Abstract
Background and aims
A considerable majority of the proteins involved in exopolysaccharide synthesis in Rhizobium leguminosarum bv. trifolii are encoded by pss genes located in a large chromosomal region named Pss-I. The aim of this work was to characterize the phenotypic and symbiotic properties of strain Rt1933, which has a mutation in pssE. This gene encodes an enzyme responsible for the second step of EPS unit assembly.
Methods
The EPS-deficient Rt1933pssE strain was obtained via random mutagenesis using the mTn5SSgusA40 transposon. The mutation site in the Rt1933 genome was identified using hybridization, PCR amplification, and sequence analysis. Complementation of the pssE mutation was performed using biparental conjugation and a set of plasmids containing different fragments of the Pss-I region. The phenotypic properties of this mutant were established in growth kinetics experiments, as well as motility, sensitivity and hydrophobicity assays. The symbiotic proficiency of the Rt1933 strain in interaction with red clover (Trifolium pratense) was determined in plant tests, whereas occupation of root nodules by this mutant was investigated using light microscopy and bacteria harboring gusA for β-glucuronidase.
Results
An exo99 mutation in Rt1933 was identified at 3′-end of the pssE gene located in region Pss-I, which resulted in a lack of 16 amino acids at the C-end of PssE. This mutation totally abolished EPS synthesis in R. leguminosarum bv. trifolii. Strain Rt1933 was characterized by considerably decreased growth kinetics and motility, and an increased sensitivity to some stress factors. Also, the hydrophobicity of the mutant cells differed significantly from that of the wild-type Rt24.2 and the complemented Rt1933 cells. Moreover, the pssE mutant showed strong disturbances in symbiosis with clover; it induced much fewer nodules on clover roots at a later time than normal, and the mass of the plants inoculated with the mutant was significantly lower than that of the plants inoculated with the wild-type strain.
Conclusions
The pssE gene plays a crucial role in EPS synthesis in R. leguminosarum bv. trifolii, and the presence of this polysaccharide affects the cell-surface properties of the bacterium and is required for both adaptation to stress conditions and the establishment of effective symbiosis with clover plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhizobium leguminosarum bv. trifolii is a Gram-negative soil bacterium which establishes symbiotic interactions with clover plants (Trifolium spp.). Nitrogen-fixing symbiosis is a complex process which involves an exchange of many signals, among which legume flavonoids and rhizobial lipochitooligosaccharides play crucial roles (Janczarek et al. 2015c; López-Baena et al. 2016). An important part in that process is also played by acidic exopolysaccharide (EPS) synthesized in large amounts by rhizobia, which is required for the formation of nitrogen-fixing nodules on legume roots (Downie 2010). Previously, it has been established that this polymer is essential for symbiosis with host plants which form indeterminate-type nodules, such as clover, vetch, pea, and alfalfa (Rolfe et al. 1996; Chen et al. 1985; van Workum et al. 1995; Cheng and Walker 1998). Furthermore, EPS plays an important role in protecting rhizobial cells against desiccation and other stress factors occurring in soil (Jaszek et al. 2014; Fujishige et al. 2006). EPS-deficient strains of R. leguminosarum bvs. trifolii and viciae and Sinorhizobium meliloti induce nodules on compatible host plants, which are inefficient in nitrogen fixation (Ivashina et al. 1994; van Workum et al. 1997; Cheng and Walker 1998; Rolfe et al. 1996; Janczarek and Urbanik-Sypniewska 2013). However, recent data indicate that the role of EPS in legume–rhizobium symbiosis is more complex and depends on the type of host plant. For example, EPS produced by S. fredii strain HH103 is not required for nodulation of Glycyrrhiza uralensis, a host plant that also forms indeterminate-type nodules (López-Baena et al. 2016; Margaret-Oliver et al. 2012; Crespo-Rivas et al. 2015). By contrast, other data indicate that EPS is required for symbiotic interactions of some legumes which form determinate-type nodules, such as Lotus corniculatus and Lotus japonicus (Kelly et al. 2013; Muszyński et al. 2016). Kawaharada et al. (2015) have described the crucial role of EPS as a signal in early stages of symbiosis. They identified a receptor-like kinase EPR3 in the root-hair membrane of L. japonicus which was involved in the recognition and binding of EPS from a compatible bacterium Mesorhizobium loti. Moreover, a conditional requirement for EPS was found in this symbiotic model, since mutants of M. loti R7A strain affected in different steps of EPS synthesis showed significant differences in the effectiveness of their symbiotic interactions with L. corniculatus and L. japonicus. For example, an exoB mutant affected in an early stage of EPS synthesis induced nitrogen-fixing nodules on these host plants (following a delay), whereas an exoU mutant, which synthesized pentasaccharide monomers, induced empty nodule-like structures on the host roots, inefficient in nitrogen fixation (Kelly et al. 2013; Muszyński et al. 2016).
In the case of R. leguminosarum, the structure of its EPS has been determined in detail. This polysaccharide is composed of octasaccharide subunits which contain d-glucose, d-glucuronic acid, and d-galactose residues in a molar ratio 5:2:1, and are additionally substituted with O-acetyl and pyruvyl groups (Robertsen et al. 1981; McNeil et al. 1986; Cremers et al. 1991; O’Neill et al. 1991; Breedveld et al. 1993; Philip-Hollingsworth et al. 1989). However, relatively little is known of the genetic control of EPS biosynthesis in R. leguminosarum. Up to now, the functions of only a few proteins participating in the five steps of EPS unit assembly have been identified experimentally. The assembly is initiated by the PssA protein which transfers glucose-1-phosphate from UDP-glucose to a C55-isoprenylphosphate carrier anchored in the bacterial inner membrane (Pollock et al. 1998). Glucuronosyl-(β1 → 4)-glucosyltransferase PssDE is engaged in the second step, whereas glucuronosyl-(β1 → 4)-glucuronosyltransferase PssC in the third step of EPS unit assembly. The fourth sugar is added to the growing subunit by glucosyl-(α1 → 4)-glucuronosyltransferase PssS (Ivashina and Ksenzenko 2012; Janczarek et al. 2015b). The PssD, PssE, PssC, and PssS proteins are encoded by genes located in a large chromosomal cluster named Pss-I (Król et al. 2007). The only exception is PssA encoded by a gene located at a long distance from this EPS synthesis region (Ivashina et al. 1994; van Workum et al. 1997). The structural and functional organization of the Pss region has been characterized in three R. leguminosarum strains (VF39, TA1, Rt24.2) (Sadykov et al. 1998; Król et al. 2007; Janczarek et al. 2015b). Mutations in pssA, pssD, and pssS genes totally abolish EPS production, whereas a mutation located in the pssC upstream region results in a nearly two-fold decrease in EPS biosynthesis in comparison to the wild-type strain (Ivashina et al. 1994; van Workum et al. 1997; Janczarek et al. 2015b). pssJ encodes a protein which is probably engaged in the last step of subunit assembly, since the exo344::Tn5 strain carrying a mutation in this gene has been found to synthesize only residual amounts of EPS, whose units lack a terminal d-galactose (Breedveld et al. 1993). pssM, which is also located in the Pss-I region, encodes a ketal pyruvate transferase responsible for one of the non-sugar modifications of EPS, i.e. pyruvylation in the subunit of the subterminal d-glucose (Ivashina et al. 2010). A mutation in pssM has been observed to negatively affect symbiosis of R. leguminosarum bv. viciae with compatible host plants. So far, genes encoding proteins involved in the remaining steps of EPS unit assembly have not been identified. Proteins encoded by pssT, pssN, pssO, pssP, and pssL genes, which are also located in the Pss-I region, are engaged in the polymerization and export of EPS (Mazur et al. 2005; Marczak et al. 2013, 2014).
Regulation of EPS biosynthesis in R. leguminosarum has been shown to involve several proteins, such as PsiA, PsrA, ExoR, and RosR, and environmental factors (e.g., phosphate and nitrogen limitation, carbon source, legume root exudates) (Borthakur and Johnston 1987; Borthakur et al. 1988; Reeve et al. 1997; Janczarek and Skorupska 2011; Rachwał et al. 2015). Among these proteins, PsiA, PsrA, and ExoR participate in negative regulation of EPS synthesis, whereas RosR is a positive regulator of this process.
Two very interesting Pss proteins involved in EPS biosynthesis of R. leguminosarum are PssD and PssE, which, as reported by Pollock and others (Pollock et al. 1998), are required for the addition of a second sugar residue (UDP-glucuronic acid) to growing EPS repeating units. PssD and PssE are homologous to N-terminal and C-terminal regions, respectively, of glycosyltransferase SpsK of Sphingomonas S88, as well as to proteins Cps14F and Cps14G of Streptococcus pneumoniae (van Workum et al. 1997; Sadykov et al. 1998). SpsK is involved in sphingan synthesis in Sphingomonas S88, whereas Cps14G is responsible for the second step of CPS synthesis in S. pneumoniae (Kolkman et al. 1996; Yamazaki et al. 1996). Moreover, van Workum and others (1997) have identified a sequence motif in the central part of R. leguminosarum PssE which showed a similarity to the QX13TX2 GX7LX4 PX4 PX3DQX4A consensus proposed as a UDP-binding site (Hundle et al. 1992). However, the similarity found in this motif was not high, since only three out of the nine conserved amino acids were identical (these residues are underlined).
In this study, we report that a mutation in pssE influences some of the physiological properties and symbiotic behavior of R. leguminosarum bv. trifolii cells. We characterize the Rt1933pssE mutant strain obtained via a random mutagenesis and show that the mutation in the 3′-end of this gene affects EPS synthesis, cell-surface properties and the effectiveness of its symbiosis with clover.
Materials and methods
Strains, plasmids and growth conditions
Strains, plasmids and oligonucleotide primers used in this study are listed in Table 1. E. coli strains were grown in LB medium at 37 °C (Bertani 1951), whereas R. leguminosarum bv. trifolii strains were cultured at 28 °C in two energy-rich media (tryptone-yeast extract [TY] and 79CA with 1% glycerol as a carbon source) and minimal M1 medium with 1% glycerol (Beringer 1974; Kowalczuk and Lorkiewicz 1979). When required, antibiotics were used at the following final concentrations: rifampicin, 40 μg ml−1; kanamycin, 40 μg ml−1; tetracycline, 10 μg ml−1; ampicillin, 100 μg ml−1; and spectinomycin, 40 μg ml−1. To compare growth kinetics of Rt24.2, Rt1933, and Rt1933(pM3), the strains were cultured over 72 h in 79CA, TY, and M1 media. After 0, 24, 48, and 72 h, the optical density (OD600) of these cultures was measured, and 100-μl aliquots were taken and their serial dilutions were placed onto agar plates, incubated at 28 °C and the colonies appearing were counted (CFU - colony-forming units). The experiment was repeated twice with three biological replicates for each strain and medium tested.
DNA methods and sequence analysis
Standard techniques were used for genomic and plasmid DNA isolation, restriction enzyme digestion, conjugation, hybridization, PCR reaction, and sequencing (Sambrook et al. 1989). For PCR amplifications, REDTaq Ready PCR Reaction Mix (Sigma-Aldrich, St. Louis, MO, USA) was used. PCR products obtained were sequenced using the BigDye terminator cycle sequencing kit and the ABI Prism 310 sequencer (Applied Biosystems, Waltham, MA, USA). Database searches were done with the BLAST and FASTA programs available from the National Center for Biotechnology Information (Bethesda, MD, USA) and the European Bioinformatic Institute (Hinxton, UK). Amino acid sequence analyses were performed using the BLASTP program (http://blast.ncbi.nlm.nih.gov/) and the CAZy database ( www.cazy.org/).
Mutagenesis of the pssE gene
To obtain derivatives of R. leguminosarum bv. trifolii Rt24.2 defective in EPS production, a random mutagenesis of this strain was performed using S17–1 which contains the mTn5SSgusA40 transposon with a promoterless gusA gene (Wilson et al. 1995). As a result of several biparental conjugation experiments, a total of nearly 2000 colonies were obtained, among them a mutant strain characterized by a non-mucoid phenotype, named Rt1933. The localization of mTn5SSgusA40 in the Rt1933 genome was determined using hybridization, PCR, and sequencing analyses. Primers complementary to different pss genes of the Pss-I region and primer MJ-gusR2 complementary to the 3′-end of gusA in the mTn5SSgusA40 transposon were used in several PCR reactions. Only in the case of two primer pairs, pssE9/MJ-gusR2 and prsD11/MJ-gusR2, PCR products (0.5 kb- and 3.0 kb-long, respectively) were obtained. On the basis of a sequencing analysis of these products, mTn5SSgusA40 was identified in pssE of the Pss-I region (13,600/13,601 nt, GenBank acc. no. KP067320).
Complementation of the pssE mutation
For complementation of the pssE mutation in the Rt1933 strain, pARF136, pARF1368, and pM3 plasmids containing different fragments of the Pss-I region were used. pARF136 and pARF1368 had been constructed previously by Król et al. (1998), whereas pM3 was obtained in this present study. To construct pM3, the pBBR1MCS-2 vector and a 1.4-kb fragment obtained in a PCR reaction with primers Hind124 and Bam113 complementary to the pssD and pssE genes, respectively, were used. The PCR product was digested with HindIII and BamHI enzymes, cloned into the corresponding sites in pBBR1MCS-2, and the insert of the resulting plasmid pM3 was verified by sequencing. The pARF136, pARF1368, and pM3 plasmids were introduced into E. coli S17–1 by transformation and subsequently into Rt1933 by biparental conjugation according to Sambrook et al. (1989).
EPS isolation and quantification
EPS isolation and quantification were performed using bacterial cultures of Rt24.2, Rt1933, and Rt1933(pM3). These strains were grown in 10 ml of 79CA for 72 h at 28 °C. The cultures after 24, 48 and 72 h of growth were centrifuged for 20 min at 14,000 rpm, and EPS was precipitated from supernatants with 4 vol. of 96% cold ethanol. After centrifugation, the EPS obtained was dissolved in water and analyzed according to Loewus (1952). The total sugar content was calculated as glucose equivalents. The experiment was repeated twice with three replicates for each strain analyzed.
Cell hydrophobicity assay
The hydrophobicity of Rt24.2, Rt1933, and Rt1933(pM3) cells was determined using a two-phase method and dodecane (Sigma-Aldrich, St. Louis, MO, USA) according to Neu and Poralla (1990) with minor modifications (Rachwał et al. 2016). Bacteria were collected from 79CA agar plates and suspended in PUM buffer (22.2 g K2HPO4 × H2O, 7.26 g KH2PO4, 1.8 g urea, 0.2 g MgSO4 × 7H2O, 1 l water) to an optical density of ~0.5 (OD1) at 405 nm. Then, 100 μl of dodecane was added to 200-μl aliquots of bacterial suspensions and vigorously vortexed for 2 min. After 15-min equilibration at room temperature, the lower phase of these samples was collected, and optical density was measured (ODa) in a microplate reader (Biochrom Asys UVM 340). The degree of hydrophobicity was calculated as follows: % hydrophobicity = 100–100(ODa/OD1). The experiment was repeated three times with three replicates for each strain analyzed.
LPS profile
The LPS profiles of Rt24.2, Rt1933, and Rt1933(pM3) strains were determined as previously described (Janczarek et al. 2010). Briefly, 48-h cultures in 79CA were centrifuged, and the bacterial pellets obtained were washed twice with 0.9% NaCl to remove EPS. Then, the pellets were used to obtain LPS profiles according to Lesse et al. (1990). The samples were separated by 12.5% Tricine SDS-PAGE, and LPS was visualized by silver staining.
Motility assay
The motility of the rhizobial strains was determined as previously described (Janczarek and Rachwał 2013). For this purpose, 5-μl aliquots of bacterial suspensions of an optical density OD600 = 0.2 prepared in sterile water were stabbed into plates containing 0.3% and 0.7% 79CA, 0.3% and 0.7% TY, and 0.3% and 0.7% M1 media. The bacteria were incubated at 28 °C for 7 days, and then the migration distance from the injection site was measured. The assay was repeated three times with three replicates for each strain and each condition tested.
Assay of sensitivity to stress factors
To establish the sensitivity of the Rt24.2, Rt1933, and Rt1933(pM3) strains to sodium dodecyl sulfate, sodium deoxycholate, and ethanol, the minimal inhibitory concentrations of the tested factors were determined. To this end, 10-μl aliquots of bacterial suspensions of OD600 = 0.2 prepared in sterile water were placed on 79CA agar plates containing defined concentrations of the individual compounds (SDS: 0.05–1% w/v, DOC: 0.05–1% w/v, ethanol: 0.05–6% v/v). The plates were incubated for 4 days at 28 °C, and bacterial growth was established. The experiment was repeated three times with three replicates for each strain and factor tested.
Plant experiments
The symbiotic properties of strains Rt24.2, Rt1933 and Rt1933(pM3) were established using red clover (Trifolium pratense cv. Diana) as a host plant. Clover seedlings were transferred onto Fåhraeus agar slants (Vincent 1970), and, after 4 days, inoculated with bacterial suspensions (OD600 = 0.2, 100-μl aliquot plant−1). Clover plants were subsequently grown for 4 weeks in a greenhouse under natural light supplemented with artificial light (14 h at 24 °C and 10 h at 18 °C), and root nodules were counted after each week. 28-day-old plants were harvested and weighted. The experiment was done in triplicate using 40 plants in each experiment for each strain tested.
The influence of EPS on the symbiotic behavior of the pssE mutant was established using clover seedlings inoculated with Rt1933 and Rt24.2 at a 1:1 ratio (live strains) or live Rt1933 and dead Rt24.2 (previously killed by heating for 10 min at 95 °C) (1:1 ratio) or live Rt1933 together with added EPS, which was isolated from Rt24.2 cultures (50 μg plant−1), and individual live Rt1933 and Rt24.2 as control strains. All bacterial suspensions had an optical density OD600 = 0.2 and were used in 100-μl aliquots per seedling. Clover plants were grown for 4 weeks, and nodules were counted after each week. Then, the plants were harvested and weighed. The experiment was repeated three times using 20 plants for each strain and condition tested. The results obtained were subjected to an analysis of variance (a linear ANOVA model) with three replicates using Microsoft® Excel 2000 for Windows. P values ˂ 0.05 were considered significant.
In addition, nodule occupation by the bacteria tested in these conditions was established according to a method described earlier (Janczarek et al. 2009). Briefly, 12 nodules of a similar size were chosen for each condition tested. They were surface sterilized, crushed, suspended in 1 ml of 79CA, placed in serial dilutions on agar plates, and incubated at 28 °C, after which the colonies appearing were counted.
Visualization of nodule occupation using light microscopy
To determine nodule occupation by Rt1933, we used β-glucuronidase activity encoded by gusA present in mTn5SSgusA40. Rt24.2 harboring the pJBA21Tc plasmid with gusA and Rt1933(pM3) were used as control strains. Clover seedlings inoculated with the mutant and control strains were grown up to 4 weeks, and nodules appearing on the roots were stained for 3 h using 50 mM sodium phosphate buffer (pH 7.2) containing 50 μg ml−1 of 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (Janczarek and Rachwał 2013) and analyzed under a Nikon light microscope (OPTIPHOT2).
Results
Genetic characterization of mutant strain Rt1933 and complementation of a pssE mutation
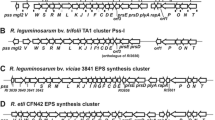
Previously, we had performed a sequence analysis of the Pss-I region involved in EPS synthesis of R. leguminosarum bv. trifolii Rt24.2 and established its genetic organization and transcriptional activity of the pss genes located in it (Janczarek et al. 2015b). In this study, we characterized a mutant strain named Rt1933 obtained via a random mutagenesis of the Rt24.2 strain with the use of the mTn5SSgusA40 transposon (Wilson et al. 1995). This mutant formed small non-mucoid colonies on 79CA agar plates, which considerably differed from those formed by the wild-type strain (data not shown). Based on hybridization, PCR amplifications with several primers complementary to different pss genes, and sequencing analyses, the localization of the mTn5SSgusA40 transposon in Rt1933, designated as the exo99 mutation, was identified inside the Pss-I region in the pssE gene (a position between 13,600 and 13,601 nt, GenBank Acc. no. KP067320) (Fig. 1). In addition, the presence of one copy of mTn5SSgusA40 in the Rt1933 genome was confirmed using Southern hybridization and a gusA probe (data not shown). The pssE gene encodes a 179-aa long protein (a coding region from 13,112 to 13,651 nt), which shows a homology to the glycosyltransferases from the GTB-type superfamily, GT family 28 (EC 2.4, CAZy database). The mTn5SSgusA40 insertion in the Rt1933 genome found 50 nt upstream of the 3′-end of the pssE gene, resulted in a lack of 16 aa at the C-end in the encoded PssE protein.
Genetic organization of the Pss-I region of R. leguminosarum bv. trifolii Rt24.2. Arrows below the map indicate the direction of the transcription of pss genes. Selected restriction sites are marked with capital letters: E – EcoRI, H – HindIII, B – BamHI. Location of the mTn5SSgusA40 transposon in the Pss-I region of the R. leguminosarum bv. trifolii mutant Rt1933 (exo99) is marked by a large white vertical arrow. Complementation analysis of the pssE mutation in Rt1933 using a set of plasmids (pARF136, pARF1368, and pM3) harboring different fragments of the Pss-I region. The plus sign (+) indicates a positive result of complementation (i.e., recovery of EPS production)
Next, we performed a complementation analysis of the exo99 mutation in Rt1933 using a set of plasmids: pARF136 containing a large part of the Pss-I region (i.e., genes from pssH to pssN), pARF1368 containing a 5.6-kb fragment with pssCDEprsDE genes, and pM3 containing a 1.4-kb fragment with pssDE genes and their promoter sequence (Fig. 1). In our earlier work, we had established that the pssDE genes were transcribed together from one common promoter (Janczarek et al. 2015b). As a result of this complementation analysis, the mucoid phenotype of the Rt1933 mutant was recovered after introduction of all of the plasmids used (pARF136, pARF1368 and pM3). This indicated that the shortest fragment of the Pss-I region present in pM3 was sufficient for complementation of the exo99 mutation (Fig. 1). We determined the amount of EPS synthesized by the wild-type Rt24.2, Rt1933 mutant and the complemented Rt1933(pM3) strains during 72-h bacterial growth (Table 2). Both control strains, Rt24.2 and Rt1933(pM3), synthesized large amounts of EPS, the production of which increased during prolonged bacterial growth. By contrast, Rt1933 did not produce any EPS even after 72 h of growth, indicating that the pssE mutation totally abolished the synthesis of this polysaccharide. These data confirm that pssE plays an essential role in EPS synthesis in R. leguminosarum.
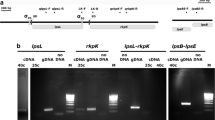
In addition, we checked whether the pssE mutation affected the synthesis of another surface polysaccharide, lipopolysaccharide (LPS), in Rt1933. For this purpose, LPS profiles of Rt24.2, Rt1933 and Rt1933(pM3) were obtained and compared (Fig. 2). The analysis showed that the LPS profiles of these strains were very similar, indicating that the mutation in pssE did not affect the synthesis of this polysaccharide.
Silver-stained Tricine SDS-PAGE profiles of LPS from the wild-type R. leguminosarum bv. trifolii and its derivatives. The LPSs (4 μg) were loaded into each well. Lanes: 1 – Salmonella enterica sv. Typhimurium (Sigma), 2 – Rt24.2, 3 – Rt1933, and 4 – Rt1933(pM3). LPS I, high-molecular-weight LPS; LPS II, low-molecular-weight LPS
The pssE mutation affects bacterial growth, motility, and surface properties of R. leguminosarum bv. trifolii
Next, we characterized other phenotypic properties of the pssE mutant. First, the ability of Rt1933 to grow in two energy-rich media (79CA and TY) and the minimal M1 medium was compared with that of the Rt24.2 and Rt1933(pM3) strains (Fig. 3). In order to eliminate the influence of EPS on this parameter, CFU (colony-forming unit) values for these strains were determined, instead of the optical density of their bacterial cultures. The growth kinetics of Rt24.2, Rt1933(pM3) and Rt1933 cells were established after 24, 48, and 72 h. Rt1933 showed slower growth in all of the tested growth media in comparison to both control strains. In 72-h cultures, the growth rate observed for the pssE mutant was approx. two- or three-fold lower than that of the Rt24.2 and Rt1933(pM3) strains, depending on the type of medium tested. This was especially visible in the case of the minimal M1 medium.
The growth of R. leguminosarum bv. trifolii Rt24.2 and its Rt1933 and Rt1933(pM3) derivatives in energy-rich 79CA a, TY b, and minimal M1 medium c. The data are mean ± SD for three experiments. Differences statistically significant for tested strains in the particular time point are marked by different letters (P < 0.05; ANOVA, post hoc Tukey’s test)
Moreover, the motility of the pssE mutant and wild-type cells was determined. The ability of these bacteria to migrate was tested using 79CA, TY, and M1 media containing 0.3% and 0.7% agar (Table 3). Rt24.2 and Rt1933(pM3) showed similarly high levels of motility in both energy-rich media. The largest migration distance for these strains was found in media containing 0.3% agar. The migration distance for Rt24.2 and Rt1933(pM3) in M1 was slightly lower than in 79CA and TY. Compared to the control cells, Rt1933 cells exhibited significantly reduced motility in all the tested growth media. The largest differences in migration between the mutant Rt1933 and the Rt24.2 and Rt1933(pM3) strains were found in 0.3% M1 (~2.5-fold) and 0.3% TY (~3.3-fold).
Furthermore, the sensitivity of Rt24.2, Rt1933 and Rt1933(pM3) to stress factors such as SDS, DOC, and ethanol was established. The Rt1933 mutant was characterized by an increased sensitivity to SDS and DOC in comparison to the wild-type strain (Table 4).
Next, the hydrophobicity of pssE mutant cells was determined and compared with that of the wild-type cells using suspensions of bacteria collected from 79CA agar plates and a two-phase method with dodecane. In this assay, the hydrophobicity of the wild-type and the complemented pssE mutant strains whose cells were surrounded by a polysaccharide capsule was similar [% hydrophobicity for Rt24.2 = 16.12% ±1.82 and Rt1933(pM3) = 15.89% ±1.74, respectively]. By contrast, Rt1933 cells were characterized by a nearly 4-fold lower hydrophobicity than the cells of the Rt24.2 and Rt1933(pM3) strains [Rt1933 = 4.12% ± 1.13; P < 0.05, ANOVA, post hoc Tukey’s test].
All these data indicate that the pssE mutation affected bacterial growth, motility, and surface properties of R. leguminosarum bv. trifolii.
The pssE mutation leads to disturbances in the symbiosis of R. leguminosarum bv. trifolii with clover
In further experiments, we investigated whether the changes observed in the phenotypic properties of the Rt1933 mutant affected its symbiotic interaction with the host plant. Red clover (Trifolium pratense) seedlings were inoculated with strains Rt24.2, Rt1933 and Rt1933(pM3) and grown for four weeks (Fig. 4). The control strains Rt24.2 and Rt1933(pM3) were highly effective in invading clover plants. On day 7 post inoculation (dpi), ~30% of the plants inoculated with these bacteria had nodules on their roots, and after 21 dpi nodules were visible on all of the tested plants (100%) (Fig. 4a). By contrast, the capacity of the pssE mutant to infect clover roots was considerably reduced with the first nodules appearing as late as two weeks into the experiment (Fig. 4b). Moreover, only ~50% of the tested plants were infected and had nodules even after four weeks post inoculation. Apart from the delay in nodule formation, also the total number of nodules elicited on clover roots by the pssE mutant was essentially lower (~3-fold) than for Rt24.2 and Rt1933(pM3) (Fig. 4b). Plants inoculated with this mutant were small, had yellow leaves, and formed non-numerous small, often irregularly shaped, white nodule-like structures, whose morphology suggested they were ineffective in nitrogen fixation. This was additionally confirmed by the mass of the upper part of the clover plants inoculated with Rt1933, which was nearly 2-fold lower than the mass of the plants inoculated with Rt24.2 and Rt1933(pM3), and similar to that of uninoculated plants (Fig. 4c). This was in contrast to the clover plants inoculated with Rt24.2 and Rt1933(pM3), which were tall, green, and had many pink, elongated nodules on their roots.
Symbiotic properties of the wild-type R. leguminosarum bv. trifolii Rt24.2 and its Rt1933 and Rt1933(pM3) derivatives determined in a 4-week experiment using red clover (Trifolium pratense) as a host plant. a The percent of plants containing root nodules; b the average number of nodules elicited on clover roots by the tested strains; c the mass of shoots and roots of plants inoculated with the Rt24.2, Rt1933 and Rt1933(pM3) strains. The data are mean ± SD for three experiments. Differences statistically significant for tested strains are marked by different letters (P < 0.05; ANOVA, post hoc Tukey’s test)
Next, the occupation of root nodules by the pssE mutant and wild-type R. leguminosarum cells was examined. For this experiment, Rt24.2 harboring the pJBA21Tc plasmid with gusA for β-glucuronidase was used. In the case of Rt1933 and Rt1933(pM3), β-glucuronidase activity was encoded by gusA located in the mTn5SSgusA40 transposon. In this experiment, we observed that both Rt24.2(pJBA21Tc) and Rt1933(pM3) occupied nodules very effectively, and these bacteria were found in both young (7-dpi) (Fig. 5a and c) as well as older (21-dpi) nodules (Fig. 5b and d). β-glucuronidase activity was detected in all zones of the 21-dpi nodules (i.e., infection zone II, interzone II–III, and nitrogen-fixing zone III). The only exception was the meristem (zone I). By contrast, occupation of clover root nodules by the Rt1933 mutant was drastically affected. Mutant cells were visible only on the surface of roots and nodules, and plant root tissues inside the nodules were not occupied by this strain (Fig. 5e–i). These observations indicated strong disturbances already in early stages of the symbiosis between the pssE mutant and its host plant (i.e., infection of host roots). As a consequence of these abnormalities, the nodules formed were not occupied by the microsymbiont and were unable to fix nitrogen.
Light microscopy of nodules elicited on clover roots (Trifolium pratense) by the wild-type R. leguminosarum bv. trifolii Rt24.2, the pssE mutant Rt1933, and the Rt1933(pM3) strains harboring the gusA reporter gene for β-glucuronidase. a, b Rt24.2 wild-type nodules at 7 and 21 days post inoculation (dpi), respectively; c, d 7-dpi and 21-dpi nodules containing Rt1933(pM3), respectively; (e - i) nodules induced by the Rt1933 mutant: e – a root after 7 dpi, f – a nodule primordium after 10 dpi, g – a 14-dpi nodule, h – a 21-dpi nodule, i – a 26-dpi nodule, respectively; bars = 0.2 mm (a and c), bars = 0.5 mm (b and d), bars = 0.3 mm (e - i). The nodules were stained for GUS activity
Another problem that we investigated was how nodulation of clover by Rt1933 was improved by adding EPS isolated from the Rt24.2 culture and by co-inoculation with Rt24.2. Co-inoculations of host plant roots were performed using bacterial suspensions containing Rt1933 and live or temperature-killed Rt24.2 at a 1:1 ratio (Table 5). It was observed that when the clover roots were inoculated with a mixture containing live cells of both Rt1933 and Rt24.2 strains, the plants looked similar to those inoculated exclusively with Rt24.2 (i.e., they formed a large number of nodules effective in nitrogen fixation, which was confirmed by their high shoot mass). Furthermore, all the nodules were occupied by wild-type cells alone. These data confirm that the competitiveness of the pssE mutant was strongly decreased in comparison to that of the wild-type strain. In the case of co-inoculation of clover roots with the bacterial suspension containing live Rt1933 and dead Rt24.2 cells, significantly higher numbers of infected plants and nodules induced by Rt1933 were observed. These nodules, however, were white, irregularly shaped and ineffective in nitrogen fixation, as a consequence of which the plants were small, yellow and had a shoot mass similar to that of the plants inoculated with Rt1933 alone. Similar effects were observed for the plants inoculated with the bacterial suspension containing Rt1933 and EPS isolated from Rt24.2. The addition of purified EPS complemented the defect of the lack of EPS in the pssE mutant by improving the strain’s effectiveness of nodule induction on host roots. This was confirmed by significantly increased numbers of plants forming nodules, nodules formed and mutant cells inside the nodules. However, also in this case, addition of the external EPS did not fully restore the wild-type symbiotic phenotype of the Rt1933 mutant. These data indicate that EPS derived from the wild-type strain complemented the defects of the EPS-deficient mutant pssE related to the only early stages of symbiotic interactions (i.e., induction of nodule formation and infection of host roots).
Discussion
Rhizobia comprise a diverse group of soil bacteria which establish nitrogen-fixing symbiosis with legumes (Fabaceae), a large and economically important group of plants that are a source of food, feed, and biofuels and are used in several other industrial applications (Olivares et al. 2013; Janczarek et al. 2015c; Gresshoff et al. 2015). Symbiotic interactions of this type are important for the functioning of the biosphere, since they provide large amounts of nitrogen to plants, increase soil fertility, and restore damaged ecosystems (Yanni et al. 1997; Kennedy et al. 1997; Qin et al. 2011; Angus et al. 2013). Recently, it has been established that rhizobial EPS plays a crucial role as a signal molecule in early stages of symbiosis (Kawaharada et al. 2015). Moreover, this polysaccharide is essential for adhesion, biofilm formation, and protection of bacterial cells against several stress conditions (Fujishige et al. 2006; Janczarek and Rachwał 2013). EPS synthesis in rhizobia is a multi-step process which involves the activity of several enzymes. In R. leguminosarum, a great majority of pss genes encoding proteins which participate in this biosynthetic pathway are located in the large chromosomal cluster Pss-I (Król et al. 2007; Sadykov et al. 1998; Janczarek et al. 2015b).
In this work, we demonstrated that the exo99 mutation located in pssE of the Pss-I region affected EPS synthesis and cell-surface properties of R. leguminosarum bv. trifolii, and the observed changes resulted in disturbances in the symbiotic interaction of this bacterium with clover plants (Fig. 1, Table 2).
It was established earlier that pssD and pssE genes were expressed from the same promoter and that large fragments of their ORFs overlapped (Sadykov et al. 1998; van Workum et al. 1997; Król et al. 2007; Janczarek et al. 2015b). Sadykov et al. (1998) have reported for R. leguminosarum bv. viciae strain VF39, which effectively infects Vicia faba, V. sativa, and Pisum sativum, that a mutation located at the 5′-end of pssE disturbs EPS synthesis. However, neither the precise site of this mutation nor the phenotypic properties of the mutant strain have been characterized. pssD mutations obtained earlier in strains of R. leguminosarum bv. viciae VF39, R. leguminosarum bv. trifolii RBL5599 and TA1 resulted in similar effects as the pssE mutation (i.e., a lack of EPS production and strong disturbances in nodule infection) (van Workum et al. 1997; Sadykov et al. 1998; Król et al. 1998).
The R. leguminosarum pssD and pssE genes encode proteins homologous to glycosyltransferases from the GTB-type superfamily. The amino acid sequences of PssD and PssE are similar to the N-terminal and C-terminal halves of the glucuronosyl-(β1,4)-glucosyltransferase SpsK of Sphingomonas S88, respectively (Sadykov et al. 1998; Ivashina and Ksenzenko 2012). This observation suggested that these proteins might represent two subunits of the same enzyme. As was proposed by Ivashina and Ksenzenko (2012), a catalytic domain was located in the PssE protein, whereas PssD was an integral inner membrane protein. They also found that integration of PssE into a bacterial membrane strongly depended on the presence of PssD and vice versa. In addition, Pollock et al. (1998) used complementation of the Sphingomonas S88 spsK mutant by pssDE of R. leguminosarum and an analysis of the sugars incorporated into lipid-linked disaccharides in permeabilized cells to demonstrate that these genes encoded proteins of glucuronosyl-(β1 → 4)-glucosyltransferase activity. They also showed that glycosyltransferases which formed alternative sugar linkages to the same substrate inhibited polysaccharide synthesis or were deleterious or even lethal to the bacterium. This confirms the very high specificity of the enzymes participating in the synthesis of various EPS. Bacterial glycosyltransferases comprise a very large group of enzymes whose chief role in the biosynthesis of disaccharides, oligosaccharides, and polysaccharides is to catalyze the transfer of sugar moieties from activated donor molecules to other sugar molecules (Schmid et al. 2016).
We also showed in the present study that other properties of the pssE mutant were considerably changed in relation to the wild type (Fig. 3, Tables 3 and 4). This suggested that the lack of EPS in the mutant affected its cell-surface properties, whereas the slower growth of the mutant may have led to decreased motility, delayed root hair invasion, and consequently, a decreased nodule occupancy. Similarly to our findings, a higher sensitivity to SDS and DOC and oxidative stress factors (e.g., hydrogen peroxide) has been described for other rhizobial mutants defective in EPS synthesis, such as the pssA and pssD mutants of R. leguminosarum and an ORS571-X15 mutant of Azorhizobium caulinodans (Wielbo et al. 2004; D'Haeze et al. 2004; Janczarek and Rachwał 2013; Jaszek et al. 2014). These data indicate that the EPS surrounding bacterial cells plays an important protective role against several stress factors.
Moreover, we observed that the hydrophobicity of the pssE mutant was considerably changed in comparison with the wild type. This finding is in congruence with our previous data for an EPS-deficient pssA mutant, which showed significant differences in surface properties (i.e., electrokinetic mobility, surface charge, and hydrophobicity) compared to the wild-type strain (Janczarek et al. 2015a; Cieśla et al. 2016). Also, other researchers indicate that the presence of extracellular polysaccharides produced by various microorganisms has an important impact on their cell-surface properties (Lage et al. 2012; Planchon et al. 2013; Jttawuttipoka et al. 2013; Postma et al. 1991).
Furthermore, the pssE mutation affected the growth and motility of R. leguminosarum bv. trifolii. Decreased growth kinetics and swarming of Rt1933 in relation to Rt24.2 were observed in all of the tested growth media (Fig. 2, Table 3). This led us to the conclusion that slower migration of the pssE mutant in comparison to the wild type was probably caused by the decreased growth kinetics of the mutant cells. Tambalo et al. (2010) showed for two R. leguminosarum bv. viciae strains VF39SM and 3841 that parameters such as the type of growth medium, agar concentration, and temperature were essential for rhizobial motility. The EPS-deficient pssA mutant of R. leguminosarum bv. trifolii was also characterized by decreased motility in relation to the wild type. Similar findings have been reported in a study by Vanderlinde et al. (2009), in which a mutant strain of R. leguminosarum bv. viciae 3841 which did not contain the 27-hydroxyoctacosanoic acid modification in LPS was impaired in motility and biofilm formation. Moreover in S. meliloti, the presence of a regulatory network linking rhizobial EPS synthesis, motility and quorum sensing has been confirmed (Daniels et al. 2006; Sourjik et al. 2000; Glenn et al. 2007).
EPS is very important not only for free-living rhizobia, but also for the establishment of successful symbiosis with many legumes (e.g., clover, pea, vetch, and alfalfa) (Ivashina et al. 1994; Cheng and Walker 1998; Janczarek and Rachwał 2013). As reported recently for M. loti–L. japonicus symbiosis, this polysaccharide plays a crucial role as a signal which is recognized and bound by a plant receptor (Kawaharada et al. 2015; Kelly et al. 2013; Muszyński et al. 2016).
Finally, we found that the lack of EPS and changes in cell-surface properties caused by the pssE mutation significantly affected the symbiosis of R. leguminosarum bv. trifolii with red clover (Figs. 4 and 5). The effectiveness of induction of root nodules by the Rt1933 mutant was strongly decreased. The non-numerous nodules appeared on the roots with a large delay, and even four weeks into the experiment, only ~50% of the plants had nodules. Moreover, these nodules were not occupied by pssE mutant cells, and, in consequence, did not fix nitrogen. The symbiotic phenotype of Rt1933 confirms that the PssE protein plays an essential role in EPS synthesis and symbiotic interactions.
In our further experiments, we studied the influence of EPS on symbiotic interactions of the pssE mutant with red clover (Table 5). The results we obtained showed that the addition of wild-type EPS, either in the form of purified EPS isolated from the Rt24.2 culture or by co-inoculation with temperature-killed Rt24.2 cells, significantly affected the early stages of symbiosis, as manifested by the higher numbers of plants forming nodules and higher number of nodules induced by the EPS-supplemented Rt1933 in comparison to the plants inoculated with Rt1933 alone. However, the addition of EPS did not restore the full symbiotic phenotype of this mutant (i.e., its ability to fix nitrogen). In contrast, Djordjevic et al. (1987) showed for some EPS-deficient mutants of S. fredii NGR234 that the addition of a homologous EPS restored not only the infection effectiveness but also the nitrogen fixation ability of these strains in symbiosis with Leucaena and Macroptilium atropurpureum (25% to 40% of the plants formed Fix+ nodules, depending on the type of mutant studied). Those authors also observed partial restoration of the symbiotic properties of the EPS-deficient mutant of R. leguminosarum bv. trifolii ANU843 when wild-type EPS was added (25% of the tested white clover plants formed Fix+ nodules). These data suggest that the ability of EPS to complement symbiotic defects of EPS-deficient mutants may depend on various factors, such as the type of mutation, the kind of bacterium, and the kind of host plant. On that basis, we hypothesized that in the case of the pssE mutant, apart from the absence of EPS, its effectiveness in host root infection and nodule occupation was additionally negatively affected by the slower growth and cell motility of this strain.
EPS plays an essential role in the attachment of rhizobia to plant roots, the formation of biofilm on the root surface and host plant infection (Jones and Walker 2008; Williams et al. 2008; Rinaudi and Giordano 2010). This polysaccharide forms the most external layer of cells with little or no associations with the bacterial envelope, in contrast to capsular polysaccharide, which usually has a very similar structure to EPS, but is associated with the bacterial outer membrane (Downie 2010). Both of these polysaccharides (and especially EPS) are required for the initiation and propagation of infection threads – special structures via which rhizobial cells invade host roots and reach nodule tissues. EPS is an essential component of the infection thread matrix and forms a protective layer which prevents penetration of toxic H2O2 generated by host plants during their invasion (D'Haeze et al. 2004; Jones and Walker 2008). D'Haeze et al. (2004) showed that an ORS571-X15 mutant of Azorhizobium caulinodans which produced considerably less EPS than the wild type, was more sensitive to oxidative stress and induced ineffective nodules on its host plant, Sesbania rostrata.
It has traditionally been accepted that rhizobial EPS plays a much more important role in symbiotic relationships in which macrosymbionts form indeterminate nodules, such as Medicago–S. meliloti, R. leguminosarum bv. trifolii–Trifolium and R. leguminosarum bv. viciae–Vicia/Pisum (Cheng and Walker 1998; Rolfe et al. 1996; Ivashina et al. 1994, van Workum et al. 1997). In these cases, EPS-deficient mutants of these rhizobial species have proved to be defective in nodule invasion, and the nodules formed were unable to fix nitrogen. However, this is not always the case, since an EPS-deficient exoA mutant of S. fredii HH103 has been shown to be able to induce nitrogen-fixing nodules on two plants which formed indeterminate nodules (G. uralensis and pigeon pea) (López-Baena et al. 2016).
It had been reported previously that mutations in pssA, pssD, and pssS genes of R. leguminosarum bvs. trifolii and viciae totally abolished EPS synthesis and led to strong disturbances in symbiosis with compatible host plants (sporadically formed nodules were small and hardly occupied by bacterial cells) (Ivashina et al. 1994; Rolfe et al. 1996; van Workum et al. 1997; Sadykov et al. 1998; Ivashina and Ksenzenko 2012; Janczarek et al. 2015b). Similarly, EPS-deficient mutants of S. meliloti harboring mutations in genes which encode glycosyltransferases (exoY, exoA, exoL, exoM, exoO and exoW) induced non-nitrogen-fixing nodules on alfalfa (Cheng and Walker 1998; Reuber et al. 1991; Becker et al. 1993a, b). However, not only the absence of EPS but also alterations in its structure might lead to severe impairment in symbiosis. For example, a lack of pyruvyl residues in subunits caused by a pssV mutation abolished succinoglycan production in S. meliloti and negatively affected its symbiosis with the host plant (Glucksmann et al. 1993; Becker et al. 1993b). Mutations in exoC and exoB genes involved in the synthesis of the sugar precursors glucose-1-phosphate and UDP-galactose, respectively, abolished the production of succinoglycan and other polysaccharides containing galactose (galactoglucan and LPS) (Glucksmann et al. 1993; Becker et al. 1993a, b). Furthermore, recent data have shown that succinyl groups in succinoglycan are critical for successful host root invasion by S. meliloti, whereas the chain length of this polymer has much less effect on its role in symbiosis (Mendis et al. 2016). An exoH mutant, which produced large amounts of succinoglycan but lacked succinyl residues, was unable to invade alfalfa roots.
In conclusion, all these data show that rhizobial EPS is essential for successful invasion of legume roots and effective nitrogen fixation. Mutations in the genes encoding glycosyltransferases which participate in EPS unit assembly and proteins responsible for sugar precursor synthesis cause similar defects, i.e. a disruption in EPS synthesis and disturbances in the establishment of symbiotic interactions.
References
Angus AA, Lee A, Lum MR, Shehayeb M, Hessab R, Fujishige NA, Yerrapragada S, Kano S, Song N, Yang P, de los Santos PE, de Faria SM, Dakora FD, Weinstock G, Hirsch AM (2013) Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a nodulating and plant growth promoting beta-proteobacterium, are influenced by environmental factors. Plant Soil 369:543–562
Becker A, Kleickmann A, Keller M, Arnold W, Pühler A (1993a) Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet 241:367–379
Becker A, Kleickmann A, Küster H, Keller M, Arnold W, Pühler A (1993b) Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol Plant-Microbe Interact 6:735–744
Beringer JE (1974) R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84:188–198
Bertani G (1951) Studies on lysogenesis. I The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300
Borthakur D, Johnston AW (1987) Sequence of psi, a gene on the symbiotic plasmid of Rhizobium phaseoli which inhibits exopolysaccharide synthesis and nodulation, and demonstration that its transcription is inhibited by psr, another gene on the symbiotic plasmid. Mol Gen Genet 207:149–154
Borthakur D, Barker RF, Latchford JW, Rossen L, Johnston AW (1988) Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol Gen Genet 213:155–162
Breedveld MW, Zevenhuizen LPTM, Canter Cremers HCJ, Zehnder AJB (1993) Influence of growth conditions on production of capsular and extracellular polysaccharides by Rhizobium leguminosarum. Antonie Van Leeuwenhoek 64:1–8
Chen HM, Redmond BJ, Rolfe BG (1985) Alteration of the effective nodulation properties of a fast growing broad host range Rhizobium due to changes in exopolysaccharide synthesis. J Plant Physiol 120:331–349
Cheng HP, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180:5183–5191
Cieśla J, Kopycińska M, Łukowska M, Bieganowski A, Janczarek M (2016) Surface properties of wild-type Rhizobium leguminosarum bv. trifolii strain 24.2 and its derivatives with different extracellular polysaccharide content. PLoS One 11(10):e0165080. doi:10.1371/journal.pone.0165080
Cremers HC, Batley M, Redmond JW, Wijfjes AH, Lugtenberg BJ, Wijffelman CA (1991) Distribution of O-acetyl groups in the exopolysaccharide synthesized by Rhizobium leguminosarum strains is not determined by the Sym plasmid. J Biol Chem 266:9556–9564
Crespo-Rivas JC, Guefrachi I, Mok KC, Villaecija-Aguilar JA, Acosta-Jurado S, Pierre O, Ruiz-Sainz JE, Taga ME, Mergaert P, Vinardell JM (2015) Sinorhizobium fredii HH103 bacteroids are not terminally differentiated and show altered O-antigen in nodules of the inverted repeat-lacking clade legume Glycyrrhiza uralensis. Environ Microbiol doi:10.1111/1462-2920.13101
Daniels R, Reynaert S, Hoekstra H, Verreth C, Janssens J, Braeken K, Fauvart M, Beullens S, Heusdens C, Lambrichts I, De Vos DE, Vanderleyden J, Vermant J, Michiels J (2006) Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci U S A 103:14965–14970
D'Haeze W, Glushka J, De Rycke R, Holsters M, Carlson RW (2004) Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol Microbiol 52:485–500
Djordjevic S, Chen C, Batley M, Redmond JW, Rolfe BG (1987) Nitrogen fixation ability of exopolysaccharide synthesis mutants of Rhizobium sp. strain NGR234 and Rhizobium trifolii is restored by the addition of homologous exopolysaccharides. J Bacteriol 169:53–60
Downie JA (2010) The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev 34:150–170
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206
Glenn SA, Gurich N, Feeney MA, Gonzalez JE (2007) The ExpR/sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J Bacteriol 189:7077–7088
Glucksmann MA, Reuber TL, Walker GC (1993) Genes needed for the modification, polimerization, export and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol 175:7045–7055
Gresshoff PM, Hayashi S, Biswas B, Mirzaei S, Indrasumunar A, Reid D, Samuel S, Tollenaere A, van Hameren B, Hastwell A, Scott P, Ferguson BJ (2015) The value of biodiversity in legume symbiotic nitrogen fixation and nodulation for biofuel and food production. J Plant Physiol 172:128–236
Hundle BS, O’Brien DA, Alberti M, Beyer P, Hearst JE (1992) Functional expression of zeaxanthin glucosyltransferase from Erwinia herbicola and a proposed uridine diphosphate binding site. Proc Natl Acad Sci U S A 89:9321–9325
Ivashina T, Ksenzenko VN (2012) Exopolysaccharide biosynthesis in Rhizobium leguminosarum: from genes to functions. In: The complex world of polysaccharides. InTech, Rijeka, pp 99–127
Ivashina TV, Khmelnitsky MI, Shlyapnikov MG, Kanapin AA, Ksenzenko VN (1994) The pss4 gene from Rhizobium leguminosarum biovar viciae VF39: Cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene 50:111–116
Ivashina TV, Fedorova EE, Ashina NP, Kalinchuk NA, Druzhinina TN, Shashkov AS, Shibaev VN, Ksenzenko VN (2010) Mutation in the pssM gene encoding ketal pyruvate transferase leads to disruption of Rhizobium leguminosarum bv. viciae-Pisum sativum symbiosis. J Appl Microbiol 109:731–742
Janczarek M, Rachwał K (2013) Mutation in the pssA gene involved in exopolysaccharide synthesis leads to several physiological and symbiotic defects in Rhizobium leguminosarum bv. trifolii. Int J Mol Sci 14:23711–23735
Janczarek M, Skorupska A (2011) Modulation of rosR expression and exopolysaccharide production in Rhizobium leguminosarum bv. trifolii by phosphate and clover root exudates. Int J Mol Sci 12:4132–4155
Janczarek M, Urbanik-Sypniewska T (2013) Expression of the Rhizobium leguminosarum bv. trifolii pssA gene involved in exopolysaccharide synthesis is regulated by RosR, phosphate and the carbon source. J Bacteriol 195:3412–3423
Janczarek M, Jaroszuk-Ściseł J, Skorupska A (2009) Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Anton Leeuw 96:471–486
Janczarek M, Kutkowska J, Piersiak T, Skorupska A (2010) Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol 10:284
Janczarek M, Rachwał K, Cieśla J, Ginalska G, Bieganowski A (2015a) Production of exopolysaccharide by Rhizobium leguminosarum bv. trifolii and its role in bacterial attachment and surface properties. Plant Soil 388:211–227
Janczarek M, Rachwał K, Kopcińska J (2015b) Genetic characterization of the Pss region and the role of PssS in exopolysaccharide production and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Plant Soil 396:257–275
Janczarek M, Rachwał K, Marzec A, Grządziel J, Palusińska-Szysz M (2015c) Signal molecules and cell-surface components involved in early stages of the legume-rhizobium interactions. Appl Soil Ecol 85:94–113
Jaszek M, Janczarek M, Kuczynski K, Piersiak T, Grzywnowicz K (2014) The response of the Rhizobium leguminosarum bv. trifolii wild-type and exopolysaccharide-deficient mutants to oxidative stress. Plant Soil 376:75–94
Jones K, Walker GC (2008) Responses of the model legume Medicago truncatula to the rhizobial exopolysaccharide succinoglycan. Plant Signal Behav 3:888–890
Jttawuttipoka T, Planchon M, Spalla O, Benzerara K, Guyot F, Cassier-Chauvat C, Chauvat F (2013) Multidisciplinary evidences that Synechocystis PCC6803 exopolysaccharides operate in cell sedimentation and protection against salt and meta stress. PLoS One 8:e55564
Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, Vinther M, Andersen SU, Krusell L, Thirup S, Jensen KJ, Ronson CW, Blaise M, Radutoiu S, Stougaard J (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312
Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW (2013) Conditional requirement for exopolysaccharide in the Mesorhizobium – Lotus symbiosis. Mol Plant-Microbe Interact 26:319–329
Kennedy IR, Pereg-Gerk LL, Wood C, Deaker R, Gilchrist K, Katupitiya S (1997) Biological nitrogen fixation in non-leguminous field crops: facilitating the evolution of an effective association between Azospirillum and wheat. Plant Soil 194:65–79
Kolkman MAB, Morrison DA, van der Zeijst BAM, Nuijten PJM (1996) The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol 178:3736–3741
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kowalczuk E, Lorkiewicz Z (1979) Transfer of RP4 and R68.45 factors to Rhizobium. Acta Microbiol Pol 28:221–229
Król JE, Wielbo J, Mazur A, Kopcińska J, Łotocka B, Golinowski W, Skorupska A (1998) Molecular characterization of pssCDE genes of Rhizobium leguminosarum bv. trifolii strain TA1: pssD mutant is affected in exopolysaccharide synthesis and endocytosis of bacteria. Mol Plant-Microbe Interact 11:1142–1148
Król JE, Mazur A, Marczak M, Skorupska A (2007) Syntenic arrangements of the surface polysaccharide biosynthesis genes in Rhizobium leguminosarum. Genomics 89:237–247
Lage OM, Bondoso J, Catita JAM (2012) Determination of zeta potential in Planctomycetes and its application in heavy metals toxicity assessment. Arch Microbiol 194:847–855
Lesse AJ, Campagnari AA, Bittner WE, Apicella MA (1990) Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods 126:109–117
Loewus FA (1952) Improvement in the anthrone method for determination of carbohydrates. Anal Chem 24:219
López-Baena FJ, Ruiz-Sainz JE, Rodríguez-Carvajal MA, Vinardell JM (2016) Bacterial molecular signals in the Sinorhizobium fredii-soybean Symbiosis. Int J Mol Sci 17:755. doi:10.3390/ijms17050755
Marczak M, Dźwierzyńska M, Skorupska A (2013) Homo- and heterotypic interactions between Pss proteins involved in the exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii. Biol Chem 394:541–559
Marczak M, Matysiak P, Kutkowska J, Skorupska A (2014) PssP2 is a polysaccharide co-polymerase involved in exopolysaccharide chain-length determination in Rhizobium leguminosarum. PLoS One 30(9):e109106
Margaret-Oliver I, Lei W, Parada M, Rodriguez-Carvajal MA, Crespo-Rivas JC, Hidalgo A, Gil-Serrano A, Moreno J, Rodríguez-Navarro DN, Buendía-Clavería A, Ollero J, Ruiz-Sainz JE, Vinardell JM (2012) Sinorhizobium fredii HH103 does not strictly require KPS and/or EPS to nodulate Glycyrrhizauralensis, an indeterminate nodule-forming legume. Arch Microbiol 194:87–102
Mazur A, Marczak M, Król J, Skorupska A (2005) Topological and transcrptional analysis of pssL gene product: a putative Wzx-like exopolysaccharide translocase in Rhizobium leguminosarum bv. trifolii TA1. Arch Microbiol 184:1–10
McNeil M, Darvill J, Darvill A, Albersheim P, van Veen R, Hooykaas P, Schilperoort R, Dell A (1986) The discernible structural features of the acidic exopolysaccharides secreted by different Rhizobium species are the same. Carbohydr Res 146:307–326
Mendis HC, Madzima TF, Queiroux C, Jones KM (2016) Function of succinoglycan polysaccharide in Sinorhizobium meliloti host plant invasion depends on succinylation, not molecular weight. mBio 7:e00606-16
Muszyński A, Heiss C, Hjuler CT, Sullivan JT, Kelly SJ, Thygesen MB, Stougaard J, Azadi P, Carlson RW, Ronson CW (2016) Structures of exopolysaccharides involved in receptor-mediated perception of Mesorhizobium loti by Lotus japonicus. J Biol Chem. doi:10.1074/jbc.M116.743856
Neu TR, Poralla K (1990) Emulsifying agents from bacteria isolated during screening for cells with hydrophobic surfaces. Appl Microbiol Biotechnol 32:521–525
O’Neill MA, Darvill AG, Albersheim P (1991) The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J Biol Chem 266:9549–9555
Olivares J, Bedmar EJ, Sanjuan J (2013) Biological nitrogen fixation in the context of global change. Mol Plant-Microbe Interact 26:486–494
Philip-Hollingsworth S, Hollingsworth RI, Dazzo FB (1989) Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. J Biol Chem 264:1461–1466
Planchon M, Jittawuttipoka T, Cassier-Chauvat C, Guyot F, Chauvat F, Spalla O (2013) Influence of exopolysaccharides on the electrophoretic properties of the model cyanobacterium Synechocystis. Colloids Surf B 110:171–177
Pollock TJ, Workum WA, Thorne L, Mikolajczak MJ, Yamazaki M, Kijne JW, Armentrout RW (1998) Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol 180:586–593
Postma J, Hok-A-Hin CH, Schotman JMT, Wijffelman CA, van Veen JA (1991) Population dynamics of Rhizobium leguminosarum Tn5 mutants with altered cell surface properties introduced into sterile and non-sterile soils. Appl Environ Microbiol 57:649–654
Qin L, Jiang H, Tian J, Zhao J, Liao H (2011) Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant Soil 349:25–36
Rachwał K, Matczyńska E, Janczarek M (2015) Transcriptome profiling of a Rhizobium leguminosarum bv. trifolii rosR mutant reveals the role of the transcriptional regulator RosR in motility, synthesis of cell-surface components, and other cellular processes. BMC Genomics 16:1111
Rachwał K, Boguszewska A, Kopcińska J, Karaś M, Tchórzewski M, Janczarek M (2016) The regulatory protein RosR affects Rhizobium leguminosarum bv. trifolii protein profiles, cell surface properties, and symbiosis with clover. Front Microbiol 7:1302, doi: 10.3389/fmicb.2016.01302
Reeve WG, Dilworth MJ, Tiwari RP, Glenn AR (1997) Regulation of exopolysaccharide production in Rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology 143:1951–1958
Reuber TL, Reed J, Glazebrook J, Glucksmann MA, Ahmann D, Marra A, Walker GC (1991) Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance. Biochem Soc Trans 19:636–641
Rinaudi LV, Giordano W (2010) An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett 304:1–11
Robertsen BK, Aman P, Darvill AG, McNeil M, Albersheim P (1981) Host - symbiont interactions. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol 67:389–400
Rolfe BG, Carlson RW, Ridge RW, Dazzo RW, Mateos FB, Pankhurst CE (1996) Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust J Plant Physiol 23:285–303
Sadykov MR, Ivashina TV, Kanapin AA, Shlyapnikov MG, Ksenzenko VN (1998) Structural and functional organization of the exopolysaccharide biosynthesis genes in Rhizobium leguminosarum bv. viciae VF39. Mol Biol (Mosk) 32:797–804
Sambrook J, Fitsch EF, Maniatis T (1989) Molecular Cloning: a laboratory manual; cold Spring Harbor laboratory press. Cold Spring Harbor, NY
Schmid J, Fariña J, Rehm B, Sieber V (2016) Editorial: microbial exopolysaccharides: from genes to applications. Front Microbiol doi: 10.3389/fmicb.2016.00308
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784–791
Sourjik V, Muschler P, Scharf B, Schmitt R (2000) VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J Bacteriol 182:782–788
Tambalo DD, Yost CK, Hynes MF (2010) Characterization of swarming motility in Rhizobium leguminosarum bv. viciae. FEMS Microbiol Lett 307:165–174
Vanderlinde EM, Muszyński A, Harrison JJ, Koval SF, Foreman DL, Ceri H, Kannenberg EL, Carlson RW, Yost CK (2009) Rhizobium leguminosarum biovar viciae 3841, deficient in 27-hydroxyoctacosanoate-modified lipopolysaccharide, is impaired in desiccation tolerance, biofilm formation and motility. Microbiology 155:3055–3069
Vincent JM (1970) A manual for the practical study of root nodule bacteria. In: international biological program handbook no. 15; Blackwell scientific publications ltd.: Oxford
Wielbo J, Skorupska A (2001) Construction of improved vectors and cassettes containing gusA and antibiotic resistance genes for studies of transcriptional activity and bacterial localization. J Microbiol Methods 45:197–205
Wielbo J, Mazur A, Krol JE, Marczak M, Skorupska A (2004) Environmental modulation of the pssTNOP gene expression in Rhizobium leguminosarum bv. trifolii. Can J Microbiol 50:1–11
Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA (2008) Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol 190:4706–4715
Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA (1995) β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691–1705
van Workum WAT, van Brussel AAN, Tak T, Wijffelman CA, Kijne JW (1995) Ethylene prevents nodulation of Vicia sativa ssp. nigra by exopolysaccharide-deficient mutants of Rhizobium leguminosarum bv. viciae. Mol Plant-Microbe Interact 8:278–285
van Workum WA, Canter Cremers HC, Wijfjes AH, van der Kolk C, Wijffelman CA, Kijne JW (1997) Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant-Microbe Interact 10:290–301
Yamazaki M, Thorne L, Mikolajczak M, Armentrout RW, Pollock TJ (1996) Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J Bacteriol 178:2676–2687
Yanni YG, Rizk RY, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, Orgambide G, De Bruijn F, Stoltzfus J, Buckley D, Schmidt TM, Mateos PF, Ladha JK, Dazzo FB (1997) Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 194:99–114
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Katharina Pawlowski.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Janczarek, M., Rachwał, K. & Turska-Szewczuk, A. A mutation in pssE affects exopolysaccharide synthesis by Rhizobium leguminosarum bv. trifolii, its surface properties, and symbiosis with clover. Plant Soil 417, 331–347 (2017). https://doi.org/10.1007/s11104-017-3262-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3262-5