles. Specifically, in PanNENs, a selected set of 30 differentially expressed miRNA has been used as a signature to classify patients in relation to tumor differentiation state [123]. In other study, miRNA signatures were able to distinguish between PanNENs tumoral and healthy tissue [63]. Moreover, the use of next generation sequencing unveiled miR-328 as a marker to specifically discriminate low from intermediate grade PanNENs [62].
Comparative studies were also performed among different types of NENs, such as PanNENs and SI-NENs, which can be distinguished measuring the expression of a limited set of miRNAs, being miR-375 and its cluster highly expressed in both types of NENs [62]. Other comparative studies have demonstrated that SI-NENs and pulmonary carcinoids share similar miRNAs profiles, which may suggest a common histological origin. In striking contrast, miRNA profiles of SCLC and pulmonary carcinoids are clearly different, despite both pathologies originating in the same organ [124]. In fact, the expression of a set of 8 miRNAs in cytologic samples was able to effectively distinguish among four types of lung neoplasms, including the SCLC and pulmonary carcinoids [125]. Other studies showed that specific miRNAs such as miR-21 and miR-155 have higher expressions in SCLC and LCNEC than in pulmonary carcinoids [80]. Also, miR-21/miR-375 ratio is known to be lower in TC and AC than in SCLC and LCNEC [78].
The potential use of miRNAs as biomarkers for diagnosis or prognosis has led to systematically test the putative associations of miRNAs aberrantly expressed in tumors with clinical parameters. In PanNENs, some miRNAs, like miR-642, were found to be differentially expressed according to tumor stage or Ki-67 index, suggesting their value as an indicator of aggressiveness of the neoplasm [63]; miR-196a, associated with tumor stage, Ki-67 score and mitotic count [64]; and miR-21, miR-10a and miR-106b showed higher expression in proliferative neoplasms compared to grade 1 tumors, the highest expression being associated to worst overall survival [65]. As well, miR-449a may play a key role in PanNEN proliferation and could be a putative prognostic factor for poor survival [66]. Moreover, some miRNAs may have differential expression in metastatic tumors, such as miR-210, or miR-3653 which were found to interact with ATRX, a chromatin remodeling gene and one of the most frequently mutated genes in PanNENs [63, 67]. Similarly, miR-431 was overexpressed in metastatic PanNENs, activating Ras/Erk signaling and promoting epithelial-mesenchymal transition both in vitro and in vivo [68].
In SI-NENs, miR-96 expression was higher in liver metastasis than in primary neoplasms, while miR-133a displayed a lower expression in metastasis [81]. miR-885-5p was upregulated in tumors with lymphovascular invasion as compared to those without it [83]. Other miRNAs were reported to be upregulated in metastatic tumors compared to primary (miR-183, -488, -19a and -19b), while some miRNAs were downregulated in metastatic tumors (miR-133a, -145, -146, -222 and -10b) [82]. In LungNENs, miR-29 family expression decreased as tumor grade increased when comparing carcinoids, LCNEC and SCLC [85]. Also, high expression of miR-100 was associated with aggressiveness features in pulmonary carcinoids [86]. When comparing both types of pulmonary carcinoids, several miRNAs were found to be upregulated in less aggressive typical carcinoids comparing with atypical ones (miR-129-5p, -409-3p, -409-5p, -185 and -497). Also, in both carcinoids, miR-409-3p, miR-409-5p and miR-431-5p were found downregulated when lymph node invasion was present [87]. Moreover, miR-21 expression in pulmonary carcinoids with lymph node invasion was also higher than in those without lymph node invasion [80].
3.1.1 miRNA function and emerging mechanistic understanding in NENs
Different studies have aimed to explore how the dysregulation of miRNAs affects NEN cells. An integrative study examining the interaction between miRNAs and mRNAs in PanNENs showed 28 miRNAs that were differentially expressed and were linked to different mRNAs [69], being miR-7-2-3p the miRNA most linked to other genes. Interestingly, in the PanNEN cell line BON-1, Menin, which is encoded by MEN1, the primary mutated gene in multiple endocrine neoplasia type 1 syndrome, was found to bind to pri-miR-24-1, facilitating its processing to miR-24-1, which, in turn, simultaneously acts as a positive regulator of MEN1 mRNA stability [70]. Some miRNAs are known to stimulate malignant processes such as cell proliferation, like the miR-144/451 cluster, which were shown to promote beta cell proliferation activating AKT pathway [71]. miR-137, miR-23b cluster, and miR-130/301 are also upregulated in PanNENs and act stimulating tumor growth and local invasion, enabling metastasis or evading apoptosis. By combining an elegant set of biocomputational and functional methods, including a genetically engineered mouse model, these miRNAs were shown to interact, respectively, with tumor suppressor Sorl1; metastatic suppressors Acvr1c/ALK7 and Robo2 and P2ry1; and Activin B [72]. miR-30a-3p is known to act in PanNENs by binding the tumor suppressor p27 mRNA and repressing its translation, competing with a p27 effector, the RNA binding protein HuD [73]. HuD is also known to bind INSM1 mRNA negatively regulating its expression in cooperation with miR-203a in an insulinoma cell line [74]. In SCLC, bioinformatic approaches also suggest that certain miRNAs could contribute to the dysregulation of pathways conveying malignancy features by acting on specific mRNA hubs [126]. In atypical carcinoids, hsa-let-7f-5p is overexpressed, showing an inverse correlation with its predicted target, HMGA2 [88]. In pulmonary carcinoids, miR-375 is highly expressed, which seems to downregulate YAP expression, inducing neuroendocrine differentiation and cell proliferation [79]. In SCLC, miR-518d-5p is known to be a negative regulator of DLL3 mediated cell proliferation and migration [89].
Changes in cell function induced by specific miRNAs provide novel therapeutic opportunities, in that altering miRNAs expression can impart antitumoral properties. In the PanNEN BON-1 cell line, expression of miR-224 promotes silencing of PCSK9, an effective regulator of low-density lipoprotein cholesterol, thereby decreasing cell proliferation and invasion and promoting apoptosis [75]. Moreover, in a PanNEN mouse model, anti-miR-214 drugs impairs tumor growth and metastasis [76]. In LungNENs, the overexpression of miR-886-3p inhibits SCLC cell proliferation, migration and colony formation and induce mesenchymal-epithelial transition by suppressing TGF-β1 synthesis in vitro and in vivo [90]. Likewise, overexpression of miR-26b in SCLC cell lines may inhibit proliferation, colony formation and migration, inducing apoptosis, probably acting by suppression of MCL1 [91]. Furthermore, miR-485-5p overexpression in SCLC cell lines, which was downregulated in SCLC tissue, decreased proliferation, migration, and invasion, possibly through FLOT2 reduction [92].
Interestingly, miRNAs may not only comprise direct actionable targets but could also be altered by and be involved in the response of other treatments and even in the development of resistance. Thus, in SI-NENs, somatostatin analogs therapy alters miRNA expression profile, which can be one of the mechanisms of response of tumor cells to this treatment [127]. In lung carcinoids, high miR-100 expression was associated with aggressiveness features and showed an inverse correlation with mTOR expression, while miR-100 inhibition in vitro increased the sensitivity to mTOR targeted treatments [86]. In SCLC, miR-7-5p, which regulates the expression of poly ADP-ribose polymerase 1, PARP1, was downregulated in chemoresistant cells. Particularly, miR-7-5p evoked suppressive effects on homologous recombination repair and thus, it might be used to avoid chemoresistance in patients [93]. Similarly, miR-22-3p, a negative regulator of WRNIP1, a gene associated with DNA damage repair, enhanced SCLC radiosensitivity and its expression in tumors was proposed to be evaluated in patients that are going to receive radiotherapy [94]. Also, miR-30a-5p negatively regulates BECN1, and thereby sensitizes drug resistant SCLC cells to chemotherapy [95].
miRNAs could be also envisaged as non-invasive biomarkers. In this sense, miR-193-b, which was upregulated in neoplastic tissue, was found to be upregulated also in PanNEN patients serum [63]. Moreover, exosomal secreted miRNAs were found to be differentially expressed among different pancreatic lesions, showing different miRNA signatures that discriminate PanNENs from chronic pancreatitis or from pancreatic adenocarcinoma [128]. Four miRNAs (miR-125b-5p, -362-5p, -425-5p and -500a-5p) were also upregulated in serum of SI-NENs patients. miR-125b-5p and miR-362-5p remained upregulated one month, and one year after surgery, respectively, in those patients with residual disease and/or recurrent disease [84]. Two plasmatic microRNAs panels were developed with high potential in the diagnosis of lung cancer, and which could discriminate between SCLC and non small-cell lung carcinoma [129].
In summary, miRNAs may be used to stratify NENs and their stability in plasma makes them good minimally invasive biomarkers. Moreover, recent evidence indicate that miRNAs are involved in diverse malignancy processes in NENs, and suggest that they could become actionable therapeutic targets. Nevertheless, further research and solid clinical evidence, including prospective studies, are still required for the use of miRNAs to arrive to NENs management.
3.2 LncRNAs and NENs
Long non-coding RNAs (lncRNAs) are a class of transcripts that were discovered more recently than miRNAs but have been also rapidly linked to disease and, particularly, to cancer. These are RNA molecules with more than 200 nucleotides in length that do not normally code into proteins. LncRNAs mainly act regulating gene expression of other RNAs, which may be coding or non-coding [130].
LncRNAs can interact with miRNAs regulating their function, usually acting as miRNA sponges. In SCLC, this phenomenon was well studied, as is the case HOTTIP, a lncRNA overexpressed in SCLC samples, where it correlates with tumor stage and shorter overall survival. HOTTIP works as a miR-574-5p sponge, regulating the expression of histone methylase EZH1 [109] and also regulating epithelial-mesenchymal transition [110]. HOTTIP may also sponge miR-216a, thus increasing the expression of the antiapoptotic protein BCL2 and enhancing chemoresistance, as miR-216a is correlated with good prognosis and suppresses chemoresistance [111]. The knockdown of HOTTIP itself in a mouse model of SCLC resulted in an inhibition of tumor growth in vivo [109]. LINC00173 is also highly expressed in SCLC, promoting chemoresistance, proliferation, migration, and invasion in vitro and in vivo. LINC00173 upregulates BMX sponging miR-218 and thereby acting as a competitive endogenous RNA (ceRNA) [112]. LncRNA LUADT1 was also overexpressed in SCLC cells, and it sponged miR-15a-3p, upregulating TWIST1 transcription factor, and thereby promoting cell invasion and migration [113]. Another example is lncRNA PPIAP43, a pseudogene from PPIA, which displayed an increase in its expression in SCLC cell lines after radiation exposure and may act as a sponge for miRNAs that bind PPIA, increasing its expression. Overexpression of PPIA, however, was associated with cancer progression [114].
LncRNAs have been widely studied since the emergence of high-throughput sequencing technologies, being also considered as disease biomarkers. Indeed, single nucleotide polymorphisms (SNPs) were found through whole genome sequencing in the lncRNAs PVT1 and HOTAIR genes. These SNPs were overrepresented in PanNENs patients, suggesting a role in the development of the disease [96]. In a recent integrative next-generation study, a lncRNA-ceRNAs network was built to find new biomarkers which could be used in SCLC treatment, integrating these genes with alternative splicing events and showing useful information in this field [131]. LncRNAs expression can also be correlated with clinical parameters, as pointed above and reported in several transcriptomic studies. LncRNA XLOC_221242 expression is increased in PanNENs compared with normal tissue, positively correlating with DNER mRNA, a key factor of the Notch signaling pathway [97]. LncRNAs HOTAIR and MALAT1 expressions were higher in PanNENs with lower tumor stage and subsequent risk of developing metastasis [98]. In contrast, HOTAIR expression is higher in gastroenteropancreatic NECs than in NETs, unveiling an apparent discrepancy in the potential prognostic role of this lncRNA to stratify these diseases [103]. In SCLC, the lncRNA SBF2-AS1 was highly expressed and correlated with several malignancy parameters, like clinical stage, metastasis, or tumor size, and, even lower overall survival in patients [115]. BLACAT1 lncRNA was overexpressed in SCLC tissue, being associated with clinical stage, lymph node and distant metastasis, tumor size and poor prognosis. Intracellularly, BLACAT1 may play a role in cell proliferation and motility regulation [116].
Numerous lncRNAs may contribute to neoplasm progression in NENs. In PanNENs, 2080 lncRNAs were found to be differentially expressed between tumor tissue and adjacent tissue, which were related to cancer biology through diverse cell processes and signaling pathways [132]. In a different study, with a lower number of PanNENs samples, 363 lncRNAs were found differentially expressed between tumor and adjacent tissue, which serve to build a lncRNA-mRNA coexpression network [97]. In physiological conditions, MEN1 activates the lncRNA MEG3, which acts as a tumor suppressor gene through downregulation of the expression of the protooncogene c-Met. In contrast, in MEN1 syndromic PanNENs, caused by a MEN1 mutation, MEG3 is not activated, and therefore it is unable to reduce c-Met expression [106]. The molecular mechanism underlying c-Met downregulation by MEG3 could involve histone modifications or transcriptional repression through triplex formation [108]. In gastric NENs, lncNEN885 expression was severely decreased in tumor tissue and its silencing in cell models increased cell migration and invasion and enhanced epithelial-mesenchymal transition, which led to propose it as a potential metastasis biomarker [99]. In PanNENs patients, high expression of the lncRNA H19 was associated with poorer survival, while in vitro overexpression promoted tumor cell growth and metastasis features, through a mechanism likely involving VGF nerve growth factor inducible and activation of PI3K/AKT/CREB signaling [100]. Similarly to MEG3 in PanNETs, in SCLC the lncRNA HOTAIR regulates H3K27me3 methylation, which in turn induced multidrug resistance, possibly acting in a feedback mechanism, as a negative regulator of HOTAIR [104].
LncRNAs may also be used as non-invasive biomarkers as they can be secreted to circulation. Thus, lncRNA CASC11 expression was upregulated in SCLC patients’ serum, in association to TGF-β1 expression, which is stimulated by CASC11, increasing cancer cell stemness in SCLC [117]. In contrast, LNCNEF was downregulated in SCLC patients and its expression in plasma was associated with distant tumor metastasis [118].
Hence, available evidence clearly supports that the expression of numerous lncRNAs is altered in NENs, and evidence is also growing that modification of their expression may entail therapeutic benefits. Indeed, in PanNENs cells, overexpression of MEG3 in vitro decreased cell proliferation and invasion by decreasing miR-183 [107]. Likewise, overexpression of HNF1A-AS1 also had antitumor properties in gastroenteropancreatic NENs models in vitro and in vivo [102]. A natural antisense of somatostatin receptor subtype 5, SSTR5-AS1 has recently been reported to be upregulated in PanNEN tissue (as compared to non-tumoral adjacent tissue) where its levels were directly and strongly associated with SSTR5 expression in both tumor and non-tumor tissue. Moreover, in vitro assays revealed that SSTR5-AS1 influences both SSTR5 and its own expression, while its silencing enhances NEN cell aggressiveness features, including proliferation, migration, and colony formation, and, most importantly, influences the modest response of PanNEN cells to the SST5-preferring somatostatin analog pasireotide [101]. LncRNA KCNQ1OT1 was highly expressed in SCLC chemoresistant cells and its knockdown inhibited cell malignancy parameters in vitro through the activation of JAK2/STAT3 pathway [119]. LncRNAs HOTAIR, MEG3 and PCA3 displayed higher expression in SCLC than in other LungNENs, and knocking-down MEG3 and PCA3 decreased cell proliferation in SCLC cell lines [105]. Also, the in vitro overexpression of LNCNEF resulted in a reduction in migration and invasion capacities of the SCLC cells, probably mediated by TGF-β1 downregulation [118].
Therefore, although lncRNAs are still less well studied than miRNAs in NENs, probably due to their more recent discovery, there is solid evidence that these RNA molecules are also involved in key processes of NENs malignancy, especially in LungNENs. Moreover, given their involvement in the regulation of gene expression at different levels, lncRNAs are likely to become useful tools as future biomarkers and therapeutic targets in NENs.
3.3 Other non-coding RNAs
Rapid expansion of improved sequencing technologies has led to the recent discovery of other RNA molecules. Despite their recent identification, dysregulations in these new RNA species have been described and rapidly linked to cancer, although their precise implications are just starting to be studied. Some of these RNAs include other interfering RNA which, like miRNAs, are likely involved in gene expression regulation, as is the case of PIWI-interacting RNAs (piRNAs). These piRNAs are also known to be implied in other processes like chromatin rearrangement or protein regulation, and, of course, some of them are dysregulated in cancer contributing to cancer cell malignancy [133]. However, whether piRNAs are dysregulated in NENs remains to be defined.
Similarly, circular RNAs (circRNAs) have been identified recently from different synthesis processes and their role in cancer biology possible therapeutic value looks promising. CircRNAs are covalently closed RNA molecules which can act as miRNAs or even as a sponge for other types of small RNAs [134]. In contrast with the numerous studies relating circRNAs with cancer, only a few publications have explored their role in NENs, specifically in SCLC. Particularly, the circRNA cESRP1 was downregulated in chemoresistant SCLC cells, where it enhanced chemotherapy sensitivity, sponging miR-93-5p and thereby inhibiting TGF-β pathway [120]. FLI1 circRNAs were upregulated in SCLC tissues, were they directly correlated with lymph node metastasis. Also, one of them was found in exosomes isolated from serum and associated with poor survival. Silencing of these circRNAs decreased in vitro migration and reduced metastasis in vivo [121].
There are also RNA molecules which are involved in processing other RNAs, the small nuclear (snRNAs) and small nucleolar RNAs (snoRNAs). snRNAs are implied in mRNA maturation, whilst the snoRNAs (which are in turn classified according to their function in H/ACA box snoRNAs, C/D box snoRNAs and small cajal RNAs [135]) participate in synthesis and maturation of rRNA. These RNAs often interact with proteins forming ribonucleoprotein complexes (snRNPs and snoRNPs) to carry out their function, and misfunction of these complexes is linked to diseases and cancer [136,137,138]. In ileal NENs and PanNENs, several small RNA molecules were studied, but just as normalizers [77]. In fact, this study revealed that SNORD61 and SNORD95, among others, were suitable normalizers for miRNA expression studies, at least in ileal NETs. Also, SNORD95 and miR-93 were suitable for miRNAs analyses comparing ileal and PanNENs [77]. Nevertheless, the function of snRNAs and their relationship with altered splicing and the role of snoRNAs in NENs are vastly unexplored and may lead to open a wide research field.
4 Conclusion
Recent evidence reveals the emerging role of RNA biology in cancer, from alternative splicing and RNA surveillance to newly discovered non-coding RNAs species. In NENs, RNA biology studies are still limited as compared to other cancers (Fig. 3) and, indeed, not all NENs are equally studied, being GI-NENs much less explored on this particular field. However, convincing data supports the growing notion that alternative splicing is altered in NENs, both at the level of its operating machinery, with dysregulated splicing factors profiles, and at the outcome of spurious splicing variants with oncogenic potential. Likewise, increasing evidence reveals the important role of disrupted levels of specific non-coding RNA species, such as miRNAs and lncRNAs, in diverse NENs. Nevertheless, additional RNA biology processes, like RNA surveillance, and other RNA species, such as circRNAs, piRNAs, snRNAs or snoRNAs, all of which have already been linked to other cancers, are still to be explored in detail in these tumors.
The relevance of studying RNA biology and its defects in NENs lays in its potential to generate basic knowledge to better understand these complex and heterogeneous tumors. But this new knowledge also opens novel translational opportunities. Indeed, altered splicing machinery components and splicing variants, as well as dysregulated miRNAs and lncRNAs unveil new candidate biomarkers and identifies novel actionable targets with therapeutic potential. The implications of a more profound comprehension of RNA biology in NENs include the possibility to add new “omics” layers of molecular information, from miRNA-omics to “spliceosomics”, which may help to better understand NENs heterogeneity and, ultimately, facilitate their complex management. In this manner, advances in RNA knowledge can pave the way to bring personalized precision medicine to NENs, by providing new biomarkers to refine classification and stratification, improve prognosis prediction and identify novel potential actionable therapeutic targets.
The unprecedented uprise of RNA as a central player in translational biomedical research cannot be best explained than by the example of the RNA vaccines developed in record time to fight the COVID-19 pandemic. This beam illuminates the path to be followed to keep a successful quest to research new RNA functions and species, elucidate their role in tumor cell biology, and devise their use to fight cancer.
Abbreviations
- NEN:
-
Neuroendocrine Neoplasm
- NET:
-
Neuroendocrine Tumor
- NEC:
-
Neuroendocrine Carcinoma
- GI-NEN:
-
Gastrointestinal Neuroendocrine Neoplasm
- LungNEN:
-
Lung Neuroendocrine Neoplasm
- PanNEN:
-
Pancreatic Neuroendocrine Neoplasm.
- SI-NEN:
-
Small Intestine Neuroendocrine Neoplasm
- mTOR:
-
Mammalian Target Of Rapamycin
- mRNA:
-
Messenger Ribonucleic Acid
- TC:
-
Typical Carcinoid
- AC:
-
Atypical Carcinoid
- LCNEC:
-
Large Cell Neuroendocrine Carcinoma
- SCLC:
-
Small Cell Lung Cancer
- rRNA:
-
Ribosomal Ribonucleic Acid
- tRNA:
-
Transfer Ribonucleic Acid
- ncRNA:
-
Non Coding Ribonucleic Acid
- TERC:
-
Telomerase RNA Component
- aRNA:
-
Antisense RNA
- PCR:
-
Polymerase Chain Reaction
- snRNA:
-
Small Nuclear Ribonucleic Acid
- TMD:
-
Transmembrane Domain
- miRNA:
-
Micro Ribonucleic Acid
- lncRNA:
-
Long Non Coding Ribonucleic Acid
- circRNA:
-
Circular Ribonucleic Acid
- piwiRNA or piRNA:
-
PIWI-Interacting Ribonucleic Acid
- snRNA:
-
Small Nuclear Ribonucleic Acid
- snoRNAs:
-
Small Nucleolar Ribonucleic Acid
- RNP:
-
Ribonucleoprotein
References
Cives M, Strosberg JR. Gastroenteropancreatic neuroendocrine tumors. CA: a Cancer Journal for Clinicians. 2018; 68(6), 471–487, doi:https://doi.org/10.3322/caac.21493.
Fisseler-Eckhoff A, Demes M. Neuroendocrine tumors of the lung. Cancers. 2012;4(3):777–98. doi:https://doi.org/10.3390/cancers4030777.
Dasari A, et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol. 2017;3(10):1335–42. doi:https://doi.org/10.1001/jamaoncol.2017.0589.
Rindi G, et al. A common classification framework for neuroendocrine neoplasms: an International Agency for Research on Cancer (IARC) and World Health Organization (WHO) expert consensus proposal. Mod Pathol. 2018;31(12):1770–86. doi:https://doi.org/10.1038/s41379-018-0110-y.
Ito T, et al. Carcinoid-syndrome: recent advances, current status and controversies. Curr Opin Endocrinol Diabetes Obes. 2018;25(1):22–35. doi:https://doi.org/10.1097/MED.0000000000000376.
Pedraza-Arévalo S, et al. Multilayered heterogeneity as an intrinsic hallmark of neuroendocrine tumors. Reviews in Endocrine & Metabolic Disorders. 2018;19(2):179–92. doi:https://doi.org/10.1007/s11154-018-9465-0.
Oberg K, et al. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncol. 2015;16(9):e435–46. doi:https://doi.org/10.1016/S1470-2045(15)00186-2.
Herrera-Martínez AD, et al. Targeted systemic treatment of neuroendocrine tumors: current options and future perspectives. [Review] Drugs. 2019;79(1):21–42. doi:https://doi.org/10.1007/s40265-018-1033-0.
Capdevila J, et al. Translational research in neuroendocrine tumors: pitfalls and opportunities. Oncogene. 2017;36(14):1899–907. doi:https://doi.org/10.1038/onc.2016.316.
Mafficini A, Scarpa A. Genetics and Epigenetics of Gastroenteropancreatic Neuroendocrine Neoplasms. Endocr Rev. 2019;40(2):506–36. doi:https://doi.org/10.1210/er.2018-00160.
Derks JL, et al. New Insights into the molecular characteristics of Pulmonary Carcinoids and large cell neuroendocrine carcinomas, and the impact on their clinical management. J Thorac Oncol. 2018;13(6):752–66. doi:https://doi.org/10.1016/j.jtho.2018.02.002.
Scarpa A, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65–71. doi:https://doi.org/10.1038/nature21063.
Banck MS, et al. The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest. 2013;123(6):2502–8. doi:https://doi.org/10.1172/JCI67963.
George J, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi:https://doi.org/10.1038/nature14664.
Alcala N, et al. Integrative and comparative genomic analyses identify clinically relevant pulmonary carcinoid groups and unveil the supra-carcinoids. Nat Commun. 2019;10(1):3407. doi:https://doi.org/10.1038/s41467-019-11276-9.
Fernandez-Cuesta L, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun. 2014;5:3518. doi:https://doi.org/10.1038/ncomms4518.
Group PTC, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578(7793):129–36. doi:https://doi.org/10.1038/s41586-020-1970-0.
Crick F. Central dogma of molecular biology. Nature. 1970;227(5258):561–3. doi:https://doi.org/10.1038/227561a0.
Brosius J, Raabe CA. What is an RNA? A top layer for RNA classification. RNA Biology. 2016;13(2):140–4. doi:https://doi.org/10.1080/15476286.2015.1128064.
Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi:https://doi.org/10.1016/j.cell.2014.03.008.
Lieberman J. Unveiling the RNA world. N Engl J Med. 2018;379(13):1278–80. doi:https://doi.org/10.1056/NEJMcibr1808725.
Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science. 2019;366(6467):822–7. doi:https://doi.org/10.1126/science.aax2957.
Coulon A, et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife. 2014; 3, doi:https://doi.org/10.7554/eLife.03939.
Corley M, et al. How RNA-Binding proteins interact with RNA: molecules and mechanisms. Mol Cell. 2020;78(1):9–29. doi:https://doi.org/10.1016/j.molcel.2020.03.011.
Asthana S, et al. The Physiological Roles of the Exon Junction Complex in Development and Diseases. Cells. 2022;11(7), 1192. doi:https://doi.org/10.3390/cells11071192.
Cooper TA, et al. RNA and disease. Cell. 2009;136(4):777–93. doi:https://doi.org/10.1016/j.cell.2009.02.011.
Hansen TB, et al. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–12. doi:https://doi.org/10.1158/0008-5472.CAN-13-1568.
Yang G, et al. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–109. doi:https://doi.org/10.1016/j.bbagrm.2014.08.012.
Scott AT, et al. Gene expression signatures identify Novel therapeutics for metastatic pancreatic neuroendocrine tumors. Clin Cancer Res. 2020;26(8):2011–21. doi:https://doi.org/10.1158/1078-0432.CCR-19-2884.
Keck KJ, et al. Changes in gene expression in small bowel neuroendocrine tumors associated with progression to metastases. Surgery. 2018;163(1):232–9. doi:https://doi.org/10.1016/j.surg.2017.07.031.
Simbolo M, et al. Gene expression profiling of Lung atypical carcinoids and large cell neuroendocrine Carcinomas identifies three Transcriptomic subtypes with specific genomic alterations. J Thorac Oncol. 2019;14(9):1651–61. doi:https://doi.org/10.1016/j.jtho.2019.05.003.
Diedisheim M, et al. Prognostic transcriptome classes of duodenopancreatic neuroendocrine tumors. Endocrine-related Cancer. 2021;28(8):563–71. doi:https://doi.org/10.1530/ERC-21-0051.
Modlin IM, et al. A multianalyte PCR blood test outperforms single analyte ELISAs (chromogranin A, pancreastatin, neurokinin A) for neuroendocrine tumor detection. Endocrine-related Cancer. 2014;21(4):615–28. doi:https://doi.org/10.1530/ERC-14-0190.
Modlin IM, et al. Molecular genomic Assessment using a blood-based mRNA signature (NETest) is cost-effective and predicts neuroendocrine tumor recurrence with 94% accuracy. Ann Surg. 2021;274(3):481–90. doi:https://doi.org/10.1097/SLA.0000000000005026.
Modlin IM, et al. Early identification of residual Disease after neuroendocrine tumor resection using a Liquid Biopsy Multigenomic mRNA signature (NETest). Ann Surg Oncol. 2021;28(12):7506–17. doi:https://doi.org/10.1245/s10434-021-10021-1.
Wagner RE, Frye M. Noncanonical functions of the serine-arginine-rich splicing factor (SR) family of proteins in development and disease. BioEssays. 2021;43(4):e2000242. doi:https://doi.org/10.1002/bies.202000242.
Marasco LE, Kornblihtt AR. The physiology of alternative splicing. Nat Reviews: Mol Cell Biology. 2022. doi:https://doi.org/10.1038/s41580-022-00545-z.
Obeng EA, et al. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discovery. 2019;9(11):1493–510. doi:https://doi.org/10.1158/2159-8290.CD-19-0399.
Pedraza-Arevalo S, et al. Spliceosomic dysregulation unveils NOVA1 as a candidate actionable therapeutic target in pancreatic neuroendocrine tumors. Translational Research: The Journal of Laboratory and Clinical Medicine. 2022. doi:https://doi.org/10.1016/j.trsl.2022.07.005.
Shimojo M, et al. A gapmer antisense oligonucleotide targeting SRRM4 is a novel therapeutic medicine for lung cancer. Sci Rep. 2019;9(1):7618. doi:https://doi.org/10.1038/s41598-019-43100-1.
Loiselle JJ, et al. RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5. PLoS ONE. 2017;12(6):e0180258. doi:https://doi.org/10.1371/journal.pone.0180258.
Zheng M, et al. ESRP1 regulates alternative splicing of CARM1 to sensitize small cell lung cancer cells to chemotherapy by inhibiting TGF-beta/Smad signaling. Aging. 2021;13(3):3554–72. doi:https://doi.org/10.18632/aging.202295.
Gao CC, et al. NUDT21 suppresses the growth of small cell lung cancer by modulating GLS1 splicing. Biochem Biophys Res Commun. 2020;526(2):431–8. doi:https://doi.org/10.1016/j.bbrc.2020.03.089.
Günther T, et al. International Union of Basic and Clinical Pharmacology. CV. Somatostatin receptors: structure, function, ligands, and new nomenclature. Pharmacol Rev. 2018;70(4):763–835. doi:https://doi.org/10.1124/pr.117.015388.
Fuentes-Fayos AC, et al. Somatostatin receptor splicing variant sst5TMD4 overexpression in Glioblastoma is Associated with Poor Survival, increased aggressiveness features, and somatostatin Analogs Resistance. Int J Mol Sci. 2022; 23(3), doi:https://doi.org/10.3390/ijms23031143.
Hormaechea-Agulla D, et al. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J. 2017;31(11):4682–96. doi:https://doi.org/10.1096/fj.201601264RRR.
Mole D, et al. The expression of the truncated isoform of somatostatin receptor subtype 5 associates with aggressiveness in medullary thyroid carcinoma cells. Endocrine. 2015;50(2):442–52. doi:https://doi.org/10.1007/s12020-015-0594-x.
Luque RM, et al. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett. 2015;359(2):299–306. doi:https://doi.org/10.1016/j.canlet.2015.01.037.
Puig-Domingo M, et al. The truncated isoform of somatostatin receptor5 (sst5TMD4) is associated with poorly differentiated thyroid cancer. PloS One. 2014;9(1):e85527. doi:https://doi.org/10.1371/journal.pone.0085527.
Durán-Prado M, et al. The new truncated somatostatin receptor variant sst5TMD4 is associated to poor prognosis in breast cancer and increases malignancy in MCF-7 cells. Oncogene. 2012;31(16):2049–61. doi:https://doi.org/10.1038/onc.2011.389.
Durán-Prado M, et al. Identification and characterization of two novel truncated but functional isoforms of the somatostatin receptor subtype 5 differentially present in pituitary tumors. J Clin Endocrinol Metab. 2009;94(7):2634–43. doi:https://doi.org/10.1210/jc.2008-2564.
Sampedro-Nuñez M, et al. Presence of sst5TMD4, a truncated splice variant of the somatostatin receptor subtype 5, is associated to features of increased aggressiveness in pancreatic neuroendocrine tumors. Oncotarget. 2016;7(6):6593–608. doi:https://doi.org/10.18632/oncotarget.6565.
Sanchez C, et al. Characterization of a novel five-transmembrane domain cholecystokinin-2 receptor splice variant identified in human tumors. Mol Cell Endocrinol. 2012;349(2):170–9. doi:https://doi.org/10.1016/j.mce.2011.10.010.
Johannessen LE, et al. Upregulation of INS-IGF2 read-through expression and identification of a novel INS-IGF2 splice variant in insulinomas. Oncol Rep. 2016;36(5):2653–62. doi:https://doi.org/10.3892/or.2016.5132.
Xu X, et al. Actinin-4 splice variant - a complementary diagnostic and prognostic marker of pancreatic neuroendocrine neoplasms. J Cancer. 2020;11(8):2318–28. doi:https://doi.org/10.7150/jca.37503.
Chen X, et al. RHAMM(B)-mediated bifunctional nanotherapy targeting Bcl-xL and mitochondria for pancreatic neuroendocrine tumor treatment. Mol Ther Oncolytics. 2021;23:277–87. doi:https://doi.org/10.1016/j.omto.2021.10.002.
Vander Borght A, et al. The 180 splice variant of NCAM-containing exon 18-is specifically expressed in small cell lung cancer cells. Transl Lung Cancer Res. 2018;7(3):376–88. doi:https://doi.org/10.21037/tlcr.2018.03.03.
Alam SK, et al. ASCL1-regulated DARPP-32 and t-DARPP stimulate small cell lung cancer growth and neuroendocrine tumour cell proliferation. Br J Cancer. 2020;123(5):819–32. doi:https://doi.org/10.1038/s41416-020-0923-6.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. doi:https://doi.org/10.1038/nrd.2016.246.
Bhan A, et al. Long noncoding RNA and Cancer: a New Paradigm. Cancer Res. 2017;77(15):3965–81. doi:https://doi.org/10.1158/0008-5472.CAN-16-2634.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Reviews: Cancer. 2021;21(1):22–36. doi:https://doi.org/10.1038/s41568-020-00306-0.
Panarelli N, et al. Evaluating gastroenteropancreatic neuroendocrine tumors through microRNA sequencing. Endocrine-related Cancer. 2019;26(1):47–57. doi:https://doi.org/10.1530/ERC-18-0244.
Thorns C, et al. Global microRNA prolifiling of pancreatic neuroendocrine neoplasias. Anticancer Res. 2014;34(5):2249–54.
Lee YS, et al. High expression of MicroRNA-196a indicates poor prognosis in Resected pancreatic neuroendocrine tumor. Med (Baltim). 2015;94(50):e2224. doi:https://doi.org/10.1097/MD.0000000000002224.
Grolmusz VK, et al. Prognostic relevance of proliferation-related miRNAs in pancreatic neuroendocrine neoplasms. Eur J Endocrinol. 2018;179(4):219–28. doi:https://doi.org/10.1530/EJE-18-0305.
Klieser E, et al. HDAC-Linked “Proliferative” miRNA expression pattern in pancreatic neuroendocrine tumors. Int J Mol Sci 2018; 19(9), doi:https://doi.org/10.3390/ijms19092781.
Gill P, et al. MiRNA-3653 is a potential tissue biomarker for increased metastatic risk in pancreatic neuroendocrine tumours. Endocr Pathol. 2019;30(2):128–33. doi:https://doi.org/10.1007/s12022-019-9570-y.
Zhang T, et al. miR-431 promotes metastasis of pancreatic neuroendocrine tumors by targeting DAB2 interacting protein, a ras GTPase activating protein tumor suppressor. Am J Pathol. 2020;190(3):689–701. doi:https://doi.org/10.1016/j.ajpath.2019.11.007.
Zhou HQ, et al. Integrative microRNA-mRNA and protein-protein interaction analysis in pancreatic neuroendocrine tumors. Eur Rev Med Pharmacol Sci. 2016;20(13):2842–52.
Luzi E, et al. An autoregulatory network between menin and pri-mir-24-1 is required for the processing of its specific modulator mir-24-1 in BON1 cells. Mol Biosyst. 2016;12(6):1922–8. doi:https://doi.org/10.1039/c6mb00118a.
Jiang X, et al. miR-144/451 promote cell proliferation via Targeting PTEN/AKT pathway in Insulinomas. Endocrinology. 2015;156(7):2429–39. doi:https://doi.org/10.1210/en.2014-1966.
Michael IP, et al. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc Natl Acad Sci USA. 2019;116(48):24184–95. doi:https://doi.org/10.1073/pnas.1913307116.
Kim C, et al. Reduced expression of the RNA-binding protein HuD in pancreatic neuroendocrine tumors correlates with low p27(Kip1) levels and poor prognosis. J Pathol. 2018;246(2):231–43. doi:https://doi.org/10.1002/path.5135.
Kim C, et al. RNA binding protein HuD and microRNA-203a cooperatively regulate insulinoma-associated 1 mRNA. Biochem Biophys Res Commun. 2020;521(4):971–6. doi:https://doi.org/10.1016/j.bbrc.2019.11.030.
Bai J, et al. A retrospective study of NENs and miR-224 promotes apoptosis of BON-1 cells by targeting PCSK9 inhibition. Oncotarget. 2017;8(4):6929–39. doi:https://doi.org/10.18632/oncotarget.14322.
Dettori D, et al. Therapeutic silencing of miR-214 inhibits Tumor Progression in multiple mouse models. Mol Ther. 2018;26(8):2008–18. doi:https://doi.org/10.1016/j.ymthe.2018.05.020.
Sperveslage J, et al. Establishment of robust controls for the normalization of miRNA expression in neuroendocrine tumors of the ileum and pancreas. Endocrine. 2014;46(2):226–30. doi:https://doi.org/10.1007/s12020-014-0202-5.
Wong JJM, et al. Classifying lung neuroendocrine neoplasms through MicroRNA sequence data mining. Cancers. 2020; 12(9), doi:https://doi.org/10.3390/cancers12092653.
Yang X, et al. A miR-375/YAP axis regulates neuroendocrine differentiation and tumorigenesis in lung carcinoid cells. Sci Rep. 2021;11(1):10455. doi:https://doi.org/10.1038/s41598-021-89855-4.
Lee HW, et al. Altered expression of microRNA miR-21, miR-155, and let-7a and their roles in pulmonary neuroendocrine tumors. Pathol Int. 2012;62(9):583–91. doi:https://doi.org/10.1111/j.1440-1827.2012.02845.x.
Mandal R, et al. Analysis of miR-96 and miR-133a expression in gastrointestinal neuroendocrine neoplasms. Endocr Pathol. 2017;28(4):345–50. doi:https://doi.org/10.1007/s12022-017-9504-5.
Ruebel K, et al. MicroRNA expression in ileal carcinoid tumors: downregulation of microRNA-133a with tumor progression. Mod Pathol. 2010;23(3):367–75. doi:https://doi.org/10.1038/modpathol.2009.161.
Mitsuhashi K, et al. Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget. 2015;6(26):22114–25. doi:https://doi.org/10.18632/oncotarget.4294.
Malczewska A, et al. Circulating MicroRNAs in small-bowel neuroendocrine tumors: a potential Tool for diagnosis and Assessment of Effectiveness of Surgical Resection. Ann Surg. 2019. doi:https://doi.org/10.1097/SLA.0000000000003502.
Mairinger FD, et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: results of a profiling study. Mod Pathol. 2014;27(12):1632–40. doi:https://doi.org/10.1038/modpathol.2014.74.
Rapa I, et al. High miR-100 expression is associated with aggressive features and modulates TORC1 complex activation in lung carcinoids. Oncotarget. 2018;9(44):27535–46. doi:https://doi.org/10.18632/oncotarget.25541.
Rapa I, et al. Identification of MicroRNAs differentially expressed in lung carcinoid subtypes and progression. Neuroendocrinology. 2015;101(3):246–55. doi:https://doi.org/10.1159/000381454.
Di Fazio P, et al. Expression of hsa-let-7b-5p, hsa-let-7f-5p, and hsa-mir-222-3p and their putative targets HMGA2 and CDKN1B in typical and atypical carcinoid tumors of the lung. Tumour Biol. 2017;39(10):1010428317728417. doi:https://doi.org/10.1177/1010428317728417.
Huang J, et al. DLL3 is regulated by LIN28B and miR-518d-5p and regulates cell proliferation, migration and chemotherapy response in advanced small cell lung cancer. Biochem Biophys Res Commun. 2019;514(3):853–60. doi:https://doi.org/10.1016/j.bbrc.2019.04.130.
Shen J, et al. MicroRNA-886-3P functions as a tumor suppressor in small cell lung cancer. Cancer Biol Ther. 2018;19(12):1185–92. doi:https://doi.org/10.1080/15384047.2018.1491505.
Yu JG, et al. MicroRNA-26b suppresses tumorigenicity and promotes apoptosis in small cell lung cancer cells by targeting myeloid cell leukemia 1 protein. Kaohsiung J Med Sci. 2018;34(11):593–605. doi:https://doi.org/10.1016/j.kjms.2018.06.005.
Gao F, et al. MicroRNA-485-5p suppresses the proliferation, migration and invasion of small cell lung cancer cells by targeting flotillin-2. Bioengineered. 2019;10(1):1–12. doi:https://doi.org/10.1080/21655979.2019.1586056.
Lai J, et al. MiR-7-5p-mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer. 2019;19(1):602. doi:https://doi.org/10.1186/s12885-019-5798-7.
Jiang W, et al. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J Cell Biochem. 2019;120(10):17650–61. doi:https://doi.org/10.1002/jcb.29032.
Yang X, et al. Intensified Beclin-1 mediated by low expression of Mir-30a-5p promotes Chemoresistance in Human Small Cell Lung Cancer. Cell Physiol Biochem. 2017;43(3):1126–39. doi:https://doi.org/10.1159/000481754.
Moschovis D, et al. Association between genetic polymorphisms in long non-coding RNAs and pancreatic cancer risk. Cancer Biomarkers. 2019;24(1):117–23. doi:https://doi.org/10.3233/CBM-181959.
He R, et al. Transcriptional profiling reveals the Regulatory Role of DNER in promoting pancreatic neuroendocrine neoplasms. Front Genet. 2020;11:587402. doi:https://doi.org/10.3389/fgene.2020.587402.
Chu YH, et al. In situ hybridization analysis of long non-coding RNAs MALAT1 and HOTAIR in gastroenteropancreatic neuroendocrine neoplasms. Endocr Pathol. 2019;30(1):56–63. doi:https://doi.org/10.1007/s12022-018-9564-1.
Wei YL, et al. LncNEN885 inhibits epithelial-mesenchymal transition by partially regulation of Wnt/beta-catenin signalling in gastroenteropancreatic neuroendocrine neoplasms. Cancer Sci. 2018;109(10):3139–48. doi:https://doi.org/10.1111/cas.13747.
Ji M, et al. lncRNA H19 binds VGF and promotes pNEN progression via PI3K/AKT/CREB signaling. Endocrine-related Cancer. 2019;26(7):643–58. doi:https://doi.org/10.1530/ERC-18-0552.
Pedraza-Arevalo S, et al. Epigenetic and post-transcriptional regulation of somatostatin receptor subtype 5 (SST5) in pituitary and pancreatic neuroendocrine tumors. Mol Oncol. 2022;16(3):764–79. doi:https://doi.org/10.1002/1878-0261.13107.
Xue J, et al. TCF-3-mediated transcription of lncRNA HNF1A-AS1 targeting oncostatin M expression inhibits epithelial-mesenchymal transition via TGFbeta signaling in gastroenteropancreatic neuroendocrine neoplasms. Aging. 2021;13(10):14065–77. doi:https://doi.org/10.18632/aging.203024.
Di Mauro A, et al. Aberrant expression of Long non coding RNA HOTAIR and de-regulation of the paralogous 13 HOX genes are strongly Associated with Aggressive Behavior of Gastro-Entero-Pancreatic neuroendocrine tumors. Int J Mol Sci 2021; 22(13), doi:https://doi.org/10.3390/ijms22137049.
Fang S, et al. H3K27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR. Ann Transl Med. 2018;6(22):440. doi:https://doi.org/10.21037/atm.2018.10.21.
Narayanan D, et al. Long non-coding RNAs in pulmonary neuroendocrine neoplasms. Endocr Pathol. 2020;31(3):254–63. doi:https://doi.org/10.1007/s12022-020-09626-1.
Modali SD, et al. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol. 2015;29(2):224–37. doi:https://doi.org/10.1210/me.2014-1304.
Zhang YY, Feng HM. MEG3 suppresses human pancreatic neuroendocrine tumor cells growth and metastasis by Down-Regulation of Mir-183. Cell Physiol Biochem. 2017;44(1):345–56. doi:https://doi.org/10.1159/000484906.
Iyer S, et al. Long noncoding RNA MEG3 is an epigenetic determinant of Oncogenic Signaling in Functional pancreatic neuroendocrine tumor cells. Mol Cell Biol. 2017; 37(22), doi:https://doi.org/10.1128/MCB.00278-17.
Sun Y, et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16(1):162. doi:https://doi.org/10.1186/s12943-017-0729-1.
Sun Y, et al. Mir-574-5p mediates epithelial-mesenchymal transition in small cell lung cancer by targeting vimentin via a competitive endogenous RNA network. Oncol Lett. 2021;21(6):459. doi:https://doi.org/10.3892/ol.2021.12720.
Sun Y, et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018;9(2):85. doi:https://doi.org/10.1038/s41419-017-0113-5.
Zeng F, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate etk expression. Oncogene. 2020;39(2):293–307. doi:https://doi.org/10.1038/s41388-019-0984-2.
Wang D, et al. LncRNA LUADT1 sponges miR-15a-3p to upregulate Twist1 in small cell lung cancer. BMC Pulm Med. 2019;19(1):246. doi:https://doi.org/10.1186/s12890-019-0991-7.
Wang S, Yu J. Long non-coding RNA transcribed from pseudogene PPIAP43 is associated with radiation sensitivity of small cell lung cancer cells. Oncol Lett. 2019;18(5):4583–92. doi:https://doi.org/10.3892/ol.2019.10806.
Zhang Y, et al. SBF2-AS1: an oncogenic lncRNA in small-cell lung cancer. J Cell Biochem. 2019;120(9):15422–8. doi:https://doi.org/10.1002/jcb.28809.
Chen W, et al. BLACAT1 predicts poor prognosis and serves as oncogenic lncRNA in small-cell lung cancer. J Cell Biochem. 2018. doi:https://doi.org/10.1002/jcb.27548.
Fu Y, et al. LncRNA CASC11 promotes TGF-beta1, increases cancer cell stemness and predicts postoperative survival in small cell lung cancer. Gene. 2019;704:91–6. doi:https://doi.org/10.1016/j.gene.2019.04.019.
Wu L, Wang P. Long non-coding RNA-neighboring enhancer of FOXA2 inhibits the migration and invasion of small cell lung carcinoma cells by downregulating transforming growth factor-beta1. Oncol Lett. 2019;17(6):4969–75. doi:https://doi.org/10.3892/ol.2019.10152.
Zhu Y, et al. KCNQ1OT1 lncRNA affects the proliferation, apoptosis, and chemoresistance of small cell lung cancer cells via the JAK2/STAT3 axis. Ann Transl Med. 2021;9(10):891. doi:https://doi.org/10.21037/atm-21-1761.
Huang W, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging mir-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2020;27(5):1709–27. doi:https://doi.org/10.1038/s41418-019-0455-x.
Li L, et al. FLI1 Exonic Circular RNAs as a Novel Oncogenic driver to promote Tumor Metastasis in Small Cell Lung Cancer. Clin Cancer Res. 2019;25(4):1302–17. doi:https://doi.org/10.1158/1078-0432.CCR-18-1447.
Krol J, et al. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi:https://doi.org/10.1038/nrg2843.
Sadanandam A, et al. A Cross-Species Analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discov. 2015;5(12):1296–313. doi:https://doi.org/10.1158/2159-8290.CD-15-0068.
Yoshimoto T, et al. Pulmonary carcinoids and Low-Grade Gastrointestinal Neuroendocrine Tumors Show Common MicroRNA expression profiles, different from Adenocarcinomas and small cell carcinomas. Neuroendocrinology. 2018;106(1):47–57. doi:https://doi.org/10.1159/000461582.
Gilad S, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14(5):510–7. doi:https://doi.org/10.1016/j.jmoldx.2012.03.004.
Mao Y, et al. Bioinformatics analysis of mRNA and miRNA microarray to identify the key miRNAgene pairs in smallcell lung cancer. Mol Med Rep. 2019;20(3):2199–208. doi:https://doi.org/10.3892/mmr.2019.10441.
Bosch F, et al. Treatment with somatostatin analogs induces differentially expressed let-7c-5p and mir-3137 in small intestine neuroendocrine tumors. BMC Cancer. 2019;19(1):575. doi:https://doi.org/10.1186/s12885-019-5794-y.
Vicentini C, et al. Exosomal miRNA signatures of pancreatic lesions. BMC Gastroenterol. 2020;20(1):137. doi:https://doi.org/10.1186/s12876-020-01287-y.
Lu S, et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer. 2018;123:44–51. doi:https://doi.org/10.1016/j.lungcan.2018.06.027.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:https://doi.org/10.1016/j.cell.2018.01.011.
Lei Y, et al. Identification of alternative splicing and lncRNA genes in pathogenesis of small cell lung cancer based on their RNA sequencing. Adv Clin Exp Med. 2019;28(8):1043–50. doi:https://doi.org/10.17219/acem/94392.
Ji M, et al. Long noncoding RNA-mRNA expression profiles and validation in pancreatic neuroendocrine neoplasms. Clin Endocrinol. 2020;92(4):312–22. doi:https://doi.org/10.1111/cen.14156.
Liu Y, et al. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18(1):123. doi:https://doi.org/10.1186/s12943-019-1052-9.
Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91. doi:https://doi.org/10.1038/s41576-019-0158-7.
Huang ZH, et al. snoRNAs: functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discov. 2022;8(1):259. doi:https://doi.org/10.1038/s41420-022-01056-8.
Dvinge H, et al. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019;29(10):1591–604. doi:https://doi.org/10.1101/gr.246678.118.
Liu Y, et al. The genetic and pharmacogenomic landscape of snoRNAs in human cancer. Mol Cancer. 2020;19(1):108. doi:https://doi.org/10.1186/s12943-020-01228-z.
Wood KA, et al. The role of the U5 snRNP in genetic Disorders and Cancer. Front Genet. 2021;12:636620. doi:https://doi.org/10.3389/fgene.2021.636620.
Acknowledgements
This work has been supported by Spanish Ministry of Economy [MINECO; BFU2016–80360-R (to JPC)] and Ministry of Science and Innovation [MICINN; PID2019‐105201RB‐I00, AEI/https://doi.org/10.13039/501100011033 (to JPC)]. Instituto de Salud Carlos III, co‐funded by European Union (ERDF/ESF, “Investing in your future”) [Postdoctoral Grant Sara Borrell CD19/00255 (to AIC); Predoctoral contract FI17/00282 (to EAP)]. Society for Endocrinology Early Career Grant (to AIC). Spanish Ministry of Universities Predoctoral contracts FPU18/02275 (to R.B-E.) and FPU20/03958 (to V.G.V). Junta de Andalucía (BIO‐0139); FEDER UCO-202099901918904 (to JPC and AIC). Grupo Español de Tumores Neuroendocrinos y Endocrinos (GETNE2016 and GETNE2019 Research grants, to JPC). Fundación Eugenio Rodríguez Pascual (FERP2020 Grant to JPC). CIBERobn Fisiopatología de la Obesidad y Nutrición. CIBER is an initiative of Instituto de Salud Carlos III.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors report there are no competing interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Blázquez-Encinas, R., Moreno-Montilla, M., García-Vioque, V. et al. The uprise of RNA biology in neuroendocrine neoplasms: altered splicing and RNA species unveil translational opportunities. Rev Endocr Metab Disord 24, 267–282 (2023). https://doi.org/10.1007/s11154-022-09771-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11154-022-09771-4