Abstract
Background and aims
Interactions between species invasions and climate change have the potential to drive changes in plant communities more than either factor alone. One pathway through which these effects can occur is via changes to the rhizosphere microbial community. Invasive plants can alter these microbial communities affecting natives’ abilities to compete with invaders. At the same time, climate change is leading to more frequent extreme wet and dry events. Understanding the response of plant communities to these combined global change drivers requires a comprehensive approach that assesses the relationship between plant competition and belowground rhizosphere microbial community responses.
Methods
Here we use a field experiment in a California grassland with a set of six native annual forbs (i.e., wildflowers) and three invasive annual grasses to test how competition with invasive plants alters both identity and function in the native rhizosphere microbiome, and whether competition between these groups interacts with rainfall to amplify or ameliorate microbial shifts.
Results
Metagenomics of rhizosphere communities revealed that drought combined with competition from invaders altered a higher number of functions and families in the native rhizosphere compared to invasive competition alone or drought alone. Watering combined with invasion led to fewer shifts.
Conclusion
This suggests invasion-driven shifts in the microbial community may be involved in weakening natives’ ability to cope with climate change, especially drought. Understanding the role of the microbial community under invasion and climate change may be critical to mitigating the negative effects of these interacting global change drivers on native communities.
Graphical abstract
Understanding plant community response to global change drivers requires a comprehensive approach that assesses the relationship between plant competition and belowground rhizosphere microbial community responses. (a) In this work, we use a field experiment in a California grassland with a set of native forbs (purple) and invasive grasses (teal) to assess the combined effects of competition and water availability (drought, control, watered) on the rhizosphere microbiome. (b) Drought combined with competition from invaders altered the relative abundance of 36 functions (white) and 22 microbial families (blue) in the native rhizosphere compared to the effects of competition (3 functions, 16 families) or drought alone on natives (not shown: 5 functions, 0 families). (c) Additionally, regardless of watering treatment, invasive grasses sourced more of the taxonomic community in native-invasive mixes and this was exacerbated during drought. Overall, these results suggest invasion-driven shifts in the microbiome may be involved in weakening natives’ ability to cope with climate change, especially drought.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The invasion of novel species can drive a multitude of cascading effects on the structure and function of native communities, but climate change combined with invasion has the potential to amplify and worsen these effects. Invasive plants alter soil chemical and microbial composition, outcompete natives for light, water and nutrient resources, and change the pollinator community, often to the advantage of invaders over native residents (Reinhart and Callaway 2006; Ehrenfeld 2010; Traveset and Richardson 2014; Rodríguez-Caballero et al. 2020). The effects of invasive species, however, inherently depend on the availability of resources - thus, under a rapidly changing climate, the strength of these effects may shift and novel pathways of invasive dominance may arise. One of the least understood pathways contributing to invasive dominance over natives lies in the rhizosphere microbiome. While there is ample evidence that invaders can shift the rhizosphere microbiome, and some evidence suggesting that these invader-induced changes may aid in the dominance of invasives over natives (LaForgia et al. 2022), how climate interacts with invasive species to alter the rhizosphere microbiome remains poorly understood.
The microbial community that inhabits the rhizosphere, the region of soil directly surrounding plant roots, often forms close associations with host plants (Edwards et al. 2015). Members of this community can be detrimental (pathogenic), neutral (commensalistic) or beneficial (mutualistic) to host plant fitness (Thrall et al. 2007). For example, beneficial microbes can help host plants with amelioration of biotic and abiotic stress (Yang et al. 2009; Doubková et al. 2012), acquisition of nutrients (Richardson et al. 2009), promotion of plant growth (Qu et al. 2020), and pathogen resistance (Ritpitakphong et al. 2016). In return, host plants provide benefits to the microbial community, often through the release of sugar compounds into rhizosphere soil; these sugars double as attractants to entice microbes into beneficial interactions (Paterson et al. 2007; Hartmann et al. 2008). As a result, rhizosphere microbial communities tend to be host plant-specific (Aleklett et al. 2015; Dawson et al. 2017) and are critical for the persistence of native plant species (Chung et al. 2015; David et al. 2019). When a native plant community becomes invaded, the addition of novel plant hosts may alter the microbial composition of the rhizosphere, either directly by recruiting a different set of microbes from the bulk soil than natives (Batten et al. 2006) or indirectly through the release of compounds that might select for a different community composition in the bulk soil (Ulbrich et al. 2022), affecting the availability of microbes to neighboring native plants.
We face a daunting knowledge gap about how climate change will interact with microbial communities to affect soil and plant health. In addition to being closely tied to plant host identity, the dynamic makeup of rhizosphere microbial communities is strongly influenced by abiotic soil variables including soil water availability (Fierer et al. 2012). For example, functional and taxonomic microbial diversity tend to be lower in more arid regions (Fierer et al. 2012; Maestre et al. 2015) and microbial activity tends to decrease during drought (Acosta-Martinez et al. 2014; Legay et al. 2017; Ochoa-Hueso et al. 2018). Further, changes in water availability have the capacity to shift the relationship between plants and microbes (Cheng et al. 2019), and complicating matters, both plants and microbes can also shape surrounding abiotic soil conditions. As climate change drives increases in rainfall variability in many systems, especially semi-arid and arid ones, understanding the effects on the rhizosphere microbiome and their plant hosts becomes increasingly important.
In California annual grasslands, rainfall variability mediates the composition of native forbs (i.e., wildflowers or non-grass herbaceous plants) and invasive grasses through time. In general, wet years favor competitively dominant invasive grasses, while successive drought years indirectly favor subdominant natives via decreased invasive grass competition (Dudney et al. 2017; LaForgia et al. 2018). Further, recent work suggests that native species may be resilient to changes in climate in the absence of invasive grasses (LaForgia et al. 2020). In the presence of these invasives, however, the negative effects of drought are intensified and the beneficial effects of increased rainfall are dampened (LaForgia et al. 2020). Invasive grasses in this community also drive shifts in the rhizosphere microbial community composition (Batten et al. 2006), which may be contributing to invasive dominance over natives (LaForgia et al. 2022). Neighbor-induced changes in the rhizosphere microbiome tend to be greatest under strong competition (Ulbrich et al. 2022) and may result in a microbiome assembled predominantly from invasive grasses, the dominant competitor as found elsewhere (Hortal et al. 2017; Lozano et al. 2019). In order to explore the effects of invasion on native plant communities under climate change, microbial shifts must be assessed under different levels of rainfall availability at the local level.
To understand how rainfall variability and competition from invasive grasses affect the rhizosphere microbial community of native forbs, we manipulated watering regimes and plant hosts in a field experiment. We then used metagenomics to assess rhizosphere differences between watering and plant host treatments in terms of taxonomy and function as well as host contribution to rhizosphere community assembly. Based on previous findings, we hypothesized that (1) function and taxonomy in the rhizosphere would differ between plant hosts and between watering treatments, but the combined effect of invasive competition and watering treatments on native forb rhizospheres would be larger than invasive grass competition or watering treatments alone. To test this hypothesis we investigated community dissimilarity (i.e., beta diversity) and differential (relative) abundance of key taxa and functions across treatments. (2) We also hypothesized that, while microbial functional and taxonomic communities in invaded native communities under increased watering would be assembled equally from invader and native hosts, as was seen in LaForgia et al. (2022), under drought, a majority of the community would be assembled from invaders as drought has been shown to intensify the negative effects of invasive competition on natives (LaForgia et al. 2020). To test this hypothesis we implemented a Bayesian classifier, SourceTracker2, to predict the proportion of the microbial community sourced from each host when natives compete with invasives.
Methods
Experimental set-up
In November 2017, we set up a field experiment in an annual serpentine grassland at the University of California McLaughlin Natural Reserve in the Inner North Coast Range (N 38°52’, W 122°26’). This reserve has a Mediterranean climate with cool, wet winters and hot, dry summers.
We utilized an existing rainfall manipulation experiment in a lush serpentine (high Mg, low Ca) area of the reserve, which hosts both high native annual diversity and abundant invasive annual grasses. These invasive grasses are the largest biomass contributor at the site, accounting for about 71% of total plant cover (LaForgia, unpublished data). The existing experiment consists of 30 watered plots along three watering lines emanating from a rainfall catchment system with each plot centered on a sprinkler that casts water over a 3-m radius (Mini Rotor Drip Emitters, Olson Irrigation) (Fig. 1a). Of the 30 watered plots, we randomly chose 10 for the present study. The experiment also utilized 10 drought plots interspersed with watered plots. Drought plots were each covered by a 3 × 3 m shelter constructed following the design of DroughtNet (wp.natsci.colostate.edu/droughtnet) except that the removable roofs intercepted 100% of rainfall. Roofs were placed on the shelters from approximately 1 December 2017 to 1 March 2018. In previous years, these watering treatments led to an average decrease in soil moisture of 42% in the drought plots and an average increase in soil moisture of 27% in the water addition plots (data not published). Watering was applied weekly from December 1 to February 1 to maintain total rainfall at or slightly above its 30-y average. Ten unmanipulated control plots were also interspersed with treatment plots. All plots were no less than 4 m from each other.
a Diagram of plot locations at the field site. Watered plots (blue circles) included a sprinkler along watering lines. Control plots (gray squares) and drought plots (red triangles) were interspersed within the lush serpentine area of the site. b Diagram of subplot setup within each plot. c Photograph of rhizosphere core sampling method with a native-invasive mix. A sterilized bulb planter was used to core a representative assemblage of plants within each subplot treatment
Within each of these 30 experimental plots we cleared aboveground vegetation and seeded in three treatment subplots (Fig. 1b): (1) invasive annual grasses, (2) native annual forbs, and (3) a mixture of invasive annual grasses and native annual forbs. We used two different seed mixes of forbs (mix 1: Lasthenia californica, Clarkia purpurea, and Agoseris heterophylla; mix 2: Calycadenia pauciflora, Hemizonia congesta, and Plantago erecta). All seeds were collected from the site during the previous spring/summer and subplots were hand weeded halfway through the study to maintain plant host treatments as seeded. Many of these species do maintain seed banks and although we cleared vegetation prior to seeding, we cannot say whether the resulting community came from collected seeds or the natural background seed bank.
As these species have vastly different flowering phenologies, and development can alter microbial composition (Chaparro et al. 2014; Zhang et al. 2018a; Xiong et al. 2021), we chose to sample the rhizosphere prior to any flowering so that all plants were sampled during the same life stage. This occurred after the majority of rainfall had occurred (April 5th, 2018). Plants and associated soil were removed from each subplot using a hand bulb planter (Fig. 1c), which allowed us to collect a representative of each species included in each treatment at the same time. Samples were placed into a Ziplock bag and processed immediately. We sterilized the bulb planter between subplots and recorded all species present in the core sample (Table S1). For a step-by-step field protocol see Ettinger and LaForgia (Ettinger and LaForgia 2021).
Molecular analysis
Excess soil was shaken gently from the roots associated with each subplot. We sampled the joint rhizosphere of all representatives removed by the hand bulb planter as roots were interwoven and individual species could not be distinguished. The collective roots from each subplot, with soil still attached, were then placed in a 50 mL conical tube of autoclaved nanopure water and stirred vigorously to collect the soil rhizosphere fraction associated with the plant community in each subplot similar to rhizosphere collection in Edwards et al. (Edwards et al. 2015). Samples were then transported to UC Davis for processing on ice. Conical tubes were centrifuged at 4000 g for 1 min to obtain rhizosphere soil pellets, excess water was decanted and soil pellets were transferred to new 1.5 mL tubes for storage at -80 C until further processing. We chose a subset of 66 samples (out of a total of 150 samples) for DNA extraction and subsequent metagenomic analysis based on final plant community composition within the subplots. We prioritized sequencing of rhizosphere samples that accurately represented our treatments as seeded (i.e., without representatives from alternative functional groups; Table S1).
Samples were randomized using a random number generator prior to DNA extraction. Approximately ~ 250 mg of the rhizosphere soil pellet was used for DNA extractions using the MoBio PowerSoil DNA extraction kit (MO BIO Laboratories, Inc.). Extractions were performed following manufacturer’s instructions, with one modification (samples were bead beaten on the “homogenize” setting for 1 min during step 5). We additionally extracted DNA from three replicates each of a positive technological control using the ZymoBIOMICS Microbial Community Standard (Zymo Research Inc.), four replicates of a negative technological control from autoclaved nanopure water, and three replicates of a negative technological control where no sample was added during DNA extraction. DNA from replicate controls for each control type were pooled to produce representative control samples and these were included as controls during metagenomic sequencing.
Data generation
DNA samples were randomly aliquoted into wells in a 96 well plate using a random number generator and sent to the Integrated Microbiome Resource (IMR) at Dalhousie University for metagenomic sequencing on an Illumina NextSeq 550. Briefly, the IMR used one nanogram of each sample for Nextera XT (Illumina) library preparation, per the manufacturer’s instructions except clean-up and normalization which were completed using the Just-a-Plate 96 PCR Purification and Normalization Kit (Charm Biotech). Equal amounts of all barcoded samples were then pooled and sequenced on a shared 150 + 150 bp PE Illumina NextSeq 550. The resulting libraries produced a total of 625,865,988 reads, with a median of 9,133,838 reads per sample (min: 27,026, max: 16,911,776). The raw sequence reads generated for this metagenomic project were deposited at Genbank under BioProject accession no. PRJNA925931 (Table S2).
Data processing
Quality filtering of sequence reads was performed using bbMap v. 37.68 with the following parameters: qtrim = rl, trimq = 10, maq = 10 (Bushnell 2022). MEGAHIT v.1.1.3 was then used to co-assemble the filtered reads from all samples into a single metagenome assembly (Li et al. 2015). Prior to downstream analysis, we used the anvi’o v.7 metagenomic workflow to process the metagenomic data (Eren et al. 2015). Briefly, this involved using ‘anvi-gen-contigs-database’ to make a database of contigs from our co-assembly and then predict open reading frames using Prodigal v.2.6.2, resulting in 8,829,098 putative genes (Hyatt et al. 2010). Open reading frames were exported from anvi’o using ‘anvi-get-sequences-for-gene-calls’ and functionally annotated using Eggnog-mapper v. 1.0.3 (Cantalapiedra et al. 2021) and InterProScan v. 5.52-86.0 (Jones et al. 2014). Taxonomy was predicted for each gene using Kaiju v.1.5.0 against the NCBI BLAST nonredundant (nr) protein database, including fungi and microbial eukaryotes, v.2017-05-16 (Menzel et al. 2016). Finally, Salmon v. 1.3.0 was used to quantify the number of reads from each individual sample that mapped to predicted genes (Patro et al. 2017).
Salmon read counts and Kaiju taxonomies for each gene were imported into R for analysis and visualization (R Core Team 2022). Count tables were filtered to remove genes with no read counts and those not seen more than 3 times in at least 5% of the samples. To identify genes associated with possible contaminant taxa, we used decontam’s (Davis et al. 2018) prevalence method with a threshold of 0.5 to identify genes with a higher prevalence in negative controls (e.g., no-sample technological controls). Using this threshold, we identified 143 contaminant genes which were subsequently removed prior to downstream analysis, resulting in a dataset of 128,022 genes.
Genes taxonomically assigned by Kaiju as being from eukaryotes or which were unable to be classified were further investigated using MMseqs2 v. 13.45111 (Steinegger and Söding 2017) against the NCBI non-redundant database (downloaded December 13, 2022). We then removed 10,860 genes assigned by both software with a eukaryotic, viral or unclassified taxonomic assignment in order to focus on bacterial and archaeal genes. We then further removed 13,266 genes that did not have an associated COG functional annotation. The final set after filtering totaled 104,065 genes. For downstream beta-diversity analysis, samples were rarefied to a read count of 66,880. This value was chosen after examining rarefaction curves in order to balance maximizing the number of reads per sample while minimizing the number of samples removed from downstream analysis. This resulted in the sample with the lowest read count (35,618) being excluded from further analysis.
To understand how function in the microbial community differed between watering and plant host treatments, we agglomerated gene counts based on shared COG function using tax_glom in the phyloseq package (McMurdie and Holmes 2013). To understand how the taxonomic composition of the community differed between watering and plant host treatments, we agglomerated gene counts at the genus level also using tax_glom. To assess native and invasive contribution of function and identity to community assembly in the native-invasive rhizosphere, we agglomerated gene counts to both COG function and taxonomic family. This was done on rarefied gene counts for downstream beta-diversity analysis and on raw gene counts for downstream differential abundance analysis and community assembly investigations.
Beta diversity (community dissimilarity) analyses
To assess how the functional beta diversity of rhizosphere microbiomes varied across watering and plant host treatments, we conducted a PERMANOVA on Bray-Curtis dissimilarity between samples of the rarefied and agglomerated gene function data. This was done using adonis2 in the vegan package with by = margin and 9999 permutations (Oksanen et al. 2022). We also included plot as a factor to account for our nested experimental design. We conducted a similar analysis as above to examine how taxonomic beta diversity in the rhizosphere varied across watering and plant host treatments with the agglomerated genus data. To assess which treatment combinations were driving results, we conducted pairwise adonis contrasts on focal treatment combinations.
Given that plant species composition varied within plant host treatments, we also assessed whether presence or absence of specific host species were drivers of beta diversity within invasive, native or native-invasive mixes. Taking the previously calculated Bray-Curtis dissimilarities, we used the envfit function to fit host species presence/absence onto a principal coordinates analysis ordination made with the cmdscale function, both from the vegan package. This was done for both the agglomerated functional dataset and the agglomerated taxonomy dataset. We tested for correlations between overall plant host composition and Bray-Curtis dissimilarity using Mantel tests with 9999 permutations.
Functional and taxonomic differential (relative) abundance analyses
To assess how function in the rhizosphere varied across watering and plant host treatments, we used DESeq2 (Love et al. 2014) which has been shown to be a suitable statistical method for assessment of differentially abundant genes in metagenomes (Jonsson et al. 2016). DESeq2 was run on the agglomerated and unrarefied gene function data to examine the log2fold change (i.e. differential [relative] abundance, hereafter referred to as differential abundance) of each functional category between pairwise contrasts of plant host groups (native forbs, invasive grasses, and native-invasive mixes) and watering treatment (control, watered, drought). We used Benjamini-Hochberg corrections to adjust p-values and kept functions that had an adjusted p-value < 0.05. We then summed log2fold changes to the broader COG category to assess overall patterns of functional differences.
We conducted a similar analysis as above to examine how taxonomy in the rhizosphere varied across watering and plant host treatments with the agglomerated taxonomy data, then summed the resulting significant log2fold changes to the family level.
Host plant contribution to assembly of genes in native-invasive mixes
To assess the proportion of the joint microbial community sourced from native forbs or invasive grasses in mixed native-invasive subplots (similar to LaForgia et al. (2022), we employed a Meta-SourceTracker approach using Sourcetracker2 (Knights et al. 2011; McGhee et al. 2020). SourceTracker2 is effective at predicting the proportion of a community assembled from known sources in metagenomic data, enabling better understanding of microbial community assembly under competing host plants. Sourcetracker2 was used to estimate the fraction of families or functions detected in sinks (mixed subplots) that might originate from sources (invasive grasses, native forbs, or unknown habitats). We used source and sink rarefaction depths of 25,000. We obtained the predicted proportion of each taxonomic family and of each COG function in mixed subplots originating from each source using the --per_sink_feature_assignments option. We used Wilcoxon signed-rank tests to assess differences in the SourceTracker2 predicted proportions between different sources (invasive grasses, native forbs, or unknown) for the microbial communities across different watering regimes.
Results
Taxonomic and functional beta diversity differ between watering treatments and plant hosts
As stated above, we hypothesized that function and taxonomy in the rhizosphere would differ between plant hosts and watering treatments, with the interaction between invasive competition and watering treatments showing the largest effects on native forb rhizosphere. Contrary to our hypothesis, the interaction between watering treatment and plant host treatment did not have a significant effect on functional beta diversity (p = 0.213) nor on taxonomic beta diversity (p = 0.336). Both functional beta diversity and taxonomic beta diversity did however significantly differ between watering treatments (function: p < 0.001, Fig. 2a; taxonomy: p < 0.001, Fig. S1a) and between plant host treatments (function: p = 0.044, Fig. 2b; taxonomy: p = 0.011, Fig. S1b). For both taxonomy and function, significant differences between plant hosts were found between native forbs and native-invasive mixes (function: p = 0.024; taxonomy: 0.009), while differences between watering treatments were significant between all treatment combinations (see Table S3 for full results).
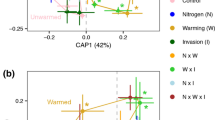
Ordination on Bray-Curtis dissimilarities of rhizosphere function from genes that appeared more than 3 times in at least 5% of the samples. a Principal coordinate analysis of functional beta diversity across watering treatments (control plots in black, watered plots in blue and drought plots in red). Functional beta diversity in the rhizosphere differed between watering treatments (p < 0.001). b Principal coordinate analysis of functional beta diversity across plant host treatments (native forbs in purple, invasive grasses in green, mixes in yellow). Functional beta diversity differed between plant host treatments (p = 0.044). c Centroids of microbial communities from each plant host (circles: native forbs, triangles: invasive grasses, squares: mixes) across watering treatments (drought: red, control: black, watered: blue). Note that scales between plots differ to allow for better visualization of patterns within each plot
Due to variation in plant species composition within host plant treatments, we assessed whether overall host composition or presence of specific host species were drivers of beta diversity within treatment groups. Host plant composition was not significantly correlated with taxonomic beta diversity within invasive, native or native-invasive mixes (Fig. S2; Mantel: p > 0.05), nor was the presence of individual host plant species (envfit for all species: p > 0.05). Similarly, overall host plant composition was not significantly correlated with functional beta diversity within invasive, natives or native-invasive mixes (Mantel: p > 0.05). While the presence of individual plant species was not a significant driver of the functional beta diversity of invasives or natives (envfit for all species: p > 0.05), the presence of two invasive grasses were significantly correlated with functional beta diversity in native-invasive mixes (envfit: Elymus caput-medusae: p = 0.039, Festuca perennis: p = 0.039).
Invasive grasses interact with watering treatments to shift function and taxonomy in the native rhizosphere
While we did not find evidence supporting our hypothesis that the interaction between watering treatments and invasion were stronger than individual effects when investigating shifts in taxonomic and functional beta diversity, we instead found evidence supporting this hypothesis when assessing the differential abundance of gene function and taxonomy.
Using DESeq2, we identified 44 COG functions that significantly varied in abundance between treatments. We found no evidence that drought or watering alone shifts function in the native rhizosphere, nor that competition with natives shifts the function in the invasive rhizosphere. Instead, all differences in rhizosphere functions were found when comparing the native forbs growing alone to native forbs growing in competition with invasive grasses (see Graphical Abstract). In all, we found that for natives, drought combined with competition from invaders altered the relative abundance of 36 functions, invasive competition alone altered 3 functions, and drought alone altered 5 functions. Watering combined with invasive competition only altered the relative abundance of 3 functions in the native rhizosphere, while watering alone did not shift any functions. Competition with natives did not shift any functions for invasives, regardless of watering treatment.
Analysis of the summed log-fold changes of these functions to the broader COG category level, revealed 16 COG categories (out of 25 total possible) that significantly varied between native rhizospheres and native-invasive mixed rhizospheres. Under control conditions, native-invasive mixes saw changes in relative abundance of four functional categories (FC) (Fig. 3d). Specifically, we saw one function increase, COG3746 (Phosphate-selective porin OprP; FC = inorganic ion transport and metabolism), and observed a strong decrease in COG3592 (Uncharacterized Fe-S cluster protein Yjdl; FC = function unknown). Mixed communities under drought, however, saw significant changes in all 16 of these FCs compared to native communities in the control (Fig. 3a), with two FCs showing significant increases and 13 FCs showing significant decreases. The one remaining FC (amino acid transport and metabolism) had a summed log-fold change close to 0. In particular, we found that FCs associated with posttranslational modification saw the largest increases in mixed communities under drought, while transcription and signal transduction mechanisms saw the largest decreases. In terms of specific COG functions, we saw the largest increases related to COG3158 (Potassium uptake protein Kup; FC = inorganic ion transport), COG1644 (Sigma 54 homolog RpoN; FC = transcription), and COG0339 (Zn-dependent oligopeptidase, M3 family Dcp; FC = post translational modification). While many functions decreased, we highlight decreases related to COG1276 (Putative copper export protein PcoD; FC = inorganic ion transport and metabolism) and COG3861 (Stress related protein YsnF; FC = function unknown).
Log2 fold changes of COG gene categories between a mixes in drought and natives in control treatment; b mixes in drought and natives in drought treatment; c mixes in watering and natives in control treatment; d mixes in control and natives in control treatment. Values greater than zero indicate higher abundance in mixes, values less than zero indicate higher abundance in natives
We also identified 3 FCs that significantly varied in mixed communities under drought compared to native communities under drought (Fig. 3b; largest increase in translation) and 3 FCs that significantly varied between natives under control and native-invasive mixes under watering (Fig. 3c). In terms of specific COG functions, native-invasive watered plots saw an increase in the COG3401 (Fibronectin type III domain FN3; FC = general function prediction only) and relatively small decreases in COG0161 (Adenosylmethionine-8-amino-7-oxononanoate aminotransferase BioA; FC = coenzyme transport and metabolism) and COG2205 (K + sensing histidine kinase KdpD; FC = signal transduction mechanisms) (Fig. S3).
Using DESeq2 we also identified 41 genera across 30 families that significantly varied in abundance between treatments with results largely mirroring functional results. That is, the majority of differences were observed between native-invasive mixes in drought and natives in control. Analysis of family-level summed log-fold changes identified 22 families that differed between mixes in drought and natives in control (Fig. 4a), 5 families that differed between mixes in watered treatments and natives in controls (Fig. 4b), and 16 families differed between mixes in controls and natives in controls (Fig. 4c). The majority of these families belong to the phyla Actinobacteria (10 families) and Proteobacteria (10 families), with the majority of Actinobacteria families decreasing (8 families) and the majority of Proteobacteria families increasing during competition (10 families), with the strongest shifts under drought. Of the many families that saw changes in abundance, Sandaracinaceae (Proteobacteria), Hungateiclostridiaceae (Firmicutes), and Burkholderiales (Proteobacteria) all showed the largest increases under invasion and drought. Contrary to functional results, no differences were found between natives under drought and mixes under drought.
Log2 fold changes of taxonomic families between a mixes in drought and natives in control treatment; b mixes in watering and natives in control treatment; c mixes in control and natives in control treatment. Values greater than zero indicate higher abundance in mixes, values less than zero indicate higher abundance in natives
Under drought, the majority of the microbial community is assembled from invasive grasses
We assessed the contribution of native forbs and invasive grasses to microbial families in native-invasive subplots using SourceTracker2. We hypothesized that invasives would source a majority of the community during drought, but that the microbial community would be equally sourced in watered plots. Instead, we observed that across all mixed subplots, invasives were the main contributors to the communities, sourcing consistently over half of community membership (Fig. 5). This was exacerbated during drought where invasives sourced on average 75.6% (± se: 6.89%) of the microbial community, a significantly higher proportion than was sourced by natives (mean ± se: 23.7% ± 6.87%; p < 0.001). Invasive and native contributions were also significantly different in control treatments (p = 0.019) and in watered plots (p = 0.015), with the contribution of the community sourced by natives highest during control treatments (native mean ± se: 35.6% ± 7.66%). Consistent with the overall community sourcing patterns, we found that differentially abundant families were on average sourced more by invasives and that this was heightened during drought (Fig. 6).
Invasives source more taxonomic families during drought. SourceTracker2 was used to predict whether genes associated with particular taxonomic families in treatments originated from invasive grasses (dark green), or native forbs (purple). Here we show the proportion of the community predicted to assemble from each source for each watering treatment. Comparisons that are significantly different from each other are notated by different letters (e.g., a vs. b). Error bars represent standard error
Bacterial families containing significant differentially abundant genera are more sourced from invasives during drought. Proportion of families predicted to colonize from each source for each taxonomic family found to have differentially abundant genera between treatments. SourceTracker2 was used to predict whether genes associated with particular bacterial families in treatments originated from invasive grasses (dark green), or native forbs (purple). A dashed line is used to designate a hypothesis of equal contribution from both sources (i.e., 50% proportion)
We also used SourceTracker2 to investigate the origin of COG functions in native-invasive subplots. Again, invasives contributed significantly more to the functional potential of the community sourcing slightly more functions than natives (Fig. S4). This was highest during drought where invasives sourced on average 50.9% (± se: 1.10%) of the functional potential of the community, a significant, but only slightly higher proportion than is sourced by natives (mean ± se: 44.9% ± 0.97%; p = 0.003). Invasive and native contributions were also significantly different in control treatments (p = 0.009) and in the watered treatments (p = 0.012), with the contribution of the community sourced by natives again highest during control treatments (native mean ± se: 46.3% ± 0.57%). Similar to the overall functional sourcing patterns, we found that differentially abundant COG functions were sourced fairly equally across treatments, with invasives providing a slightly higher proportion of the functional potential overall (Fig. S5). One notable exception to this trend is the putative copper export protein (PcoD) function, which was sourced in its entirety by natives, regardless of watering treatment.
Discussion
Our study adds to growing evidence that invasive species may affect the ability of native communities to cope with climate change and highlights that altering key functions and taxa in the rhizosphere microbiome is one pathway through which this may occur. While we did not find evidence for interactive effects of rainfall and plant host on microbial beta diversity, we found that beta diversity varied in response to the main effects of watering and host treatments and further, that drought combined with invasive competition led to shifts in abundance of more functions and families than invasive competition or drought alone. Thus, while overall community composition was not sensitive to interactive effects of invasive competition and watering, interacting global change factors have the potential to alter numerous specific functional pathways in native rhizosphere communities. Conversely, competition with native forbs did not appear to shift the abundance of specific functions or microbial families within invasive grasses regardless of watering treatment, indicating that invasive grass microbiomes are less affected by both competition and climate change in this community. Further, invasive grasses seem to be responsible for sourcing a majority of microbial families and functions in native-invasive mixes regardless of watering treatment, but with the highest sourcing of microbial families under drought. This suggests that invasives are likely exerting stronger selection on the microbial rhizosphere community than natives overall and that invasive-associated microbes may be able to outcompete native-associated microbes even during drought.
Competition with invasives shifts native microbial communities
Invasive grass competition changed the overall community similarity between native rhizospheres, and led to enrichment in the abundance of specific families and functions. In previous work utilizing the same set of species, we found that taxonomic beta diversity significantly differed between invasive grasses, native forbs, and native-invasive pairs using 16 S rRNA gene amplicon sequencing (Rodríguez-Caballero et al. 2020; LaForgia et al. 2022). While we did not find that here, it is possible that intra-group interactions (i.e., interactions between native forbs or between invasive grasses) in the field mask differences between natives and invasives as plants compete for limited nutrients (Hawkins and Crawford 2018). Despite this, our findings that invasive competition shifts native rhizosphere microbes is generally well-supported (Komatsu and Simms 2019; Rodríguez-Caballero et al. 2020; LaForgia et al. 2022), and we speculate that these invasive-associated microbial shifts play a role in the direction and magnitude of native-invasive interactions as seen elsewhere (Klironomos 2002; Emam et al. 2014). The sole function that increased in abundance under invasive competition was OprP. OprP is expressed during phosphate starvation, has an affinity for orthophosphate (the preferred form of P uptaken by plants), and genes with this annotation have been previously reported in the barley rhizosphere (Siehnel et al. 1988, 1992; Chhabra et al. 2013). While we do not know for certain that genes with this annotation perform the same function here, the enrichment of this function in native-invasive mixes may indicate competition for key nutrients like P by microbes during host competition.
Competition between natives and invasives also led to shifts in the abundance of numerous families including an enrichment of families in the phylum Proteobacteria, which tend to dominate serpentine soils (Mattarozzi et al. 2017) as well as grass rhizosphere microbiomes (Singh et al. 2007), and a decrease in families in Actinobacteria under competition. Both Actinobacteria and Proteobacteria are commonly enriched in serpentine soils, especially in the rhizospheres for species with high heavy metal tolerance (Abou-Shanab et al. 2009; Navarro-Noya et al. 2010; Mattarozzi et al. 2017; Igwe et al. 2021), thus it is interesting that these groups had divergent responses to our treatments. We speculate that, in this study, Proteobacteria are more closely associated with the fast-growing invasive grasses while Actinobacteria are more closely associated with heavy-metal tolerant natives. Thus, the differential responses observed may be due to adaptive differences of microbes within phyla - Actinobacteria tend to be slow-growing with low metabolic rates, while Proteobacteria are largely fast-growing with high metabolic rates (Mattarozzi et al. 2017).
Of particular note within Proteobacteria, we observed a greater than 5 log-fold increase in Burkholderiales in native-invasive mixes compared to native subplots. This corroborates our previous findings in which members of this family were predicted to be primarily sourced from invasive grasses and whose abundance was correlated with invasive biomass during competition (LaForgia et al. 2022). Members of the Burkholderiales are often beneficial endophytes of grasses and can provide antimicrobial, antifungal and nitrogen fixation functions (Dos Santos et al. 2012; Eberl and Vandamme 2016; Depoorter et al. 2016; Shehata et al. 2016; Banerjee et al. 2018). Given the results of this and our previous work, we hypothesize that members of the Burkholderiales may play a direct or indirect role in aiding the competitive dominance of invasive grasses in California grassland ecosystems. Together, these findings suggest that invasive grasses may be shifting rhizosphere communities to their competitive benefit.
Drought amplifies microbial effects of invasive competition
Although competition with invaders was associated with some shifts in the microbial community, the majority of changes occurred when natives competed with invasives under drought. Specifically, functions that were less abundant in native-invasive mixes under drought compared to natives in controls had annotations related to transport and metabolism, energy production, replication, mobilome, and defense. Many of these functions are related to secondary metabolism, which may provide a competitive advantage under local serpentine conditions (Haferburg et al. 2009; Porter et al. 2017), but also have a high energetic cost (Yan et al. 2018). Thus, they may be less advantageous under resource-limited conditions such as during drought when there is decreased microbial biomass and enzymatic activity (Sardans and Peñuelas 2005; Baldrian et al. 2010). Alternatively, microbial taxa associated with these functions may be simply less abundant during drought, though this seems less probable given the observed function redundancy in these communities across watering treatments. Regardless, it is possible that a decrease in genes associated with secondary metabolism under drought may relate to a higher susceptibility of natives to invasive grass competition.
Genes with a higher abundance under drought in native-invasive mixes had putative functional annotations related largely to nutrient acquisition. For example, potassium is a critical macronutrient for microbes and their plant hosts (Pandey et al. 2020), and increased abundance of genes with predicted Kup, a potassium uptake protein, annotations may relate to increased competition for limited nutrients in native-invasive mixes during drought. Another putative function that increased, RpoN, can act as a regulator of nitrogen assimilation and virulence (Kullik et al. 1991), and in both capacities could provide competitive dominance for invasives through nutrient acquisition or microbial warfare with native-associated microbes, assuming that genes with this annotation perform similarly here. Further, Dcp, which also included genes that increased in mixes under drought, is reported to be involved in metabolism in cleaving a variety of bacterial peptides, and may relate to survival during carbon deficiency (Vimr et al. 1983; Paschoalin et al. 2005; Jasilionis and Kuisiene 2015; Masuyer et al. 2016). Our findings also highlight that competing with invasive grasses under drought shifts rhizosphere function in a way that differs from the effect of drought on natives alone. Compared to natives exposed to drought, native-invasive mixes under drought displayed higher levels of genes associated with transcription and translation, possibly indicating a capacity for increased microbial activity in mixed subplots versus native subplots during drought. Thus, it is possible that invasive grasses under drought may be aided by their recruited microbiome at the expense of natives during competition.
Competition from invaders under drought was also associated with shifts in the abundance of 22 families (10 decreases, 12 increases). Largely, we saw an enrichment of families in the phylum Proteobacteria and Firmicutes, which have been previously reported to be abundant under drought conditions in grassland soils (Naylor and Coleman-Derr 2017; Ochoa-Hueso et al. 2018; Zhang et al. 2020a; Xu et al. 2021). While families in Actinobacteria saw a decrease under competition with invaders, especially under drought. Both Actinobacteria and Proteobacteria have been reported to be enriched under drought (Xu et al. 2021). Sandaracinaceae (Proteobacteria) and Hungateiclostridiaceae (Firmicutes), which were both sourced predominantly from grasses, especially under drought, include members that can degrade alternative carbon sources (Mohr et al. 2012; Zhang et al. 2018b; Rettenmaier et al. 2019, 2020). This may be particularly beneficial under drought when there are reported changes in plant litter chemistry and carbon metabolism (Su et al. 2020; Malik and Bouskill 2022). Lastly, Streptosporangiaceae (Actinobacteria) was consistently higher in native rhizospheres than mixes regardless of watering treatment, with the largest decline in abundance during competition with invasives under drought. Interestingly, despite its higher abundance in natives, this family was sourced more by invasives in mixes and this was exacerbated in drought. Members of Streptosporangiaceae have been isolated from soil and plant materials (Kudo et al. 1993; Ara and Kudo 2007; Janso and Carter 2010), and are thought to harbor novel and interesting antibiotic compounds (Lazzarini et al. 2000). These results, combined with the fact that invasive grasses sourced a higher proportion of microbial families under drought, underline the importance of investigating plant-microbe interactions across environmental gradients and add to a growing body of literature suggesting negative synergistic effects of interacting global change drivers.
Effects of invasive competition under increased watering
Invasive grass competition under well-watered conditions drove substantially fewer changes in the microbial community compared to competition under drought, with native-invasive mixed subplots having less abundance of genes with predicted annotations related to signaling and metabolism, including BioA and KdpD. BioA is required for biotin synthesis, which is an important cofactor of carboxylases for rhizobia metabolism and root colonization (Streit et al. 1996; Heinz and Streit 2003); BioA has also been predicted to confer copper resistance in Burkholderia (Higgins et al. 2020). KdpD is an osmosensitive sensor kinase related to potassium homeostasis (Lipa and Janczarek 2020), which can activate under low potassium conditions and has been previously reported in rhizobia (Prell et al. 2012; Sablok et al. 2017). Potassium has been suggested to be a key signal of environmental cues as well as plant-microbe interactions and microbial virulence (Freeman et al. 2013; Tan 2021; Feng et al. 2022). Given that both BioA and KdpD have been linked to stress response and documented roles in plant-microbe interactions, assuming they have similar functional roles here, they may be local adaptations by microbes to survival in serpentine soil, or possibly play roles in regulating native forb-microbe associations. Moreover, no families differentially varied between watered native-invasive mixes and native control subplots. High water levels may therefore ameliorate competition for below ground resources between native forbs and invasive grasses as competition for light aboveground intensifies (Coleman and Levine 2006). In another study, the presence of invasive grasses dampened the beneficial effects of watering rather than exacerbating any negative effects (LaForgia et al. 2020). Thus, drought is likely to be a stronger driver of invasive-mediated microbiome shifts under future climates than high rainfall.
Invasives drive community assembly dynamics during competition
Contrary to our hypothesis of equal contribution to the microbiome from natives and invaders, we observed that invaders sourced an overall higher proportion of the taxonomic families and functional potential in the rhizosphere microbiome. This contrasts with previous results from 16 S rRNA gene amplicon work from a well-watered greenhouse experiment which found equal contribution by natives and invasives in native-forb pairs, despite higher biomass in grasses (LaForgia et al. 2022). While there are several possible explanations for this discrepancy, including methodological differences, one intriguing hypothesis is that the inclusion of multiple invasive species may result in a synergistic or additive competitive dominance over native rhizospheres (Kuebbing et al. 2013). While the overall taxonomic community was sourced predominantly by invasives, with an enrichment in drought, some taxonomic families (e.g. unclassified Coriobacteriia [Actinobacteria]) show a more complex pattern, including relatively higher sourcing from natives during well-watered treatments compared to control and drought treatments. While our taxonomic results suggest clear shifting due to selection by drought and/or invasives, the more subtle differences in functional sourcing suggest that there is likely selective pressure for community functional redundancy. This decoupling of taxonomic and functional results has been reported previously with taxonomic composition and functional potential being shaped by different abiotic and biotic factors (Louca et al. 2018).
The dominance of invasives as the source of microbes and functions regardless of watering treatment may not be surprising as invasive grasses account for 71% of plant cover at this site in general and are the largest biomass contributor there as well (LaForgia, unpublished data). However, the fact that microbial sourcing was unrelated to plant biomass in our previous work (LaForgia et al. 2022) suggests that microbial community assembly is likely to be more complex beyond host relative abundance. Further, invasive plants are known to modify the rhizosphere microbiome to negatively impact native-microbe interactions, as well as modify the soil chemistry to impact functional activity, with the intensity of these impacts increasing with invasive density (Zhang et al. 2020b; Fahey et al. 2020). In spite of invasive dominance, in mixed subplots we do see one function with differential abundance that is consistently sourced solely by native-associated microbes. That function, PcoD, encodes an internal membrane protein pump that works in copper translocation and is required for copper resistance (Rensing and Grass 2003; Solioz 2019). Copper is one of several heavy metals prevalent in serpentine soils that both microbes and plants need to evolve tolerance of to thrive (Fernandez et al. 1999; Llugany et al. 2003). The sourcing of this function highlights the importance of native plants for sustaining microbial functionality in grassland ecosystems given their evolution with local microbes. As climate change and invasion lead to shifts in the microbiome, we may start to see the loss of beneficial microbes as other taxa with the same functional potential are selected for or are brought in by invaders. These microbes may outcompete native-associated microbes and may not provide the same benefits to native plants that native-associated microbes would.
Caveats
While metagenomics allows us to move beyond high throughput amplicon sequencing to ask questions about the functional potential of a community, it does not provide information about which genes are being actively expressed. Future work should use metatranscriptomics and/or metaproteomics to directly profile how invasion and climate change may interact to shape functional activity in the rhizosphere microbiome. Further, due to volunteer species that were not seeded, as well as species that were seeded but did not establish, we had variable plant community composition within treatments and were only able to sequence 66 of 150 total samples, thus lowering replication and the power to identify differences. While we found that host plant composition was not a significant driver within treatments here, we know from other work the importance of host plant identity in shaping the microbial community (Aleklett et al. 2015). It is also possible that sampling root endospheres directly instead of the rhizosphere would have better let us detect host-specific signatures. However, we chose to sample the rhizosphere as that is where microbial crosstalk is likely occurring between competing microbes associated with invasive and natives (Sindhu et al. 2017) and is likely best to detect effects of competition. Regardless, our results are in line with other work showing that taxonomic beta diversity of the rhizosphere microbiome is heavily affected by drought (Ochoa-Hueso et al. 2018) and less so by plant species composition (Sayer et al. 2017), including invasion (Fahey et al. 2020). Similarly, the response of taxonomy to climate (Chen et al. 2022) and to invasion (Gibbons et al. 2017) has been shown to be stronger than functional response, likely due to functional redundancy across taxa.
Conclusions
To summarize, we found that both drought and competition with invasives shift native microbial communities, and that the combined effects of drought and competition, while not observed at a community level, were seen when looking at specific functions and families. A majority of shifts were in functions associated with stress, starvation and survival in minimal nutrient conditions and include functions involved with efflux and transport of heavy metals. We saw increased abundance of Proteobacteria and decreased abundance of Actinobacteria during competition, with the strongest shifts during drought. Of the families that shifted of particular note is the invasive grass-associated Burkholderiales, which in combination with our previous work, seems to possibly be a key player in assisting invasives with their competitive dominance. Further community composition investigations revealed that invasives are the dominant source of functional potential and taxonomic identity in the microbial communities of invaded subplots regardless of rainfall variability. Altogether our results support growing literature suggesting that invasives may shift rhizosphere microbial communities for their benefit, possibly at the expense of natives and native-associated microbial communities.
Data availability
The raw sequence reads generated for this metagenomic project were deposited to Genbank under BioProject accession no. PRJNA925931. All code used in this work is openly available on GitHub (https://github.com/marinalaforgia/McL_Metagenomics/).
References
Abou-Shanab RAI, van Berkum P, Angle JS, Abou-Shanab RAI, van Berkum P, Angle JS, Delorme TA, Chaney RL, Ghozlan HA, Ghanem K, Moawad H (2009) Characterization of Ni-resistant bacteria in the rhizosphere of the hyperaccumulator Alyssum murale by 16S rRNA gene sequence analysis. World J Microbiol Biotechnol 26:101–108. https://doi.org/10.1007/s11274-009-0148-6
Acosta-Martinez V, Moore-Kucera J, Cotton J, Acosta-Martinez V, Moore-Kucera J, Cotton J, Gardner T, Wester D (2014) Soil enzyme activities during the 2011 Texas record drought/heat wave and implications to biogeochemical cycling and organic matter dynamics. Appl Soil Ecol 75:43–51. https://doi.org/10.1016/j.apsoil.2013.10.008
Aleklett K, Leff JW, Fierer N, Hart M (2015) Wild plant species growing closely connected in a subalpine meadow host distinct root-associated bacterial communities. PeerJ 3. https://doi.org/10.7717/peerj.804
Ara I, Kudo T (2007) Sphaerosporangium gen. nov., a new member of the family Streptosporangiaceae, with descriptions of three new species as Sphaerosporangium melleum sp. nov., Sphaerosporangium rubeum sp. nov. and Sphaerosporangium cinnabarinum sp. nov., and transfer of Streptosporangium viridialbum Nonomura and Ohara 1960 to Sphaerosporangium viridialbum comb. nov. Nippon Hosenkin Gakkaishi 21:11–21. https://doi.org/10.3209/saj.SAJ210102
Baldrian P, Merhautová V, Petránková M, Baldrian P, Merhautová V, Petránková M, Cajthaml T, Šnajdr J (2010) Distribution of microbial biomass and activity of extracellular enzymes in a hardwood forest soil reflect soil moisture content. Appl Soil Ecol 46:177–182. https://doi.org/10.1016/j.apsoil.2010.08.013
Banerjee S, Schlaeppi K, van der Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576. https://doi.org/10.1038/s41579-018-0024-1
Batten KM, Scow KM, Davies KF, Harrison SP (2006) Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol Invasions 8:217–230. https://doi.org/10.1007/s10530-004-3856-8
Bushnell B (2022) BBMap. https://sourceforge.net/projects/bbmap/
Cantalapiedra CP, Hernández-Plaza A, Letunic I, Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J (2021) eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. https://doi.org/10.1093/molbev/msab293
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. https://doi.org/10.1038/ismej.2013.196
Chen H, Ma K, Lu C, Chen H, Ma K, Lu C, Fu Qi, Qiu Y, Zhao J, Huang Yu, Yang Y, Schadt CW, Chen H (2022) Functional redundancy in soil microbial community based on metagenomics across the globe. Front Microbiol 13:878978. https://doi.org/10.3389/fmicb.2022.878978
Cheng YT, Zhang L, He SY (2019) Plant-microbe interactions facing environmental challenge. Cell Host Microbe 26:183–192. https://doi.org/10.1016/j.chom.2019.07.009
Chhabra S, Brazil D, Morrissey J, Chhabra S, Brazil D, Morrissey J, Burke JI, O’Gara F, N. Dowling D (2013) Characterization of mineral phosphate solubilization traits from a barley rhizosphere soil functional metagenome. Microbiologyopen 2:717–724. https://doi.org/10.1002/mbo3.110
Chung YA, Miller TEX, Rudgers JA (2015) Fungal symbionts maintain a rare plant population but demographic advantage drives the dominance of a common host. J Ecol 103:967–977. https://doi.org/10.1111/1365-2745.12406
Coleman HM, Levine JM (2006) Mechanisms underlying the impacts of exotic annual grasses in a coastal California meadow. Biol Invasions 9:65–71. https://doi.org/10.1007/s10530-006-9008-6
David AS, Quintana-Ascencio PF, Menges ES et al (2019) Soil Microbiomes Underlie Population Persistence of an Endangered Plant Species. Am Nat 194. https://doi.org/10.1086/704684
Davis NM, Proctor DM, Holmes SP, Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ (2018) Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. https://doi.org/10.1186/s40168-018-0605-2
Dawson W, Hör J, Egert M, Dawson W, Hör J, Egert M, van Kleunen M, Pester M (2017) A small number of low-abundance bacteria dominate plant species-specific responses during Rhizosphere colonization. Front Microbiol 8:975. https://doi.org/10.3389/fmicb.2017.00975
Depoorter E, Bull MJ, Peeters C, Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E (2016) Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. https://doi.org/10.1007/s00253-016-7520-x
Dos Santos PC, Fang Z, Mason SW, Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R (2012) Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. https://doi.org/10.1186/1471-2164-13-162
Doubková P, Suda J, Sudová R (2012) The symbiosis with arbuscular mycorrhizal fungi contributes to plant tolerance to serpentine edaphic stress. Soil Biol Biochem 44:56–64. https://doi.org/10.1016/j.soilbio.2011.09.011
Dudney J, Hallett LM, Larios L et al (2017) Lagging behind: have we overlooked previous-year rainfall effects in annual grasslands? J Ecol 105:484–495
Eberl L, Vandamme P (2016) Members of the genus Burkholderia: good and bad guys. F1000Res 5. https://doi.org/10.12688/f1000research.8221.1
Edwards J, Johnson C, Santos-Medellín C, Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. https://doi.org/10.1073/pnas.1414592112
Ehrenfeld JG (2010) Ecosystem consequences of biological Invasions. Annu Rev Ecol Evol Syst 41:59–80
Emam TM, Espeland EK, Rinella MJ (2014) Soil sterilization alters interactions between the native grass Bouteloua gracilis and invasive Bromus tectorum. J Arid Environ 111:91–97. https://doi.org/10.1016/j.jaridenv.2014.08.006
Eren AM, Esen ÖC, Quince C, Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO (2015) Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. https://doi.org/10.7717/peerj.1319
Ettinger C, LaForgia M (2021) Plant-associated microbiome sampling protocol for field work. protocols.io. https://doi.org/10.17504/protocols.io.buttnwnn
Fahey C, Koyama A, Antunes PM, Fahey C, Koyama A, Antunes PM, Dunfield K, Flory SL (2020) Plant communities mediate the interactive effects of invasion and drought on soil microbial communities. ISME J 14:1396–1409. https://doi.org/10.1038/s41396-020-0614-6
Feng X-Y, Tian Y, Cui W-J et al (2022) The PTS-KdpDE-KdpFABC Pathway contributes to low potassium stress adaptation and competitive nodulation of Sinorhizobium fredii. MBio 13:e0372121. https://doi.org/10.1128/mbio.03721-21
Fernandez S, Seoane S, Merino A (1999) Plant heavy metal concentrations and soil biological properties in agricultural serpentine soils. Commun Soil Sci Plant Anal 30:1867–1884. https://doi.org/10.1080/00103629909370338
Fierer N, Leff JW, Adams BJ, Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. https://doi.org/10.1073/pnas.1215210110
Freeman ZN, Dorus S, Waterfield NR (2013) The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog 9:e1003201. https://doi.org/10.1371/journal.ppat.1003201
Gibbons SM, Lekberg Y, Mummey DL et al (2017) Invasive Plants Rapidly Reshape Soil Properties in a Grassland Ecosystem. mSystems 2. https://doi.org/10.1128/mSystems.00178-16
Haferburg G, Groth I, Möllmann U, Haferburg G, Groth I, Möllmann U, Kothe E, Sattler I (2009) Arousing sleeping genes: shifts in secondary metabolism of metal tolerant actinobacteria under conditions of heavy metal stress. Biometals 22:225–234. https://doi.org/10.1007/s10534-008-9157-4
Hartmann A, Schmid M, van Tuinen D, Berg G (2008) Plant-driven selection of microbes. Plant Soil 321:235–257. https://doi.org/10.1007/s11104-008-9814-y
Hawkins AP, Crawford KM (2018) Interactions between plants and soil microbes may alter the relative importance of intraspecific and interspecific plant competition in a changing climate. AoB Plants 10:ly039. https://doi.org/10.1093/aobpla/ply039
Heinz EB, Streit WR (2003) Biotin limitation in Sinorhizobium meliloti strain 1021 alters transcription and translation. Appl Environ Microbiol 69. https://doi.org/10.1128/AEM.69.2.1206-1213.2003
Higgins S, Gualdi S, Pinto-Carbó M, Eberl L (2020) Copper resistance genes of Burkholderia cenocepacia H111 identified by transposon sequencing. Environ Microbiol Rep 12:241–249. https://doi.org/10.1111/1758-2229.12828
Hortal S, Lozano YM, Bastida F, Hortal S, Lozano YM, Bastida F, Armas C, Moreno JL, Garcia C, Pugnaire FI (2017) Plant-plant competition outcomes are modulated by plant effects on the soil bacterial community. Sci Rep 7:17756. https://doi.org/10.1038/s41598-017-18103-5
Hyatt D, Chen G-L, Locascio PF, Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. https://doi.org/10.1186/1471-2105-11-119
Igwe AN, Quasem B, Liu N, Vannette RL (2021) Plant phenology influences rhizosphere microbial community and is accelerated by serpentine microorganisms in Plantago erecta. FEMS Microbiol Ecol 97:fiab085. https://doi.org/10.1093/femsec/fiab085
Janso JE, Carter GT (2010) Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl Environ Microbiol 76. https://doi.org/10.1128/AEM.02959-09
Jasilionis A, Kuisiene N (2015) Characterization of a novel thermostable oligopeptidase from Geobacillus thermoleovorans DSM 15325. J Microbiol Biotechnol 25:1070–1083. https://doi.org/10.4014/jmb.1412.12049
Jones P, Binns D, Chang HY et al (2014) InterProScan 5: genome-scale protein function classification. Bioinf 30. https://doi.org/10.1093/bioinformatics/btu031
Jonsson V, Österlund T, Nerman O, Kristiansson E (2016) Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics 17:78. https://doi.org/10.1186/s12864-016-2386-y
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70. https://doi.org/10.1038/417067a
Knights D, Kuczynski J, Charlson ES, Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. https://doi.org/10.1038/nmeth.1650
Komatsu KJ, Simms EL (2019) Invasive legume management strategies differentially impact mutualist abundance and benefit to native and invasive hosts. Restor Ecol 28:378–386
Kudo T, Itoh T, Miyadoh S et al (1993) Herbidospora gen. nov., a new genus of the family Streptosporangiaceae Goodfellow. 1990. Int J Syst Bacteriol 43. https://doi.org/10.1099/00207713-43-2-319
Kuebbing SE, Nuñez MA, Simberloff D (2013) Current mismatch between research and conservation efforts: the need to study co-occurring invasive plant species. Biol Conserv 160:121–129. https://doi.org/10.1016/j.biocon.2013.01.009
Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer HM (1991) Bradyrhizobium japonicum has two differentially regulated, functional homologs of the sigma 54 gene (rpoN). J Bacteriol 173:1125–1138. https://doi.org/10.1128/jb.173.3.1125-1138.1991. (J Bacteriol 173)
LaForgia ML, Spasojevic MJ, Case EJ, LaForgia ML, Spasojevic MJ, Case EJ, Latimer AM, Harrison SP (2018) Seed banks of native forbs, but not exotic grasses, increase during extreme drought. Ecology 99:896–903. https://doi.org/10.1002/ecy.2160
LaForgia ML, Harrison SP, Latimer AM (2020) Invasive species interact with climatic variability to reduce success of natives. Ecology 101:e03022. https://doi.org/10.1002/ecy.3022
LaForgia ML, Kang H, Ettinger CL (2022) Invasive grass dominance over native forbs is linked to shifts in the bacterial rhizosphere Microbiome. Microb Ecol 84:496–508. https://doi.org/10.1007/s00248-021-01853-1
Lazzarini A, Cavaletti L, Toppo G, Marinelli F (2000) Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek 78(3–4):399–405
Legay N, Piton G, Arnoldi C, Legay N, Piton G, Arnoldi C, Bernard L, Binet M-N, Mouhamadou B, Pommier T, Lavorel S, Foulquier A, Clément J-C (2017) Soil legacy effects of climatic stress, management and plant functional composition on microbial communities influence the response of Lolium perenne to a new drought event. Plant Soil 424:233–254. https://doi.org/10.1007/s11104-017-3403-x
Li D, Liu CM, Luo R, Li D, Liu C-M, Luo R, Sadakane K, Lam T-W (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31:1674–1676. https://doi.org/10.1093/bioinformatics/btv033. (Bioinformatics 31)
Lipa P, Janczarek M (2020) Phosphorylation systems in symbiotic nitrogen-fixing bacteria and their role in bacterial adaptation to various environmental stresses. PeerJ 8:e8466. https://doi.org/10.7717/peerj.8466
Llugany M, Lombini A, Poschenrieder C et al (2003) Different mechanisms account for enhanced copper resistance in Silene armeria ecotypes from mine spoil and serpentine sites. Plant Soil 251:55–63. https://doi.org/10.1023/A:1022990525632
Louca S, Polz MF, Mazel F, Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, Doebeli M, Parfrey LW (2018) Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943. https://doi.org/10.1038/s41559-018-0519-1
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Lozano YM, Hortal S, Armas C, Pugnaire FI (2019) Soil micro-organisms and competitive ability of a tussock grass species in a dry ecosystem. J Ecol 107:1215–1225
Maestre FT, Delgado-Baquerizo M, Jeffries TC, Maestre FT, Delgado-Baquerizo M, Jeffries TC, Eldridge DJ, Ochoa V, Gozalo B, Quero JL, García-Gómez M, Gallardo A, Ulrich W, Bowker MA, Arredondo T, Barraza-Zepeda C, Bran D, Florentino A, Gaitán J, Gutiérrez JR, Huber-Sannwald E, Jankju M, Mau RL, Miriti M, Naseri K, Ospina A, Stavi I, Wang D, Woods NN, Yuan X, Zaady E, Singh BK (2015) Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci U S A 112:15684–15689. https://doi.org/10.1073/pnas.1516684112
Malik AA, Bouskill NJ (2022) Drought impacts on microbial trait distribution and feedback to soil carbon cycling. Funct Ecol 36:1442–1456. https://doi.org/10.1111/1365-2435.14010
Masuyer G, Cozier GE, Kramer GJ et al (2016) Crystal structure of a peptidyl-dipeptidase K‐26‐DCP from Actinomycete in complex with its natural inhibitor. FEBS J 283:4357. https://doi.org/10.1111/febs.13928
Mattarozzi M, Manfredi M, Montanini B, Mattarozzi M, Manfredi M, Montanini B, Gosetti F, Sanangelantoni AM, Marengo E, Careri M, Visioli G (2017) A metaproteomic approach dissecting major bacterial functions in the rhizosphere of plants living in serpentine soil. Anal Bioanal Chem 409:2327–2339. https://doi.org/10.1007/s00216-016-0175-8
McGhee JJ, Rawson N, Bailey BA, McGhee JJ, Rawson N, Bailey BA, Fernandez-Guerra A, Sisk-Hackworth L, Kelley ST (2020) Meta-SourceTracker: application of bayesian source tracking to shotgun metagenomics. PeerJ 8:e8783. https://doi.org/10.7717/peerj.8783
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. https://doi.org/10.1038/ncomms11257
Mohr KI, Garcia RO, Gerth K et al (2012) Sandaracinus amylolyticus gen. nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. nov. Int J Syst Evol Microbiol 62. https://doi.org/10.1099/ijs.0.033696-0
Navarro-Noya YE, Jan-Roblero J, González-Chávez MC et al (2010) Bacterial communities associated with the rhizosphere of pioneer plants (Bahia xylopoda and Viguiera linearis) growing on heavy metals-contaminated soils. Antonie Van Leeuwenhoek 97. https://doi.org/10.1007/s10482-010-9413-9
Naylor D, Coleman-Derr D (2017) Drought stress and root-associated bacterial communities. Front Plant Sci 8:2223. https://doi.org/10.3389/fpls.2017.02223
Ochoa-Hueso R, Collins SL, Delgado-Baquerizo M et al (2018) Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob Chang Biol 24:2818–2827. https://doi.org/10.1111/gcb.14113
Oksanen J, Simpson G, Blanchet F, Kindt R, Legendre P, Minchin P, O’Hara R, Solymos P, Stevens M, Szoecs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, DeCaceres M, Durand S, ... Weedon J (2022) vegan: Community Ecology Package (Version R package version 2.6-4) [Computer software]. https://CRAN.R-project.org/package=vegan
Pandey D, Kehri HK, Zoomi I et al (2020) Potassium Solubilizing Microbes: Diversity, Ecological Significances and Biotechnological Applications. In: Yadav AN, Singh J, Rastegari AA, Yadav N (eds) Plant microbiomes for sustainable agriculture. Springer Nature
Paschoalin T, Carmona AK, Oliveira V, Paschoalin T, Carmona AK, Oliveira V, Juliano L, Travassos LR (2005) Characterization of thimet- and neurolysin-like activities in Escherichia coli M3A peptidases and description of a specific substrate. Arch Biochem Biophys 441:25–34. https://doi.org/10.1016/j.abb.2005.06.011
Paterson E, Gebbing T, Abel C et al (2007) Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol 173. https://doi.org/10.1111/j.1469-8137.2006.01931.x
Patro R, Duggal G, Love MI et al (2017) Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat Methods 14:417. https://doi.org/10.1038/nmeth.4197
Porter SS, Chang PL, Conow CA et al (2017) Association mapping reveals novel serpentine adaptation gene clusters in a population of symbiotic Mesorhizobium. ISME J 11. https://doi.org/10.1038/ismej.2016.88
Prell J, Mulley G, Haufe F et al (2012) The PTS(Ntr) system globally regulates ATP-dependent transporters in Rhizobium leguminosarum. Mol Microbiol 84:117–129. https://doi.org/10.1111/j.1365-2958.2012.08014.x
Qu Q, Zhang Z, Wjgm P et al (2020) Rhizosphere Microbiome Assembly and its impact on Plant Growth. J Agric Food Chem 68. https://doi.org/10.1021/acs.jafc.0c00073
R Core Team (2022) R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/
Reinhart KO, Callaway RM (2006) Soil biota and invasive plants. New Phytol 170:445–457. https://doi.org/10.1111/j.1469-8137.2006.01715.x
Rensing C, Grass G (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27:197–213. https://doi.org/10.1016/S0168-6445(03)00049-4
Rettenmaier R, Gerbaulet M, Liebl W, Zverlov VV (2019) Hungateiclostridium mesophilum sp. nov., a mesophilic, cellulolytic and spore-forming bacterium isolated from a biogas fermenter fed with maize silage. Int J Syst Evol Microbiol 69. https://doi.org/10.1099/ijsem.0.003663
Rettenmaier R, Schneider M, Munk B, Rettenmaier R, Schneider M, Munk B, Lebuhn M, Jünemann S, Sczyrba A, Maus I, Zverlov V, Liebl W (2020) Importance of Defluviitalea raffinosedens for hydrolytic biomass degradation in co-culture with Hungateiclostridium thermocellum. Microorganisms 8:915. https://doi.org/10.3390/microorganisms8060915
Richardson AE, Barea J-M, McNeill AM, Prigent-Combaret C (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339. https://doi.org/10.1007/s11104-009-9895-2
Ritpitakphong U, Falquet L, Vimoltust A, Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux J-P, L’Haridon F (2016) The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol 210:1033–1043. https://doi.org/10.1111/nph.13808
Rodríguez-Caballero G, Caravaca F, Díaz G, Rodríguez-Caballero G, Caravaca F, Díaz G, Torres P, Roldán A (2020) The invader Carpobrotus edulis promotes a specific rhizosphere microbiome across globally distributed coastal ecosystems. Sci Total Environ 719:137347. https://doi.org/10.1016/j.scitotenv.2020.137347
Sablok G, Rosselli R, Seeman T, Sablok G, Rosselli R, Seeman T, van Velzen R, Polone E, Giacomini A, La Porta N, Geurts R, Muresu R, Squartini A (2017) Draft genome sequence of the nitrogen-fixing Rhizobium sullae type strain IS123T focusing on the key genes for symbiosis with its host Hedysarum coronarium L. Front Microbiol 8:1348. https://doi.org/10.3389/fmicb.2017.01348
Sardans J, Peñuelas J (2005) Drought decreases soil enzyme activity in a Mediterranean Quercus ilex L. forest. Soil Biol Biochem 37:455–461. https://doi.org/10.1016/j.soilbio.2004.08.004
Sayer EJ, Oliver AE, Fridley JD, Sayer EJ, Oliver AE, Fridley JD, Askew AP, Mills RTE, Grime JP (2017) Links between soil microbial communities and plant traits in a species-rich grassland under long-term climate change. Ecol Evol 7:855–862. https://doi.org/10.1002/ece3.2700
Shehata HR, Ettinger CL, Eisen JA, Raizada MN (2016) Genes required for the anti-fungal activity of a bacterial endophyte isolated from a corn landrace grown continuously by Subsistence Farmers since 1000 BC. Front Microbiol 7:1548. https://doi.org/10.3389/fmicb.2016.01548
Siehnel RJ, Worobec EA, Hancock RE (1988) Regulation of components of the Pseudomonas aeruginosa phosphate-starvation-inducible regulon in Escherichia coli. Mol Microbiol 2. https://doi.org/10.1111/j.1365-2958.1988.tb00038.x
Siehnel RJ, Egli C, Hancock RE (1992) Polyphosphate-selective porin OprO of Pseudomonas aeruginosa: expression, purification and sequence. Mol Microbiol 6. https://doi.org/10.1111/j.1365-2958.1992.tb01407.x
Sindhu SS, Sehrawat A, Sharma R et al (2017) Belowground Microbial Crosstalk and Rhizosphere Biology. In: Singh DP, Singh HB, Prabha R (eds) Plant-microbe interactions in agro-ecological perspectives: volume 1: fundamental mechanisms, methods and functions. Springer
Singh BK, Munro S, Potts JM, Millard P (2007) Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl Soil Ecol 36:147–155. https://doi.org/10.1016/j.apsoil.2007.01.004
Solioz M (2019) Copper Disposition in Bacteria. In: Kerkar N, Roberts EA (eds) Clinical and Translational Perspectives on WILSON DISEASE. Academic Press, pp 101–113
Steinegger M, Söding J (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35:1026–1028. https://doi.org/10.1038/nbt.3988
Streit WR, Joseph CM, Phillips DA (1996) Biotin and other water-soluble vitamins are key growth factors for alfalfa root colonization by Rhizobium meliloti 1021. Mol Plant Microbe Interact 9. https://doi.org/10.1094/mpmi-9-0330
Su X, Su X, Yang S et al (2020) Drought changed soil organic carbon composition and bacterial carbon metabolizing patterns in a subtropical evergreen forest. Sci Total Environ 736. https://doi.org/10.1016/j.scitotenv.2020.139568
Tan S (2021) Abundant Monovalent Ions as Environmental Signposts for Pathogens during Host Colonization. Infect Immun 89. https://doi.org/10.1128/IAI.00641-20
Thrall PH, Hochberg ME, Burdon JJ, Bever JD (2007) Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol 22:120–126. https://doi.org/10.1016/j.tree.2006.11.007
Traveset A, Richardson DM (2014) Mutualistic interactions and biological invasions. Annu Rev Ecol Evol Syst 45:89–113. https://doi.org/10.1146/annurev-ecolsys-120213-091857
Ulbrich TC, Rivas-Ubach A, Tiemann LK, Ulbrich TC, Rivas-Ubach A, Tiemann LK, Friesen ML, Evans SE (2022) Plant root exudates and rhizosphere bacterial communities shift with neighbor context. Soil Biol Biochem 172:108753. https://doi.org/10.1016/j.soilbio.2022.108753
Vimr ER, Green L, Miller CG (1983) Oligopeptidase-deficient mutants of Salmonella typhimurium. J Bacteriol 153:1259–1265. https://doi.org/10.1128/jb.153.3.1259-1265.1983
Xiong C, Singh BK, He J-Z, Xiong C, Singh BK, He J-Z, Han Y-L, Li P-P, Wan L-H, Meng G-Z, Liu S-Y, Wang J-T, Wu C-F, Ge A-H, Zhang L-M (2021) Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome 9:171. https://doi.org/10.1186/s40168-021-01118-6
Xu L, Dong Z, Chiniquy D, Xu L, Dong Z, Chiniquy D, Pierroz G, Deng S, Gao C, Diamond S, Simmons T, Wipf H-L, Caddell D, Varoquaux N, Madera MA, Hutmacher R, Deutschbauer A, Dahlberg JA, Guerinot ML, Purdom E, Banfield JF, Taylor JW, Lemaux PG, Coleman-Derr D (2021) Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat Commun 12:3209. https://doi.org/10.1038/s41467-021-23553-7
Yan Q, Lopes LD, Shaffer BT et al (2018) Secondary Metabolism and Interspecific Competition Affect Accumulation of Spontaneous Mutants in the GacS-GacA Regulatory System in. MBio 9. https://doi.org/10.1128/mBio.01845-17
Yang J, Kloepper JW, Ryu CM (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4. https://doi.org/10.1016/j.tplants.2008.10.004
Zhang J, Zhang N, Liu YX et al. (2018a) Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci China Life Sci 61. https://doi.org/10.1007/s11427-018-9284-4
Zhang X, Tu B, Dai LR et al (2018b) Petroclostridium xylanilyticum gen. nov., sp. nov., a xylan-degrading bacterium isolated from an oilfield, and reclassification of clostridial cluster III members into four novel genera in a new Hungateiclostridiaceae fam. nov. Int J Syst Evol Microbiol 68. https://doi.org/10.1099/ijsem.0.002966
Zhang B, Yuan Y, Pajerowska-Mukhtar KM et al (2020a) Microbial’s effects on invasive grass’ response to water and nutrient stress. Research Square. https://doi.org/10.21203/rs.3.rs-117418/v1
Zhang HY, Goncalves P, Copeland E, Zhang H-Y, Goncalves P, Copeland E, Qi S-S, Dai Z-C, Li G-L, Wang C-Y, Du D-L, Thomas T (2020b) Invasion by the weed Conyza canadensis alters soil nutrient supply and shifts microbiota structure. Soil Biol Biochem 143:107739. https://doi.org/10.1016/j.soilbio.2020.107739
Acknowledgements
We thank Cathy Koehler and Paul Aigner for their help setting up the watering experiment as well as Sonia L. Ghose and Andrew Latimer for their help with field sampling. We also thank members of the Hallett Lab, Gremer Lab, and Stajich Lab for their insightful feedback and valuable comments. Finally, we would like to thank Susan P. Harrison and Jonathan A. Eisen for useful conversations and suggestions related to this work.
Funding
This work was supported in part by grants from the UC Davis Center for Population Biology Collaborative Project research grant to CLE & MLL, UC Davis Center for Population Biology student grant to CLE, SIGMA XI Grant-In-Aid of Research to CLE, and UC Davis Center for Population Biology Hardman Foundation Research Award to MLL.
Author information
Authors and Affiliations
Contributions
CLE and MLL conceived of and designed the study, performed field and molecular work, undertook data analysis and generated figures, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Xinhua He.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1.82 MB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ettinger, C.L., LaForgia, M.L. Invasive plant species interact with drought to shift key functions and families in the native rhizosphere. Plant Soil 494, 567–588 (2024). https://doi.org/10.1007/s11104-023-06302-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06302-1