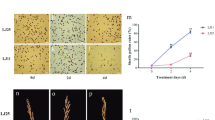
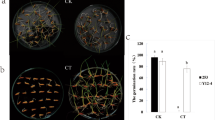
Abstract
Rice in cold region is threatened by cold stress frequently, resulting in severe losses of rice production. In the present study, two rice varieties with contrast cold tolerance in the reproductive stage (Longjing25, cold tolerant; Longjing11 cold sensitive) were compared using RNA-seq technique. A total of 14,861 and 12,148 differentially expressed genes (DEGs) were obtained by comparison of LJ25 and LJ11 after 2 days and 4 days of cold stress challenge. Subsequent Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that five significant pathways were specifically enriched in LJ11. Starch and sucrose metabolism was the only one significant pathway (containing 47 DEGs) annotated in both of varieties during two cold treatment periods. By weighted gene co-expression network analysis (WGCNA), 14 co-expression gene modules were calculated, among which turquoise, cyan, and dark green were the most concerned modules, containing 4807, 2682, and 1174 DEGs, respectively. Finally, we analyzed the transcriptional factors (TFs), and TF families of MYB, zinc finger, AP2/ERF-AP2, bHLH, NAC, bZIP, and WRKY were the biggest seven subfamilies. Collectively, our findings provided valuable insights into the molecular and genetic mechanisms responsible for rice’s response to cold stress.









Similar content being viewed by others
References
Allen DJ, Ort DR (2001) Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci 6:36–42
Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72:557–566
Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21:1453–1472
Bai B, Wu J, Sheng WT, Zhou B, Zhou LJ, Zhuang W, Yao DP, Deng QY (2015) Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress. Int J Mol Sci 16:11398–11416
Cheng C, Yun KY, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun SJ, de los Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics 8:175
Cheng H, Liu H, Deng Y, Xiao J, Li X, Wang S (2015) The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol 167:1087–1099
Cho JI, Ryoo N, Ko S, Lee SK, Lee J, Jung KH, Lee YH, Bhoo SH, Winderickx J, An G, Hahn TR, Jeon JS (2006) Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224:598–611
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Garber M, Grabherr MG, Guttman M, Trapnell C (2011) Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8:469–477
Gunawardena TA, Fukai S, Blamey FPC (2003) Low temperature induced spikelet sterility in rice. I. Nitrogen fertilization and sensitive reproductive period. Aust J Agric Res 54:937–946
Han L, Koh H, Piao Z (2002) Status and prospects of genetic and analysis for cold tolerance in rice. Chin. J. Rice Sci. 16:193–198
Heenan DP (1984) Low-temperature induced floret sterility in the rice cultivars Calrose and Inga as influenced by nitrogen supply. Anim Prod Sci 24:255–259
Hirose T, Takano M, Terao T (2002) Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol 43:452–459
Hong JY, Chae MJ, Lee IS, Lee YN, Nam MH, Kim DY, Byun MO, Yoon IS (2011) Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72:27–36
Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB (2010) The ABREbinding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167:1512–1520
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Huang JW, Chen JT, Yu WP, Shyur LF, Wang AY, Sung HY, Lee PD, Su JC (1996) Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci Biotechnol Biochem 60:233–239
Hüner NP, Bode R, Dahal K, Hollis L, Rosso D, Krol M, Ivanov AG (2012) Chloroplast redox imbalance governs phenotypic plasticity: the “grand design of photosynthesis” revisited. Front Plant Sci 3:255
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47:141–153
Ito N, Hayase H, Satake T, Nishiyama I (1970) Male sterility caused by cooling treatment at the meiotic stage in rice plants. III. Male abnormalities at anthesis. Proc. Crop Sci. Soc. Jpn. 39:60–64
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 10:2705–2222
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kim SH, Kim JY, Kim SJ, An KS, An G, Kim SR (2007) Isolation of cold stress-responsive genes in the reproductive organs, and characterization of the OsLti6b gene from rice (Oryza sativa L.). Plant Cell Rep 26:1097–1110
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Komvongsa J, Luang S, Marques JV, Phasai K, Davin LB, Lewis NG, Ketudat Cairns JR (2015) Active site cleft mutants of Os9BGlu31 transglucosidase modify acceptor substrate specificity and allow production of multiple kaempferol glycosides. Biochim Biophys Acta 1850:1405–1414
Kothari KS, Dansana PK, Giri J, Tyagi AK (2016) Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front Plant Sci 7:1057
Koumoto T, Shimada H, Kusano H, She KC, Iwamoto M, Takano M (2013) Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1,6-bisphosphatase. Plant Biotechnol. 30:47–56
Kuntothom T, Luang S, Harvey AJ, Fincher GB, Opassiri R, Hrmova M, Ketudat Cairns JR (2009) Rice family GH1 glycoside hydrolases with β-D-glucosidase and β-D-mannosidase activities. Arch Biochem Biophys 491:85–95
La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol Gen Genomics 275:374–386
Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, Luo D, Ma H, Zhang DB (2006) The rice Tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18:2999–3014
Liu J, Liu W, Guo D, Li Z (2007) Status and prospect of molecular breeding for cold tolerance in rice. Heilongjiang Agric. Sci. 4:105–108
Liu N, Song C, Wang G, Zhou X (2012) Evaluation on the cold tolerance of rice at booting stage in Sanjiang plain. Heilongjiang Agric. Sci. 10:1–7
Liu J, Chen X, Liang X, Zhou X, Yang F, Liu J, He SY, Guo Z (2016) Alternative splicing of Rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol 171:1427–1442
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods 25:402–408
Ma YY, Zhang Y, Lu JA, Shao HB (2010) Roles of plant soluble sugars and their responses to plant cold stress. Afr J Biotechnol 8:2004–2010
Mao D, Chen C (2012) Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One 7:e47275
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Masumoto C, Miyazawa S, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M (2010) Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci U S A 107:5226–5231
Matsushima S (2008) Analysis of Developmental Factors Determining Yield and Yield prediction in Lowland Rice: LVII. Effects of different concentrations of carbon dioxide in the air in different growth stages on the grain yield, yield constitutional factors and the chemical compositions of rice plants. Jpn. J. Crop Sci. 29(1):29-30
Nagao M, Minami A, Arakawa K, Fujikawa S, Takezawa D (2005) Rapid degradation of starch in chloroplasts and concomitant accumulation of soluble sugars associated with ABA-induced freezing tolerance in the moss Physcomitrella patens. J Plant Physiol 162:169–180
Nishiyama I (1982) Male sterility caused by cooling treatment at the young microspore stage in rice plants. Jap. J. Crop Sci. 51(3):386-392
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Oda S, Kaneko F, Yano K, Fujioka T, Masuko H, Park JI, Kikuchi S, Hamada K, Endo M, Nagano K, Nagamura Y, Kawagishi-Kobayashi M, Suwabe K, Suzuki G, Watanabe M (2010) Morphological and gene expression analysis under cool temperature conditions in rice anther development. Genes Genet. Syst. 85:107–120
Ogawa S, Suzuki Y, Yoshizawa R, Kanno K, Makino A (2010) Effect of individual suppression of RBCS multigene family on rubisco contents in rice leaves. Plant Cell Environ 35:546–553
Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80:135–139
Okoniewski MJ, Miller CJ (2006) Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinf. 7:276
Oliver SN, Van Dongen JT, Alfred SC, Mamun EA, Zhao X, Saini HS, Fernandes SF, Blanchard CL, Sutton BG, Geigenberger P (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28:1534–1551
Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M (2008) Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep 27:1677–1686
Royce TE, Rozowsky JS, Gerstein MB (2007) Toward a universal microarray: prediction of gene expression through nearest-neighbor probe sequence identification. Nucleic Acids Res 35:e99
Ruelland E, Zachowski A (2010) How plants sense temperature. Environ Exp Bot 69:225–232
Ruelland E, Vaultier MN, Zachowski A, Hurry V (2009) Chapter 2 cold signalling and cold acclimation in plants. Adv Bot Res 49:35–150
Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca(2+)-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol. 42:1228–1233
Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca(2+)-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42:1228–1233
Sanghera GS, Wani SH, Hussain W, Singh NB (2011) Engineering cold stress tolerance in crop plants. Curr. Genomics 12:30–43
Satake T, Hayase H (1974) Male sterility caused by cooling treatment at the young microspore stage in rice plants. IX. Revision of the classification and terminology of pollen developmental stages. Proc. Crop Sci. Soc. Jpn. 43:36–39
Satake T, Hayase H (1974) Male sterility caused by cooling treatment at the young microspore stage in rice plants. IX. Revision of the classification and terminology of pollen developmental stages. Proc Crop Sci Soc Jpn 43:36–39
Scott R, Hodge R, Paul W (1991) The molecular biology of anther differentiation. Plant Sci 80:167–191
Shao HB, Guo QJ, Chu LY, Zhao XN, Su ZL, Hu YC, Cheng JF (2007) Understanding molecular mechanism of higher plant plasticity under abiotic stress. Colloids Surf., B 54:37–45
Shi J, Cao Y, Fan X, Li M, Wang Y, Ming F (2012) A rice microsomal delta-12 fatty acid desaturase can enhance resistance to cold stress in yeast and Oryza sativa. Mol Breed 29:743–757
Shirasawa S, Endo T, Nakagomi K, Yamaguchi M, Nishio T (2012) Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxintuanheigu’, in rice, Oryza sativa L. Theor Appl Genet 124:937–946
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153:145–158
Takahashi S, Murata N (2008) How do environmental stresses accelerate photoinhibition. Trends Plant Sci 13:178–182
Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Gen Genomics 284:173–183
Tang XJ, Peng C, Zhang J, Cai Y, You XM, Kong F, Yan HG, Wang GX, Wang L, Jin J, Chen WW, Chen XG, Ma J, Wang P, Jiang L, Zhang WW, Wan JM (2016) ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci 249:70–83
Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q (2015) Characterization and functional analysis of Pyrabactin resistance-like abscisic acid receptor family in rice. Rice 8:28
Tjus SE, Møller BL, Scheller HV (1998) Photosystem I is an early target of photoinhibition in barley illuminated at chilling temperatures. Plant Physiol 116:755–764
Vaux DL, Korsmeyer SJ (1999) Cell death in development. Cell 96:245–254
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Wada T, Ogawa K, Ito T, Suzuki H, Takeoka Y (1990) Light microscopic observations on pollen and anther development in rice (Oryza sativa L.). I. Stages from pollen mother cells to tetrads. Jpn. J. Crop Sci. 59:769–777
Wan B, Lin Y, Mou T (2007) Expression of rice Ca(2+)-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett 581:1179–1189
Wan L, Zha W, Cheng X, Liu C, Lv L, Liu C, Wang Z, Du B, Chen R, Zhu L (2011) A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233:309–323
Wang YJ, Zhang ZG, He XJ, Zhou HL, Wen YX, Dai JX, Zhang JS, Chen SY (2003) A rice transcription factor OsbHLH1 is involved in cold stress response. Theor Appl Genet 107:1402–1409
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57–63
Wang E, Xu X, Zhang L, Zhang H, Lin L, Wang Q, Li Q, Ge S, Lu BR, Wang W, He Z (2010) Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication. BMC Evol Biol 10:108
Wang L, Xiang H, Wang L, Wang C, Li Z, Luo Y, Li R (2014) Analysis on susceptibility to chilling injury of different rice cultivars at booting stage. Heilongjiang Agric. Sci. 9:14–17
Xia N, Zhao HW, Lv YC, Zhao ZD, Zou DT, Liu HL, Wang JG, Jia Y (2016) Effect of cold-water stress at grainfilling stage on starch accumulation and related enzyme activities in grains of japonica rice in cold-region. Chin. J. Rice Sci. 30(1):62–74
Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S (2013) Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol 163:1868–1882
Xie G, Kato H, Sasaki K, Imai R (2009) A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Lett 583:2734–2738
Xie G, Kato H, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J 443:95–102
Yamaguchi T (2006) Analysis of genes expressed in rice anthers during the stage of maximal chilling sensitivity.Bull. natl. Agric. res. cent. tohoku Reg. 51(2):119-148
Yamori W, Noguchi K, Hikosaka K, Terashima I (2009) Cold-tolerant crop species have greater temperature homeostasis of leaf respiration and photosynthesis than cold-sensitive species. Plant Cell Physiol 50:203–215
Yang C, Wang K, Yang H, Zhou D, Li J (2009) Advances in breeding of cold-tolerance rice. Chin. Agric. Sci. Bull. 25:113–116
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Yang J, Chen X, Zhu C, Peng X, He X, Fu J, Ouyang L, Bian J, Hu L, Sun X, Xu J, He H (2015a) Using RNA-seq to profile gene expression of spikelet development in response to temperature and nitrogen during meiosis in rice (Oryza sativa L.). PLoS One 10:e0145532
Yang J, Kim SR, Lee SK, Choi H, Jeon JS, An G (2015b) An alanine aminotransferase 1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm. Plant Sci 240:79–89
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64:5085–5097
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
Zhang H, Ni L, Liu Y, Wang Y, Zhang A, Tan M, Jiang M (2012) The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice. J Integr Plant Biol 54:500–510
Zhang Q, Jiang N, Wang G-L, Hong Y, Wang Z (2013) Advances in understanding cold sensing and the coldresponsive network in rice. Adv. Crop Sci. Technol. 1:104
Zhang Q, Chen Q, Wang S, Hong Y, Wang Z (2014) Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7:24
Zhang Y, Verhoeff NI, Chen Z, Chen S, Wang M, Zhu Z, Ouwerkerk PB (2015) Functions of OsDof25 in regulation of OsC4PPDK. Plant Mol Biol 89:229–242
Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, Wu P (2012) A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol Plant 5:154–161
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Funding
This study was supported by the National Key Research and Development Plan (2016YFD0101801), the Modern Agro-industry Technology Research System (CARS-01-09), and the Science and Technology Support of the Ministry of Science and Technology (2015BAD02B01-3).
Author information
Authors and Affiliations
Contributions
ZG and CL conceived the original screening and research plans. GP and ZC supervised the experiments. ZG, RW, WX, LZ, SG, XH, XZ, JG, and XD performed most of the experiments. ZG, SZ, and LC designed the experiments and analyzed the data. ZG conceived the project and wrote the article. ZG and HL supervised and complemented the writing. All authors read and approved the final version of the paper.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Table S1
Gene expression in LJ11 and LJ25. (XLSX 2389 kb)
Table S2
All-time courses of gene expression in Longing25. (XLSX 1266 kb)
Table S3
All-time courses of gene expression in LJ11. (XLSX 1332 kb)
Table S4
DEGs in Longjng25 after 2 days and 4 days of cold stress challenge. (XLSX 653 kb)
Table S5
GO analysis of DEGs in LJ25 after 2 days and 4 days of cold stress challenge. (XLS 43 kb)
Table S6
DEGs in KEGG pathway of plant hormone signal transduction in LJ25 under cold stress. (XLSX 15 kb)
Table S7
DEGs in Longjng11 after 2 days and 4 days of cold stress challenge. (XLSX 1343 kb)
Table S8
GO analysis of DEGs in LJ11 in the entire cold stress treatment. (XLSX 24 kb)
Table S9
DEGs in KEGG pathway of carbon fixation in photosynthetic organisms in LJ11 under cold stress. (XLSX 13 kb)
Table S10
DEGs between LJ25 and LJ11 after 2 days of cold stress challenge. (XLSX 1390 kb)
Table S11
DEGs between LJ25 and LJ11 after 4 days of cold stress challenge. (XLSX 1117 kb)
Table S12
Common DEGs in KEGG pathway of starch and sucrose metabolism in LJ25 and LJ11. (XLSX 17 kb)
Table S13
DEGs in turquoise module of WGCNA. (XLSX 89 kb)
Table S14
DEGs in cyan module of WGCNA. (XLSX 54 kb)
Table S15
DEGs in darkgreen module of WGCNA. (XLSX 29 kb)
Table S16
Enrichment pathway in turquoise, cyan and darkgreen modules of WGCNA. (XLSX 12 kb)
Table S17
TFs in LJ25 and lJ11 after cold stress treatment (XLSX 160 kb)
Table S18
7 top TFs in LJ25 and LJ11 after cold stress treatment (XLSX 108 kb)
Table S19
Primers of eight DEGs for qRT-PCR validation (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Guo, Z., Liu, C., Xiao, W. et al. Comparative Transcriptome Profile Analysis of Anther Development in Reproductive Stage of Rice in Cold Region Under Cold Stress. Plant Mol Biol Rep 37, 129–145 (2019). https://doi.org/10.1007/s11105-019-01137-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-019-01137-6