Abstract
In this work, tuning oxygen tension was targeted to improve hydrogen evolution. To achieve such target, various consortia of the chlorophyte Coccomyxa chodatii with a newly isolated photosynthetic purple non-sulfur bacterium (PNSB) strain Rhodobium gokarnense were set up, sulfur replete/deprived, malate/acetate fed, bicarbonate/sulfur added at dim/high light. C. chodatii and R. gokarnense are newly introduced to biohydrogen studies for the first time. Dim light was applied to avoid the inhibitory drawbacks of photosynthetic oxygen evolution, values of hydrogen are comparable with high light or even more and thus economically feasible to eliminate the costs of artificial illumination. Particularly, the consortium of 2n− (n = 1.9 × 105 cell/ml, sulfur deprived) demonstrated its perfection for the target, i.e., the highest possible cumulative hydrogen. This consortium exhibited negative photosynthesis, i.e., oxygen uptake in the light. Most hydrogen in consortia is from bacterial origin, although algae evolved much more hydrogen than bacteria on per cell basis, but for only one day (the second 24 h), as kinetics revealed. The higher hydrogen in unibacterial culture or consortia results from higher bacterial cell density (20 times). Consortia evolved more hydrogen than their respective separate cultures, further enhanced when bicarbonate and sulfur were supplemented at higher light. The share of algae relatively increased as bicarbonate or sulfur were added at higher light intensity, i.e., PSII activity partially recovered, resulting in a transient autotrophic hydrogen evolution. The addition of acetic acid in mixture with malic acid significantly enhanced the cumulative hydrogen levels, mostly decreased cellular ascorbic acid indicating less oxidative stress and relief of PSII, relative to malic acid alone. Starch, however, decreased, indicating the specificity of acetic acid. Exudates (reducing sugars, amino acids, and soluble proteins) were detected, indicating mutual utilization. Yet, hydrogen evolution is limited; tuning PSII activity remains a target for sustainable hydrogen production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Algae, bacteria, and cyanobacteria are repeatedly studied to catalyze hydrogen evolution via their photosynthetic or fermentative pathways. Bacteria are frequently used to hydrolyze biomass and produce biohydrogen, photosynthetic PNSB (Danial et al. 2017; Abdel-Kader et al. 2022) or heterotrophic (Danial et al. 2015; Danial and Abdel-Basset 2015; Gad El-Rab et al. 2018). Algae-bacteria consortia, through various aspects linking their metabolism, may yield efficiently enhanced hydrogen production. Bacteria and algae can proceed integrative metabolism of photosynthesis and respiration (oxygen and carbon dioxide exchange), light spectra absorption and utilization of secreted metabolites, particularly acetic acid (Fakhimi et al. 2020). To induce hydrogen evolution swiftly, sulfur deprivation was developed and established in Chlamydomonas reinhardtii since Melis et al. (2000). This two-stage hydrogen evolution, at which oxygen evolution ceases along with starch accumulation/fermentation at a closed system, thus creating anaerobic conditions for hydrogen evolution. Such induction, however, is transient and typically lasts for up to a number of (3–4) days. A characteristic effect of sulfur limitation is the decline in the expression of many photosynthetic genes, encoding subunits of photosystem I (PSI), photosystem II (PSII) and ATPase, whereas the transcript levels of two particular antenna proteins, LHCBM9 and LHCSR1 are upregulated (Nguyen et al. 2011; Toepel et al. 2013). Response in gene expression occurs already after a few hours of sulfur deprivation (González-Ballester et al. 2008); during which most photosynthetic genes, Rubisco and antenna protein genes are downregulated (Toepel et al. 2013). The inactivation of PSII is assumed to be caused by the strongly oxidizing species Tyrz+ and P680+ upon illumination, leading to donor-side-induced photoinhibition, resulting in the relatively rapid degradation of PSII reaction center proteins, including PsbA, PSBO, CP43 and possibly several more subunits (reviewed by Antal et al. 2015; Nagy et al. 2018a). Upon sulfur deprivation, the expression level of the nuclear-encoded PSBO subunit, which normally has a relatively long lifetime (Nelson et al. 2014), is also downregulated (Toepel et al. 2013), possibly contributing to the decrease in PSII activity. The rapid loss of CP43 was unexpected; in the absence of this protein, PSII is destabilized (Sugimoto and Takahashi, 2003), which may also contribute to the loss of photosynthetic activity. Besides, Nagy et al. (2016, 2018a) proposed that with the overexcitation of PSII, 1O2 singlet oxygen in PSII are produced exerting oxidative stress, which triggers ascorbate biosynthesis; when accumulated to the mM concentration range, ascorbate may contribute to the inactivation of the Mn-cluster in the OECs (oxygen evolving complexes) due to its reducing capacities and may provide electrons to PSII, albeit at a low rate.
Sulfur deprivation also leads to strong starch and phosphatidylglycerol accumulation (Sugimoto et al. 2007, 2008) and the alteration of the cell wall structure (Takahashi et al. 2001). On the other hand, respiration is maintained, which, together with the loss of PSII activity, leads to the establishment of hypoxia, essential for the expression and activity of the highly O2-sensitive hydrogenases and nitrogenases (Ghirardi et al. 1997; Abdel-Basset and Bader 1997, 2008; Abdel-Basset et al. 1998; Milligan et al. 2007) which catalyze hydrogen evolution in algae and bacteria, respectively. The purple anoxygenic photosynthetic bacterium Rhodospirillum rubrum displayed lack of growth, cessation of bacteriochlorophyll and protein accumulation and inhibition of H2 evolution although most cells remained viable after 100 h of S-deprivation (Melis and Melnicki 2006; Melnicki et al. 2009). All of its nitrogenase functions including N2-fixation are promptly downregulated at multiple levels in response to S-deprivation, preventing hydrogen production. Instead, cell volume increased, and large amounts of polymer poly-b-hydroxybutyrate (PHB) were found to accumulate extracellularly (3.5-fold within 24 h of S-deprivation). Similarly, R. rubrum, as Rhodobacter sphaeroides and Rhodopseudomonas palustris exhibited a similar response to S-deprivation (Melnicki et al. 2009).
Despite such consequences of sulfur deprivation, cell sulfur decreased by only 25% (Nagy et al. 2016) and, furthermore, sulfur reserves (membrane sulfolipids) mobilize upon sulfur deprivation (Sugimoto et al. 2007, 2008, 2010).
Light and bicarbonate orchestrate photosynthetic oxygen evolution and sulfur re-addition might recover PSII to resume its activity to evolve oxygen. Other than carbon fixation in Calvin–Benson (CB) cycle and carbon concentrating mechanism (CCM) (Hong et al. (2016), bicarbonate affects PSII activity via Warburg’s effect (Govindjee and coworkers, reviewed in Shevela et al. 2012). Light intensity and quality have been studied in the field of hydrogen evolution (e.g., Jurado-Oller et al. 2015). Light might turn stressful not only to photosynthesis and hydrogen evolution by inducing high rates of oxygen evolution that inhibits hydrogenases but also to microbial life itself by production of singlet oxygen species. Down regulation is among the adaptive mechanisms that protects of PSII from photooxidation induced by high light intensity (Abdel-Basset et al. 2011).
Therefore, this work was planned to enhance the hydrogen evolution capacity of the green alga Coccomyxa chodatii SAG 216-2 and a local, newly isolated photosynthetic purple non-sulfur bacterium (PNSB) (Rhodobium gokarnense), via tuning oxygen content, the powerful inhibitor of the hydrogen evolving enzymes hydrogenases and nitrogenases. To approach the relevant oxygen partial pressure, several manipulations were applied: namely, algae-bacteria consortia, sulfur deprivation, light intensity, and organic acid (malic and acetic) supplementation. A relevant oxygen partial pressure that guarantees the balance between photosynthetic electron flow for proton reduction on the one hand but not inhibitory to the enzymes on the other hand is a highly complicated network of metabolic processes. Whether cessation of hydrogen evolution is reversible, attributed to cells death or nutrient depletion are questions to be answered in this work via supplementing more nutrients (bicarbonate or sulfur). Organic acids (malic or malic/acetic mixture) are supposed to enhance respiration of both organisms, thus, anaerobiosis in addition to electron supplementation. A minimum (dim) light intensity has been applied to avoid overreduction of the electron transport components and their subsequent deterioration as well as eliminating the costs of artificial light if applicable.
Materials and methods
Algal culture and growth
Cultures of the green alga C. chodatii SAG 216-2 (kindly offered by SAG, Germany) were cultivated under sterile conditions in modified Bold Basal medium (BBM, Bischoff and Bold 1963) at a temperature of 25 ± 2 °C, pH of 6.8 ± 0.1, illuminated continuously with white, fluorescent lamps (50 μmol/m2/s) and agitated by orbital shaker at 150 rpm for 7 days. Cultures of C. chodatii were subjected to sulfur deprivation by replacing sulfate salts in BBM by their analogous chlorides and designated throughout the whole manuscript as C+ and C− (sulfur replete and sulfur deprived, respectively).
Bacterial isolation, growth and characterization
Strains of photosynthetic PNSB were isolated from local ponds in Assiut University, Assiut, Egypt and grown anaerobically in RÄH medium (Biebl and Pfennig 1981) at 30 °C and a white light intensity of about 70 μmol/m2/s. After 1–2 weeks of incubation, a purplish red color developed in the medium and the PNSB colonies were characterized morphologically and biochemically in addition to molecular identification. They were analyzed macroscopically considering pigmentation, colony length and width; the colony size and shape were determined using light microscopy and the genotypic characterizations of the different isolates were assessed. The PNSB cultures “R” were designated throughout the whole manuscript as R+ and R− (sulfur replete and sulfur deprived, respectively).
Phylogenetic analysis of bacteria
The morphological, biochemical and molecular characteristics were used for strain characterization, according to Bergey’s Manual of Systematic Bacteriology (Brenner et al. 2005): bacterial strains were identified based according to the partial 1500 bp sequences of 16Sr RNA of the strains, and comparison in the GenBank databases.
Total genomic DNA was extracted and purified from the samples. The primer set of F (5-AGA GTT TGA TCC TGG CTC AG-3) with a GC clamp and R (5-GGT TAC CTT GTT ACG ACT T-3) at the annealing temperature of 65 °C were used for the PCR amplification of the variable region of 16S rDNA from the purified genomic DNA. Then, we made PCR clean up to the PCR product using GeneJET™ PCR Purification Kit (Thermo). Loading to 4 µl from the PCR mixture was carried out to examine the PCR product on 1% agarose gel against 1 Kb plus ladder (Fermentas). Finally, sequencing to the PCR product on GATC Company using ABI 3730xl DNA sequencer, forward and reverse primers were conducted. Sequence analysis was achieved by searching through online databases using BLAST. The phylogenetic analysis was performed using MEGA 3.1 software. The phylogenetic tree was constructed by the Neighbor Joining method. The sequences obtained were compared with available database sequences using a BLAST search.
Experimental set up for growth and hydrogen evolution
Various algal/bacterial consortia were set up by mixing different volumes of the green alga C. chodatii SAG 216-2 (C+/−) with the photosynthetic PNSB R. gokarnense (R+/−), either sulfur replete (+) or sulfur deprived (−). The consortia were subjected to sulfur deprivation by replacing sulfate salts by their analogous chlorides. These consortia are designated throughout the work as 1n+, 1n−, 2n+, 2n−, 3n+, 3n−, 4n+, 4n−; n indicates cell number of bacteria (1.8 × 105 cells/ml) and algae (0.085 × 105 cells/ml) giving together a final optical density of about 0.25 A at 0-time of cultivation. However, the consortium of 3n refers to or equals 2n(R) + n(C). The same O.D.750 nm was containing much less algal (1/20th) than bacterial cell numbers, due to the larger algal cell size. These consortia were followed for growth, hydrogen evolution and the related metabolic processes.
Bottles containing hydrogen evolving cocktails (10% phosphate buffer pH 6.8 ± 0.2, 10% a mixture of bacteria and algae at early log phase, 80% medium to obtain a total volume of 1 l) were stoppered and flushed with nitrogen for 15 min. They were then kept at a light intensity of 2 or 70 µmol/m2/s (tungsten lamp), 25 °C and shaken at 150 rpm as long as hydrogen was evolving. Two sets of experiments were identically set up, one for hydrogen collection and another set for tracing growth of algae and bacteria.
Hydrogen collection and detection
The gas evolved from the different culture cocktails was passed over NaOH solution (700 ml 1 M) to absorb carbon dioxide and then collected by replacing water in a water-filled graduated cylinder inverted in water. Gas samples at the head space of the fermentation bottles were checked for hydrogen purity using FID-TCD gas chromatography (Thermo Scientific TRACE GC Ultra, Germany).
Treatments
Algal and bacterial consortia were grown in BB medium that has been subjected to the following modifications and amendments:
Malic acid supplementation
Culture combinations, suspended in BBM, were enriched with malic acid at the same concentration of RÄH medium (Biebl and Pfennig 1981), as the basic medium to evolve hydrogen by the PNSB.
Malic and acetic acids supplementation
In addition to malic acid, acetic acid was added to BBM at 1 ml/l, the concentration conventionally supplemented in TAP medium for Chlamydomonas reinhardii.
Sulfur deprivation
Two sets of experiments, one sulfur replete (+S) and sulfur deprived (−S) of Coccomyxa codtatii, R. gokarnense and their consortia were studied at the above-described hydrogen evolution cocktails.
Bicarbonate addition
After cessation of hydrogen evolution (4 days), bicarbonate (sodium salt) at two concentrations of 5 and 10 mM were added to the cultures aiming to resume autotrophic metabolism and its impact on hydrogen evolution was recorded. Only the results of 10 mM bicarbonate are presented as they induced more hydrogen than 5 mM bicarbonate.
Sulfur re-addition
After cessation of hydrogen evolution induced by bicarbonate, sulfur (magnesium sulfate 0.30 mM), aiming to resume regular (sulfur replete) metabolism and its impact on hydrogen evolution.
Light intensity
The above-described experiments were performed at dim light of 2 µmol/m2/s (room light at winter) or higher light intensity of 70 µmol/m2/s (two tungsten lamps).
Analytical methods
Optical density and cell number
Optical density at 750 nm (O.D.750 nm) of all consortia was followed daily over the growth period. Cell number of algae was counted using an Improved Neubauer ruled hemocytometer.
Bacterial cell number counting
Using the dilution plate method, bacterial colonies developed after growth for 24–48 h at RÄH agar medium, were counted as the colony forming units (CFU).
Photosynthesis and respiration
The net photosynthetic oxygen evolution (PN) and dark respiratory oxygen uptake (RD) were monitored daily using a Clark type electrode computerized to an Oxygen Monitoring System (OMS, Hansatech Instruments Inc., donation from the Alexander von Humboldt Foundation Germany to R. Abdel Basset). Two milliliters of algal cultures and consortia were followed under continuous red-light intensity of 2 and 70 μmol/m2/s (growth lights) at 25 °C for 15 min; the rate of PN was calculated as nmole O2↑ µg−1 Chl/h. Respiration (RD) was also monitored as O2 uptake using the above system, but in the dark; the rate of RD was calculated as nmole O2↓ µg−1 Chl/h.
Estimation of metabolites
Chlorophylls and carotenoids assessment
Chlorophylls (a and b) and carotenoids contents were assessed in ethanol extracts according to Arvola (1981), calculated and expressed as µg/ml algal suspension according to Metzner et al. (1965).
Bacteriochlorophyll (B.Chl) assessment
Bacteriochlorophyll of the R. gokarnense were assessed substantially as in R. cupsulata following Sojka et al. (1970) in a series of dilutions of the bacterium intact cells based on that of Barer (1955) and Namsaraev (2009). Absorbances at 860, 660 and 775 nm were measured (in cuvettes with 1-cm light path). The A860–A660 differences were calculated and used to calculate B.Chl/ml of culture utilizing an extinction coefficient of 75 mM/cm (Clayton 1963); absorbances at 775 nm were treated the same way. Ordinary aqueous suspensions, ratios of peak-to-trough absorbances (e. g., 860 nm relative to 660 nm) are high (Sojka et al. 1970), suggesting that the light-scattering background might not contribute greatly to the total absorbance observed at the 860 nm. B.Chl peak. A correlation between cell number and bacteriochlorophyll was set up. Estimating bacteriochlorophyll in intact cells saves time, solvent costs, solvent hazards and protective from in vitro degradation.
Ascorbate assessment
Ascorbate assessment was performed substantially according Jagota and Dani (1982). Aliquots of 2 ml culture was sonicated for 2 min (Sonicator Bandelin Sonopuls HD 3200 homogenizer in Ultrasonic, Germany) in 5% TCA (trichloroacetic acid), then centrifuged at 3000 rpm for 5 min; the supernatant was used for ascorbate determination. Ascorbate contents were substantially assessed according Jagota and Dani (1982). A volume of 0.2 ml of the supernatant, 0.8 ml of 10% TCA, 1 ml H2O and 0.2 ml Folin (diluted) were mixed well and left for 10 min; the developed blue color was read at 760 nm using spectrophotometer.
Starch determination
At the end of the growth period, 2 ml of the cultures were boiled in glass tubes containing ml ethanol (80%) for 2 h.; after centrifugation, the supernatants were decanted. The collected pellets were resuspended in 5 ml of diluted perchloric acid solution (9.2 mM) for 30 min at 100 °C to dissolve starch. After centrifugation the supernatant was used for determining reducing sugars resulting from starch hydrolysis following the procedure of Miller (1959).
Cell exudations
Batches of the green alga and the bacterium consortia were grown at conditions identical to those of hydrogen photoevolution i.e., for 7 days at light intensity of 2 μmol/m2/s, once at aerobic and another at anaerobic conditions. In unialgal, unibacterial and Co-culture exudates, reducing sugars, amino acids and soluble proteins were determined as exudates into the growth media (50 ml); separated from cells by centrifugation (2000×g). The reducing sugars were determined following the procedure of Miller (1959); amino acids according to the method of Moore and Stein (1948) and soluble protein contents using the method of Lowry et al. (1951), using egg albumin as the standard protein.
Statistical analysis
Each experiment was repeated three times and the mean values of three replicates ± standard error (se) are presented. Statistical analysis of the data was conducted using ANOVA one-way test (analysis of variance) by SPSS program version 21, and Duncan values were determined at 0.05 level. Different letters (a–f) on the graphs indicate significant differences between the treatments.
Results
Identification of the bacterium
Morphological and biochemical characteristics of bacterial colonies are presented in Table 1 and the phylogenetic affiliation of the genetic material is presented in Fig. 1. According to the molecular analysis, the phylogenetic tree indicated that the strain exhibited 99% nucleotide base homology with the data in gene bank designating the strain as the PNSB R. gokarnense, the accession number of the herein studied strain is ON186790.
Growth
Rhodobium gokarnense was combined with the green alga C. chodatii to set up consortia containing successively increasing cell number (1n+, 1n−, 2n+, 2n−, 3n−, 3n−, 4n+, 4n−), in addition to the unialgal (C+/−) and unibcterial (R+/−) cultures; “n” refers to the cell number of 0.085 × 105 algal cells/ml mixed with 1.8 × 105 bacterial cells/ml giving a final O.D. of about 0.25 A at zero time of cultivation. Cultures and cocultures were followed for their growth, hydrogen evolution capacities and the cellular metabolism affecting this capacity. Growth (O.D.750 nm and cell number), light-induced oxygen evolution (photosynthesis), oxygen uptake in the dark (respiration). Cellular contents of chlorophyll, carotenoids, ascorbic acid and starch in addition to cellular exudations (reducing sugars, amino acids, soluble proteins) into the growth media were assessed.
Figure 2 shows growth as the increase in optical density (O.D.750 nm) of the unialgal culture (C), unibacterial culture (R), and the bacterial/ algal combinations. In cultures supplemented with malate as the sole carbon source (Fig. 2a), the optical density was increasing up to its highest values at 72–96 h old cultures, decreased thereafter. The highest O.D. was exhibited at 2n− culture, among all cultures or culture combinations before leveling off at 120 h. The unialgal culture of C. chodatii, either replete (C+) or deprived of sulfur (C−), displayed the lowest O.D. However, when acetate and malate were both supplemented to the consortia (Fig. 2S.c), the top optical density was exhibited at 4n+ followed by 4n− and R−. The addition of bicarbonate, followed by sulfur at higher light intensity of 70 µmol/m2/s was applied to selected cultures of R−, C−, 2n− and 4n− representative to the most pronounced treatments. Upon addition of bicarbonate (10 mM) followed by re-addition of sulfur, cultures recommenced growth and the optical density of cultures (except R−) was transiently enhanced for the next 24 h only, following each addition, i.e., at 96 h and 192 h, respectively, before leveling off again at 216 and 240 h., respectively (Fig. 2b).
Optical density (O.D. 750 nm) of variously combined consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii grown at sulfur replete (+) or deprived (−) conditions; a supplemented with malate in dim light, S.c with malate/acetate mixture in dim light, b with malate/acetate, bicarbonate addition (green arrow) and sulfur re-addition (yellow arrow) in high light; n indicates cell number of bacteria (1.8 × 105 cells/ml) combined with algae (0.085 × 105 cells/ml); algae/bacteria ratio is 1/20
Cell numbers of bacteria and algae exhibited their highest values mostly at 48–72 h old cultures, at sulfur deprived or replete combinations; thereafter, cell number was mostly decreasing with the lapse of time as shown in Fig. 3a–f . Relying on malate only, bacterial cells exhibited their highest numbers at 4n− and 2n− consortia, higher than the unibacterial cultures indicating synergism with algae (Fig. 3a). However, algal cells displayed the highest values at 4n+/−; 4n−, which is promptly decreased just after 24 h (Fig. 3c). Acetic acid mostly did not enhance bacterial (Fig. 3S.e) nor algal (Fig. 3S.f) cell multiplication relative to malic acid but enhanced decline in bacteria. Bacterial cells did not respond to the addition of bicarbonate, sulfur or high light intensity (Fig. 3b) while growth of algae (Fig. 3d) resumed its increase after each addition.
Cell number (Cell/ml) of variously combined consortia of the purple non-sulfur bacterium Rhodobium gokarnense (a, 3S e, b) and the green alga Coccomyxa chodatii (c, 3Sf, d) grown at sulfur replete (+) or deprived (−) conditions; supplemented with malate (a, c), with malate/acetate mixture (3Se, 3Sf), and bicarbonate (green arrow) and sulfur re-addition (yellow arrow) (b, d)
Chlorophylls (a, b) were assessed daily; to conserve space and get a clear conclusion, only the 0-time and the third (last) day for each experiment are shown in Fig. 4. At malate (Fig. 4a), chlorophylls increased at all consortia and in the unialgal Coccomyxa cultures; 4n+ exhibited the highest chlorophyll contents among all cultures, due to the highest cell number inoculum. Sulfur deprivation decreased chlorophyll contents in all combinations, except 3n− was slightly and 2n− was remarkably enhanced relative to their respective sulfur replete cultures. Acetate with malate induced similar chlorophyll increments with time but with lower magnitude of increase (Fig.4 S.e). Bicarbonate addition induced a sharp increase in chlorophyll contents particularly at C− and 2n− while sulfur re-addition did not (Fig. 4b). The relationship between bacteriochlorophyll assessed by absorbance of intact cells at 775, 860 and 660 nm and the cell number is presented in Fig. 4c. It shows a quantity of 4–5 µg/106 cells of the PNS bacterium R. gokarnense. The absorbance of intact cells at 775 nm, multiplied by molecular weight showed more linearity with cell number (R2 = 0.957) than the difference between 860 and 660 nm (R2 = 0.907). Carotenoid contents at malate, malate/acetate and bicarbonate/sulfur addition were determined daily and shown in Fig. 4Sf, 4Sg. In malate only, R. gokarnense exhibited the highest contents (Fig. 4Sf) that is approached in malate/acetate only in consortia of 2n− and 4n +/− (Fig. 4Sg). Bicarbonate addition extremely enhanced carotenoid contents while sulfur entirely abolished such enhancement (Fig. 4d).
Pigment contents of variously combined consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii grown at sulfur replete (+) or deprived (−) conditions; chlorophyll (a + b) contents in cultures supplemented with malate (a), with malate/acetate mixture (4Se), and bicarbonate and sulfur re-addition (b), bacteriochlorophyll (c) and carotenoids (4Sf, 4Sg, d); Patterns  refer to 0-time, 3rd-, 7th-, 7th day with HCO3−, and 7th day with HCO3− and sulfur re-addition; respectively
refer to 0-time, 3rd-, 7th-, 7th day with HCO3−, and 7th day with HCO3− and sulfur re-addition; respectively
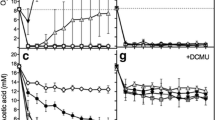
The net photosynthetic oxygen evolution (PN) is shown in Fig. 5. In malate supplemented cultures at dim light, PN was detectable in most cultures but in very low values with negative values in culture combinations of 3n−, 2n− and 4n− consortia Fig. 5a. Addition of acetate (Fig. 5Sc) absolutely inhibited net oxygen evolution in all cultures with high negative values recorded at the third day in n−, 2n− and 4n− consortia. Negative oxygen values means that respiration of both the bacterium and the alga consumed all the photosynthetically evolved oxygen in these cultures or culture combinations. Under the higher light intensity of 70 µmol/m2/s, the addition of bicarbonate and sulfur induced positive oxygen values at unialgal cultures of Coccomyxa (C−) whereas consortia remained of negative net photosynthesis (Fig. 5b). Respiratory oxygen uptake was enhanced by time, 3rd day vs 1st day in most sulfur deprived consortia supplemented with malate relative to sulfur replete, the highest at 3n followed by R− and 2n− (Fig. 6a). Respiratory oxygen uptake was higher in all cultures supplemented with malate/acetate than in malate alone (Fig. 6Sc) The addition of bicarbonate and sulfur did not accelerate respiration rates of unibacterial (R), unialgal (C), 2n− consortium but enhanced only at 4n− (Fig. 6b).
Net photosynthetic oxygen evolution (PN) as nmol O2 µg−1 Chl/h of consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii grown at sulfur replete (+) or deprived (−) conditions; supplemented with malate (a), with malate/acetate mixture (5Sc), and bicarbonate and sulfur re-addition (b); the same patterns in Fig. 4 are identically applicable in this figure
Dark respiratory oxygen uptake (RD) as nmol O2 µg−1 Chl/h of consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii grown at sulfur replete (+) or deprived (−) conditions; supplemented with malate (a), with malate/acetate mixture (6Sc), and bicarbonate and sulfur re-addition (b); the same patterns in Fig. 4 are identically applicable in this figure
Hydrogen evolution of the variously combined and treated cultures is presented in Fig. 7 for the cumulative values, Fig. 8 for the kinetics of evolution and Fig. 9 for the specific activity of algal and bacterial cells. Figure 7a shows the cumulative hydrogen evolution in malate-supplemented cultures at dim light. At such conditions, all sulfur deprived cultures were of higher hydrogen evolving capacity than the sulfur replete counterparts; the biggest volume of evolved hydrogen was recorded at the 2n− culture, followed by R− culture. The 4n− cultures, although containing double cell number, evolved only 50% hydrogen compared with the 2n−; the opposite occurred in sulfur replete cultures, i.e., 4n+ > 2n+. When acetic acid was supplemented in a mixture with malic acid to the cultures, hydrogen evolution exhibited almost an identical trend to that of malate only but enhanced to higher volumes (Fig. 7Sc). Also, its period of evolution was extended to 144 h instead of 72 h in the case of malate alone. The culture consortium 2n− remained the highest hydrogen evolving combination among all consortia, followed by R−. In malate/acetate fed cultures, 2n− evolved 830 ml H2/culture that is considerably higher than malate only (574 ml H2/culture), which indicates that acetate/malate synergistically enhanced hydrogen evolution more efficiently in consortia than in uni-cultures. Figure 7b shows hydrogen evolution in sulfur deprived unibacterial—(R−), unialgal (C−) cultures as well as in 2n− and 4n− cultures but at a higher light intensity of 70 µmol/m2/s. At such conditions, bicarbonate supplementation induced recommencement of hydrogen evolution at all cultures to higher cumulative levels. Sulfur readdition further enhanced hydrogen evolution. Also, the consortium 2n−, as usual, exhibited the highest activity, followed by 4n− while the unibacterial culture R− was also enhanced by higher light. However, the unibacterial cultures did not respond to bicarbonate supplementation nor to sulfur readdition (Fig. 7b).
Cumulative hydrogen evolution as ml/l culture of consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii at sulfur replete (+) or deprived (−) conditions; a supplemented with malate, (7Sc) with malate/acetate mixture, and b bicarbonate (yellow arrow) and sulfur re-addition (green arrow)
Figure 8 displays the kinetics of hydrogen evlution by uni-cultures and consortia. In most cultures, the major part of hydrogen was evolved within the first 24 h, continued to evolve at minor rates for the next two days, i.e. total of 72 h but at a sharply decreasing attitude; this applies to malate (Fig. 8a) or malate/acetate fed cultures (Fig. 8Sc) at dim light. While the unibacterial PNSB culture R. gokarnense (R−) evolved its major portion of hydrogen at day 1; continued for two more days but at considerably lower rates, the unialgal culture C. chodatii (C−), as always, evolved all its hydrogen at day 2 only, in -malate or malate/acetate-fed cultures (Fig. 8S). Hydrogen evolution usually ceases at 48–72 h old cells in most cultures. Howevet, at the higher light intensity, addition of bicarbonate and sulfur recpmmenced hydrogen evolution for more or two more days, depending on the type of culture or consortium (Fig. 8b). The unialgal culture, remained strict to evolve hydrogen over the second 24 h only even at additional bicarbonate and sulfur. Another form of hydrogen kinetics is presented in Fig. 8′Sa–c in the supplementary file.
Kinetics of hydrogen evolution as ml H2/day per culture of consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii at sulfur replete (+) or deprived (−) conditions; a supplemented with malate, (8Sc) with malate/acetate mixture, and b bicarbonate (yellow arrow) and sulfur re-addition (green arrow)
Specific hydrogen evolution as ml H2 106 cell/day revealed that the algal cells of C. chodatii are much more efficient in hydrogen evolution capacity than per cell of the purple non-sulfur R. gokarnense either in malate (Fig. 9a), malate/acetate (Fig. 9Sc) or when bicarbonate or sulfur were provided (Fig. 9b).
Specific hydrogen evolution as ml H2 106cells/day of consortia of the purple non-sulfur bacterium Rhodobium gokarnense and the green alga Coccomyxa chodatii at sulfur replete (+) or deprived (−) conditions; a supplemented with malate (9Sc) with malate/acetate mixture, and b bicarbonate (yellow arrow) and sulfur re-addition (green arrow)
Ascorbic acid in the bacterial strain R. gokarnense exhibited generally higher content than the alga C. chodatii (Table 2). The addition of acetic acid mostly reduced cellular contents of ascorbic acid indicating less oxidative stress relative to malic acid alone. No characteristic attitude of ascorbic acid among the various combinations or due to sulfur deprivation.
Starch content was higher in acetic/malic supplemented cultures relative to malic acid alone (Table 2), without obvious difference among the different combinations or sulfur national status, replete or deprived.
Exudates of reducing sugars, amino acids and soluble proteins from the bacterium R. gokarnense, the alga C. chodatii and their consortia were detected and presented in Table 3. Reducing sugars were almost of similar values at the various cultures or combinations. Amino acids exhibited their lowest values at unialgal cultures and their highest values in unibacterial cultures while the consortium 2n− was in between but closer to the unibacterial contents. Sulfur deprivation slightly enhanced amino acids leakage in R− and C− compared with sulfur replete R+ and C+ but the opposite was found in 2n−. Soluble proteins exhibited a trend similar to that of amino acids.
Discussion
The capacity of photosynthetic or fermentative hydrogen evolution by microorganisms (algae, cyanobacteria and bacteria) is, certainly constrained by various metabolic barriers; above which, oxygen is a powerful inhibitor of the hydrogen evolving enzymes (hydrogenase and nitrogenase). Accordingly, it is difficult to approach the theoretical hydrogen:oxygen ratio of 2:1 via PSII-photolysis of water or in sugars during fermentation. In this work, a number of manipulations tuning cultures’ oxygen content were applied seeking a breakthrough in enhancing the magnitude of hydrogen evolution. Namely, various consortia of the newly isolated PNSB strain R. gokarnense ON186790 “R” and the green alga C. chodatii “C” were set up; both microorganisms are explored for the first time in biohydrogen production field. These consortia are studied at sulfur deprivation/re-addition, malate and malate/acetate supplementation, dim/high light intensities and bicarbonate addition. Consortia grow compatibly in inorganic medium enriched with malic acid, but more with malic/acetic acids. They evolved hydrogen amounts more than their separate cultures, indicating the mutual synergism in supporting growth and hydrogen evolution of each other (R. gokarnense on C. chodatii and vice versa, i.e., algae on bacteria). In particular, the consortium 2n− (n = 1.9 × 105 cell/ml, 1:20 (Coccomyxa:Rhodobium), sulfur deprived) demonstrated its perfection with the planned aim of this work, i.e., the highest possible cumulative hydrogen evolution level more than their respective monocultures or other consortia. Such consortium is characterized by negative net photosynthesis in the light, i.e., net photosynthetic oxygen evolution was inhibited and turned oxygen uptake in the light, by respiration of both organisms. Algae and bacteria at this combination (2n−) may be at their ideal proportions to economize oxygen evolution for hypoxia with electron transport in favor of hydrogen evolution, as none of the other consortia was competent with this consortium regarding the hydrogen evolution values. The consortium 4n− resulted in less hydrogen evolution, may be due to the positive oxygen content, which started to accumulate in this consortium by the more algal cells than in 2n−. Such findings at 2n− and 4n− are indicative for synergized oxygen tuning and hydrogen evolution metabolism, same as in growth. In accordance with this, Ban et al. (2018) reported that in co-cultures of Pseudomonas sp. strain D and C. reinhardtii, the relative O2 content in the headspace plus the dissolved oxygen in the culture medium were rapidly consumed by bacterial growth, resulting in a completely anaerobic environment. These findings clue that a balanced cell number of bacteria and algae is needed for sustainable hydrogen evolution by consortia.
Kinetics of hydrogen evolution displayed that the consortia 4n− and 2n− as well as the bacterial culture R− evolved their major portion of hydrogen on the first 24 h, continued to evolve hydrogen for the next two days, i.e. 72 h but at sharply decreasing rates before ceasing. However, the sulfur deprived green alga Coccomyxa (C−) did not evove any hydrogen at the first day under any circumstance of nutrition (malate or malate/acetate), in single culture or combination; it evolved all its hydrogen over the second 24 h, i.e., at day 2 only, with a lag of the first 24 h before hydrogen becomes detectable. However, addition of bicarbonate followed by sulfur at the high light intensity recommenced hydrogen evolution for one or two more days, depending on the type of consortium, i.e. the proportion of algae to bacteria, which might lead to inhibitory concentrations of oxygen.
The specificity of hydrogen evolution as ml H2/ 106 cell/ day, of C. chodatii, is characterized by three phenomena: (a) much higher hydrogen evolution per cell of C. chodatii than per cell of R. gokarnense either in malate, malate/acetate or when bicarbonate or sulfur were provided, dim or high light intensity (b) hydrogen evolution for only 24 h and (c) a lag of 24 h before hydrogen becomes detectable. However, the higher productivity, i.e., higher cumulative hydrogen per culture resulted from bacteria (80–90%) due to higher cell density (20 times) than that of the alga, durability period, which is 2–3 days in bacteria whereas only one day in algae, in addition to an inferred integrative metabolism. If the algal cells are increased, photosynthesis concomitantly increased and oxygen tension might become inhibitory; this happened at 4n−. The share of R. gokarnense equals 2.5 times hydrogen that of the unialgal C. chodatii cultures in malate fed cultures, increased to 4.5 times with acetate. Specific hydrogen increased with light intensity (Kim et al. 2006a, b; Park and Moon 2007; Markov et al. 2006; Vijayaraghavan et al. 2010).
Starch contents of consortia were higher at acetic/malic supplementation relative to malic acid alone, without obvious differences among the different consortia or sulfur nutritional status, replete or deprived. Starch accumulation/degradation is a common feature of sulfur deprivation since discovered in C. reinhardtii by Melis et al. (2000). Also, co-culturing bacteria such as Bradyrhizobium japonicum (Xu et al. 2016), Azotobacter chroococcum (Xu et al. 2017), Pseudomonas sp. (Ban et al. 2018) and Thuomonas intermedia (He et al. 2018) favors high starch accumulation in Chlamydomonas. The fermentative bacteria such as Lactobacillus amylovorus, Vibrio fluvialis and Clostridium butyricum can degrade the Chlamydomonas biomass and excrete organic acids such as ethanol, formate, acetate, propionate and butyrate, which can be used by the partner-in consortia PNSB Rhodobacter sphaeroides, Rhodobacter capsulata, Rhodospirillum rubrum and Rhodobium marinum (Kawaguchi et al. 2001; Ike et al. 1997), Rhodobacter sphaeroides (Kim et al. 2006a, b) to photoproduce H2 via photo-fermentation. Starch-enriched Chlamydomonas biomass can be also used directly by some heterotrophic bacteria to produce H2 or in collaboration with photosynthetic PNS bacteria (Fakhimi et al. 2020). Respiratory or fermentative metabolism of sugars, derived from starch via glycolysis, supplies a significant number of electrons to the PQ-pool via the plastidial type II NAD(P)H dehydrogenase (NDA2) complex and thereby for the expressed hydrogenase, i.e. PSII-independent hydrogen evolution (Mignolet et al. 2012; Volgusheva et al. 2013; Hong et al. 2016). However, when linear electron transport is limited, neither efficient starch degradation nor high hydrogenase activity would result in strong H2 production (Nagy et al. 2016). Starch metabolism, besides being a source of electrons, accelerates oxygen uptake and thus hypoxia installation. Also, Assimilation of acetic and malic acid also participate in oxygen consumption and connected to H2 production, similar to former findings (Jurado-Oller et al. 2015; Gibbs et al. 1986; Bamberger et al. 1982; Fakhimi et al. 2020). Acetic acid probably being the metabolite linking dark H2 production with H2 photoproduction (Fakhimi et al. 2019, 2020), although the theoretical values of acetic acid-based yield are not usually reached due to different limitations as the use of photosynthetic bacteria in such integrative systems often requires two-stage bioreactors due to the growth incompatibility (Hallenbeck and Liu 2016). However, acetate uptake is greatly dependent on oxygen availability; low levels of oxygen allow for low acetate uptake rates, but paradoxically, lead to efficient and sustained production of hydrogen (Jurado-Oller et al. 2015). Organic exudates may be mutually excreted and exchanged for growth and hydrogen evolution. In this respect, Fakhimi et al. (2020) depicted some metabolites that can be potentially exchanged between algae and other microorganisms during growth and H2 production conditions. In this work, reducing sugars, amino acids and soluble proteins were detected in consortia of C. chodatii and R. gokarnense. Secreted end products can be theoretically used by bacteria as electron donors for H2 production, and some of them have been probed at an empirical level using Chlamydomonas–PNSB bacteria cultures (Miyamoto et al. 1987; Miura et al. 1992).
Light, bicarbonate and oxygen
Light, bicarbonate and sulfur are prime factors orchestrate the highly complicated process of photosynthetic oxygen evolution from PSII activity by different mechanisms. Direct photosynthetic (PSII → PSI → FDX1 → HYDA1), indirect photosynthetic (PQ → PSI → FDX1 → HYDA1) and photofermentative (P(OA)FR → PSI → FDX1 → → HYDA1) are hydrogen evolution pathways described in C. reinhardtii by González-Ballester et al. (2017); PFR is Pyruvate Ferredoxin Reductase, HYDA1 is the primary hydrogenase, OA is oxaloacetate, FDX1 is ferredoxin 1. In sulfur deprived cells, the electrons feeding H2 production originate mostly from the remaining PSII activity (Chochois et al. 2009), which decreased by only 25% (Volgusheva et al. (2013), concomitant with 25% decrease in cellular sulfur content (Nagy et al. 2018a). The electron transport from PSII was completely blocked during the anaerobic phase preceding H2 formation; hydrogenase partly removes this block, thereby permitting electron flow from water oxidation to hydrogen (Nagy et al. 2018a). Meanwhile, sulfur reserves such as sulfolipid, mostly in chloroplast membrane in the form of sulfoquinovosyl diacylglycerol (SQDG), mobilize upon sulfur deprivation, as a major internal sulfur source for protein synthesis at the early phase of sulfur starvation in Chlamydomonas reinhardti (Sugimoto et al., 2007, 2010). Based on these observations, therefore, it is very likely that the quantity of sulfur per photosynthetic reaction center decreases only moderately and may not substantially hinder PSII structure and activity. In this respect, Abdel-Basset and Bader (1998) reported that the PSII-blocker DCMU was inhibitory only at early and transition state of installing anaerobiosis. In this work, net photosynthetic oxygen evolution was only detectable in few cultures fed with malate, absolutely undetectable upon addition of acetic acid; otherwise, it was negative, i.e., respiration of consortia consumed all oxygen and established anoxic conditions necessary for hydrogen evolution. In this work, the major part of the experiments was conducted at dim light of only 2 µmol/m2/s. Dim light was applied to secure a continuous flow of electrons for proton reduction and simultaneously avoid high light intensity drawbacks, substantially the inhibitory effect of enhanced photosynthetic oxygen evolution on the hydrogen evolving enzymes (nitrogenase and hydrogenase). Drawbacks include, in addition, photooxidation and overexcitation of the redox components, oxidative stress and singlet oxygen species, and down regulation of PSII of the green alga C. chodatii as well as in its consortia with the PNSB R. gokarnense. In this respect, low oxygen levels at low light intensities in acetate‑containing media were found to elicit and improve photohydrogen production in mixotrophic non-stressed nutrient-replete Chlamydomonas cultures (Jurado-Oller et al. 2015), in a slow, continued, but sustained level of H2 production. Tsygankov et al. (2006) demonstrated possible sustained H2 photoproduction by a sulfur-deprived C. reinhardtii, under strictly photoautotrophic conditions without acetate or any other organic substrate; pre-grown with 2% CO2 under low light conditions along with a special light regime. Applying higher light intensity of 70 µmol/m2/s to the studied consortia of C. chodatii and R. gokarnense recommenced hydrogen evolution but transiently for one or two more days before ceasing again, and to a less level than the original first stage at dim light of 2 µmol/m2/s, most probably due to oxygen accumulation. In the literature, Various hydrogen evolution maxima were recorded in the literature in relation to light intensity, alternating dark/light, intermittent light or light spectra even with the same C. reinhardtii (Laurinavichene et al. 2004). Using ‘‘gain-of-function’’ mutations, Forster et al. (2005) isolated at least four such very high light resistant (VHLR) mutations in C. reinhardtii, that permit near maximal growth rates at light intensities lethal to wild type.
The addition of bicarbonate, also, induced growth recommencement of C. chodatii and R. gokarnense, but transiently over another one more day, although it did not attain the original levels of the first 48 h before finally ceasing again. Bicarbonate or sulfur addition was accompanied with positive oxygen values, i.e., the cultures become autotrophic and the oxic environment started to establish, i.e., a transient autotrophic hydrogen evolution occurred before inhibition by photosynthetic oxygen. Inorganic carbon (CO2 or bicarbonate) plays three roles by three different mechanisms in oxygenic photosynthesis: CO2 fixation into carbohydrates (Calvin cycle), CCM and Warburg’s effect (the role of bicarbonate in maintaining PSII activity). Hong et al. (2016) recorded that during H2 production phase of S deprivation, the concentrations of starch and H2 in CCM-induced cells were remarkably enhanced by 65.0% and 218.9% compared to that of CCM-uninduced cells, respectively. Warburg’s effect is a unique stimulatory role of CO2 in the Hill reaction (i.e., O2 evolution) has been studied by Govindjee and his coworkers (reviewed in Shevela et al. 2012). Wydrzynski and Govindjee (1975) discovered a definite bicarbonate effect on the electron acceptor (the plastoquinone) side of PSII. Nagy et al. (2018b), by applying a simple catalyst (glucose and glucose oxidase) to remove the evolved O2, enabled cultures to remain photosynthetically active for several days, keeping the Calvin–Benson cycle inactive by substrate limitation and preserving hydrogenase activity, with the electrons feeding the hydrogenases mostly derived from water. The amount of H2 produced is higher as compared to the sulfur-deprivation procedure and the process is photoautotrophic, ascorbic acid in R. gokarnense exhibited generally higher content than C. chodatii. No characteristic attitude of ascorbic acid content among the various consortia or due to sulfur deprivation. The addition of acetic acid, mostly reduced cellular contents of ascorbic acid indicating less oxidative stress or more relief of PSII, relative to malic acid alone. Simultaneous with reduced ascorbic acid, the cumulative hydrogen evolution, significantly enhanced. The imposed oxidative stress early on sulfur deprivation, most probably involving the formation of singlet oxygen (1O2) in PSII, which leads to an increase in the expression of GDP-l-galactose phosphorylase, exerting oxidative stress playing an essential role in ascorbate biosynthesis (Nagy et al, 2016, 2018a). In the absence of a functional donor side and sufficient electron transport, PSII reaction centers are inactivated and degraded. When accumulated to the mM concentration range, ascorbate may contribute to the inactivation of the Mn-cluster in the OECs (oxygen evolving complexes) due to its reducing capacities and may provide electrons to PSII, albeit at a low rate. The inactivation of PSII is, therefore, a complex and multistep process, which may serve to mitigate the damaging effects of sulfur limitation (Nagy et al. 2018a). Finally, At the end of the H2-producing period, sulfur limitation may restrain cell growth and viability; the entire cell system starts to degrade, and genes related to apoptosis and protein degradation are upregulated (Toepel et al. 2013).
Conclusions
-
Bacterial or algal cells were of higher numbers in consortia than in their uni-cultures, indicating no inhibitory, antagonistic or allelopathic but rather the supportive synergistic effect of algae on bacterial growth and vice versa, i.e., bacteria on algae.
The consortium revealed full perfection for the highest hydrogen evolution level is 2n− (cell density of 1.9 × 105 cell/ml sulfur deprived of the Chlorophycean Cocomyxa chodatii and the PNSB R. gokarnense at a ratio of 1:20).
-
Dim light seemed economically feasible alternative to high light, as the values of hydrogen are comparable or even more and the costs of artificial illumination can thus be eliminated.
All hydrogen evolved in this work is photofermentative; malate but more malate/acetate mixtures enhanced hydrogen evolution of algal and bacterial consortia at dim light.
-
Cells of the green alga C. chodatii evolved the highest hydrogen per cell compared with the bacterial cells but for only 24 h, with a one-day lag.
-
Bicarbonate or sulfur addition resumed hydrogen evolution for additional 24–48 h, indicating elasticity of the process and viability of the cells.
-
Yet, algal/bacterial (cyanobacterial) consortia are worth further exploration as their variables are many, e.g., different strains, cell number, proportions, light intensity and spectra, different organic acids, manipulating autotrophic vs heterotrophic life and nitrogenase/hydrogenase optimization, etc., which would improve hydrogen yield.
References
Abdel-Basset R, Bader KP (1998) Physiological analyses of hydrogen gas exchange in cyanobacteria. J Photochem Photobiol B Biol 43(2):146–151
Abdel-Basset R, Bader KP (2008) Hydrogen evolution in relation to PSI-reducible substrates in the cyanobacterium Oscillatoria chalybea assayed by means of mass spectrometry. Int J Hydrogen Energy 33:2653–2659
Abdel-Basset R, Spiegel S, Bader KP (1998) Saturation of cyanobacterial photoevolution of molecular hydrogen by photosynthetic redox components. J Photochem Photobiol B Biol 47:31–38
Abdel-Basset R, Friedl T, Mohr KI, Rybalka N, Martin W (2011) High growth, photosynthesis rate and increased hydrogen(ases) in manganese deprived cells of a newly isolated Nostoc-like cyanobacterium SAG2306. Int J Hydrogen Energy 36:12200–12210
Abdel-Basset R, Bader KP (1997) Characterization of hydrogen photoevolution in Oscillatoria chalybea detected by means of mass spectrometry. Z Nat 52c:775–781
Abdel-Kader HAA, Abdel-Basset R, Danial AW (2022) Yeast and enzymatic hydrolysis in converting Chlorella biomass into hydrogen gas by Rhodobacter sp. and Rhodopseudomonas palustris. Int J Hydrogen Energy 47:1516–1528
Antal TK, Krendeleva TE, Tyystjärvi E (2015) Multiple regulatory mechanisms in the chloroplast of green algae: relation to hydrogen production. Photosynt Res 125:357–381
Arvola L (1981) Spectrophotometric determination of chlorophyll-a and phaeopigments in ethanol extractions. Ann Bot Fenn 18:221–227
Bamberger ES, King D, Erbes DL, Gibbs M (1982) H2 and CO2 evolution by anaerobically adapted Chlamydomonas reinhardtii F-60. Plant Physiol 69:1268–1273
Ban S, Lin W, Wu F, Luo J (2018) Algal-bacterial cooperation improves algal photolysis-mediated hydrogen production. Bioresour Technol 251:350–357
Barer R (1955) Spectrophotometry of clarified cell suspensions. Science 121:709–715
Biebl H, Pfennig N (1981) Isolation of members of the family Rhodospirillaceae. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG (eds) The prokaryotes: a handbook on habitats, isolation, and identification of bacteria, vol 1. Springer, New York, pp 267–273
Bischoff HW, Bold HC (1963) Phycological studies IV. Some soil algae from enchanted rock and related algal species, vol 6318. University of Texas, Austin, pp 1–95
Brenner DJ, Kreig N, Staley JT, Garrity George M (eds) (2005) Bergey’s manual of systematic bacteriology, 2nd edn, vol. 2, Part A introductory essays. Springer, New York. https://doi.org/10.1007/0-387-28021-9
Chochois V, Dauvillée D, Beyly A, Tolleter D, Cuiné S, Timpano H, Ball S, Cournac L, Peltier G (2009) Hydrogen production in Chlamydomonas: photosystem II-dependent and -independent pathways differ in their requirement for starch metabolism. Plant Physiol 151:631–640
Clayton RK (1963) Toward the isolation of a photochemical reaction center in Rhodopseudomonas spheroides. Biochim Biophys Acta 75:312–323
Danial AW, Abdel-Basset R (2015) Orange peel inhibited Hup and enhanced hydrogen evolution in some purple non-sulfur bacteria. Int J Hydrogen Energy 40:941–947
Danial AW, Abdel Wahab AM, Arafat HH, Abdel Basset R (2015) Identification and enhanced hydrogen evolution in two alginate-immobilized strains of Brevundimonas diminuta isolated from sludge and waterlogged soil. Ecohydrol Hydrobiol 15:81–88
Danial AW, Abdel Wahab AM, Arafat HH, Abdel-Basset R (2017) Bioenergetics of lactate vs. acetate outside TCA enhanced the hydrogen evolution levels in two newly isolated strains of the photosynthetic bacterium Rhodopseudomonas. Z Nat C 72(3–4):99–105
Fakhimi N, Dubini A, Tavakoli O, Gonzalez-Ballester D (2019) Acetic acid is key for synergetic hydrogen production in Chlamydomonas-bacteria co-cultures. Bioresour Technol 289:121648
Fakhimi N, Gonzalez-Ballester D, Fernández E, Galván A, Dubini A (2020) Algae-bacteria consortia as a strategy to enhance H2 production. Cells 9:1353
Forster B, Osmond C, Pogson B (2005) Improved survival of very high light and oxidative stress is conferred by spontaneous gain-of-function mutations in Chlamydomonas. Biochim Biophys Acta 1709:45–57
Gad El-Rab SM, Hifney AF, Abdel-Basset R (2018) Costless and huge hydrogen yield by manipulation of iron concentrations in the new bacterial strain Brevibacillus invocatus SAR grown on algal biomass. Int J Hydrogen Energy 43:18896–18907
Ghirardi ML, Togasaki RK, Seibert M (1997) Oxygen sensitivity of algal hydrogen production. Appl Biochem Biotechnol 63:141–151
Gibbs M, Gfeller RP, Chen C (1986) Fermentative metabolism of Chlamydomonas reinhardtii.: III. Photoassimilation of acetate. Plant Physiol 82:160–166
González-Ballester D, Pollock SV, Pootakham W, Grossman AR (2008) The central role of a SNRK2 kinase in sulfur deprivation responses. Plant Physiol 147:216–227
González‑Ballester D, Jurado‑Oller JL, Galván A, Fernández E, Dubini A (2017) H2 production pathways in nutrient‑replete mixotrophic Chlamydomonas cultures under low light. Response to the commentary article “On the pathways feeding the H2 production process in nutrient‑replete, hypoxic conditions,” by Alberto Scoma and Szilvia Z. Tóth Biotechnol. Biofuels 10:117. https://doi.org/10.1186/s13068-017-0801-5
Hallenbeck PC, Liu Y (2016) Recent advances in hydrogen production by photosynthetic bacteria. Int J Hydrogen Energy 41:4446–4454
He J, Xi L, Sun X, Ge B, Liu D, Han Z, Pu X, Huang F (2018) Enhanced hydrogen production through co-cultivation of Chlamydomonas reinhardtii CC-503 and a facultative autotrophic sulfide-oxidizing bacterium under sulfurated conditions. Int J Hydrogen Energy 43:15005–15013
Hong ME, Shin YS, Kim BW, Sim SJ (2016) Autotrophic hydrogen photoproduction by operation of carbon-concentrating mechanism in Chlamydomonas reinhardtii under sulfur deprivation condition. J Biotechnol 221:55–61
Ike A, Toda N, Tsuji N, Hirata K, Miyamoto K (1997) Hydrogen photoproduction from CO2 -fixing microalgal biomass: application of halotolerant photosynthetic bacteria. J Ferment Bioeng 84:606–609
Jagota SK, Dani HM (1982) A new colorimetric technique for the estimation of vitamin C using Folin phenol reagent. Anal Biochem 15127(1):178–182. https://doi.org/10.1016/0003-2697(82)90162-2
Jurado-Oller JL, Dubini A, Galván A, Fernández E, González-Ballester D (2015) Low oxygen levels contribute to improve photohydrogen production in mixotrophic non-stressed Chlamydomonas cultures. Biotechnol Biofuels 8:149
Kawaguchi H, Hashimoto K, Hirata K, Miyamoto K (2001) H2Production from algal biomass by a mixed culture of Rhodobium marinum A-501 and Lactobacillus amylovorus. J Biosci Bioeng 91:277–282
Kim J, Kang C, Park T, Kim M, Sim S (2006a) Enhanced hydrogen production by controlling light intensity in sulfur-deprived Chlamydomonas reinhardtii culture. Int J Hydrogen Energy 31:1585–1590
Kim M, Baek J, Yun Y, Junsim S, Park S, Kim S (2006b) Hydrogen production from Chlamydomonas reinhardtii biomass using a two-step conversion process: Anaerobic conversion and photosynthetic fermentation. Int J Hydrogen Energy 31:812–816
Laurinavichene T, Tolstygina I, Tsygankov A (2004) The effect of light intensity on hydrogen production by sulfur-deprived Chlamydomonas reinhardtii. J Biotechnol 114:143–151
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin phenol reagent. J Biol Chem 193:265–275
Markov SA, Eivazova ER, Greenwood J (2006) Photostimulation of hydrogen production in the green alga Chlamydomonas reinhardtii upon photoinhibition of its oxygen evolving system. Int J Hydrogen Energy 31:1314–1317
Melis A, Melnicki MR (2006) Integrated biological hydrogen production. Int J Hydrogen Energy 31(11):1563–1573. https://doi.org/10.1016/j.ijhydene.2006.06.038)
Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol 122:127–135
Melnicki MR, Ela Eroglu E, Melis A (2009) Changes in hydrogen production and polymer accumulation upon sulfur-deprivation in purple photosynthetic bacteria. Int J Hydrogen Energy 34:6157–6170
Metzner H, Rau H, Singer H (1965) Untersuchungen zur Synchronisierbarkeit einzelner Pigmentmangel Mutanten von Chlorella. Planta 65:186–194
Mignolet E, Lecler R, Ghysels B, Remacle C, Franck F (2012) Function of the chloroplastic NAD(P)H dehydrogenase Nda2 for H2 photoproduction in sulfur-deprived Chlamydomonas reinhardtii. J Biotechnol 162:81–88
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Milligan AJ, Berman-Frank I, Gerchman Y, Dismukes GC, Falkowski PG (2007) Light-dependent oxygen consumption in nitrogen-fixing Cyanobacteria plays a key role in nitrogenase protection. J Phycol 43(5):845–852. https://doi.org/10.1111/j.1529-8817.2007.00395.x
Miura Y, Saitoh C, Matsuoka S, Miyamoto K (1992) Stably sustained hydrogen production with high molar yield through a combination of a marine green alga and a photosynthetic bacterium. Biosci Biotechnol Biochem 56:751–754
Miyamoto K, Ohta S, Nawa Y, Mori Y, Miura Y (1987) Hydrogen production by a mixed culture of a green alga, Chlamydomonas reinhardtii and a photosynthetic bacterium, Rhodospirillum rubrum. Agric Boil Chem 51:1319–1324
Moore S, Stein WW (1948) Amino acid free photometric ninhydrin method for use in chromatography of amino acids. J Biol Chem 176:367–388
Nagy V, Vidal-Meireles A, Tengölics R, Rákhely G, Garab G, Kovács L, Tóth SZ (2016) Ascorbate accumulation during sulfur deprivation and its effects on photosystem II activity and H2 production of the green alga Chlamydomonas reinhardtii. Plant Cell Environ 39:1460–1472. https://doi.org/10.1111/pce.12701
Nagy V, Podmaniczki A, Vidal-Meireles A, Tengölics R, Kovács L, Rákhely G, Scoma A, Tóth SZ (2018a) Water-splitting-based, sustainable and efficient H2 production in green algae as achieved by substrate limitation of the Calvin–Benson–Bassham cycle. Biotechnol Biofuels 11:69. https://doi.org/10.1186/s13068-018-1069-0
Nagy V, Vidal-Meireles A, Podmaniczki A, Szentmihalyi K, Rakhely G, Zsigmond L, Kovacs L, Tóth ST (2018b) The mechanism of photosystem-II inactivation during sulphur deprivation-induced H2 production in Chlamydomonas reinhardtii. Plant J 94:548–561. https://doi.org/10.1111/tpj.13878
Namsaraev ZB (2009) Application of extinction coefficients for quantification of chlorophyll and bacteriochlorophyll. Microbiology 78(6):794–797
Nelson CJ, Alexova R, Jacoby RP, Millar AH (2014) Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiol 166:91–108
Nguyen AV, Toepel J, Burgess S, Uhmeyer A, Blifernez O, Doebbe A, Hankamer B, Nixon P, Wobbe L, Kruse O (2011) Time-course global expression profiles of Chlamydomonas reinhardtii during photobiological H2 production. PLoS ONE 6:e29364
Park W, Moon I (2007) A discrete multi states model for the biological production of hydrogen by phototrophic microalga. Biochemical Engineering J 36(1):19–27. https://doi.org/10.1016/j.bej.2006.06.013
Shevela D, Eaton-Rye JJ, Shen JR, Govindjee (2012) Photosystem II and the unique role of bicarbonate: A historical perspective. Biochem Biophys Acta 1817:1134–1151
Sojka GA, Freeze HH, Gest H (1970) Quantitative estimation of bacteriochlorophyll in situ. Arch Biochem Biophys 136(2):578–580. https://doi.org/10.1016/0003-9861
Sugimoto I, Takahashi Y (2003) Evidence that the PsbK polypeptide is associated with the photosystem II core antenna complex CP43.J. Biol Chem 278:45004–45010
Sugimoto K, Sato N, Tsuzuki M (2007) Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett 581:4519–4522
Sugimoto K, Midorikawa T, Tsuzuki M, Sato N (2008) Upregulation of PG synthesis on sulfur-starvation for PS I in Chlamydomonas. Biochem Biophys Res Commun 369:660–665. https://doi.org/10.1016/j.bbrc.2008
Sugimoto K, Tsuzuki M, Sato N (2010) Regulation of synthesis and degradation of a sulfolipid under sulfur-starved conditions and its physiological significance in Chlamydomonas reinhardtii. New Phytol 185:676–686
Takahashi H, Braby CE, Grossman AR (2001) Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol 127:665–673
Toepel J, Illmer-Kephalides M, Jaenicke S, Straube J, May P, Goesmann A, Kruse O (2013) New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNAseq transcriptomics. Plant Biotechnol J 11:717–733
Tsygankov AA, Kosourov SN, Tolstygina IV, Ghirardi ML, Seibert M (2006) Hydrogen production by sulfur-deprived Chlamydomonas reinhardtii under photoautotrophic conditions. Int J Hydrogen Energy 31:1574–1584
Vijayaraghavan K, Karthik R, Nalini SPK (2010) Hydrogen generation from algae: a review. J Plant Sci 5:1–19. https://doi.org/10.3923/jps.2010.1.19
Volgusheva A, Styring S, Mamedov F (2013) Increased photosystem II stability promotes H2 production in sulfur-deprived Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110(18):7223–7228. https://doi.org/10.1073/pnas.1220645110
Wydrzynski T, Govindjee (1975) New site of bicarbonate effect in photosystem II of photosynthesis—evidence from chlorophyll fluorescence transients in spinach-chloroplasts. Biochim Biophys Acta 387:403–408
Xu L, Li D, Wang Q, Wu S (2016) Improved hydrogen production and biomass through the co-cultivation of Chlamydomonas reinhardtii and Bradyrhizobium. Int J Hydrogen Energy 41(22):9276–9283
Xu L, Cheng X, Wang Q (2017) Effect of co-cultivation of Chlamydomonas reinhardtii with Azotobacter chroococcum on hydrogen production. Int J Hydrogen Energy 42:22713–22719
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This study was supported by Assiut University.
Author information
Authors and Affiliations
Contributions
The first author (AWD) and the third author (HAA) implemented the experiments, data recording and analysis, statistical analysis. The second author (RA) suggested the problem, designed the experiments and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent to publication
The authors agree to publish this paper in photosynthesis research journal.
Research involving human and animal rights
Neither humans nor animals have been used in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Danial, A.W., Abdel-Basset, R. & Abdel-Kader, H.A.A. Tuning photosynthetic oxygen for hydrogen evolution in synergistically integrated, sulfur deprived consortia of Coccomyxa chodatii and Rhodobium gokarnense at dim and high light. Photosynth Res 155, 203–218 (2023). https://doi.org/10.1007/s11120-022-00961-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-022-00961-4