Abstract
Background and Aims
Laboratory and greenhouse experiments have shown that root-associated bacteria have beneficial effects on grapevine growth; however, these effects have not been tested in the field. Here, we aimed to demonstrate whether bacteria of different geographical origins derived from different crop plants can colonize grapevine to gain a beneficial outcome for the plant leading to promote growth at the field scale.
Methods
To link the ecological functions of bacteria to the promotion of plant growth, we sorted fifteen bacterial strains from a larger isolate collection to study in vitro Plant Growth Promoting (PGP) traits. We analysed the ability of these strains to colonise the root tissues of grapevine and Arabidopsis using green-fluorescent-protein-labelled strain derivatives and a cultivation independent approach. We assessed the ability of two subsets randomly chosen from the 15 selected strains to promote grapevine growth in two field-scale experiments in north and central Italy over two years. Parameters of plant vigour were measured during the vegetative season in de novo grafted vine cuttings and adult productive plants inoculated with the bacterial strains.
Results
Beneficial bacteria rapidly and intimately colonized the rhizoplane and the root system of grapevine. In the field, plants inoculated with bacteria isolated from grapevine roots out-performed untreated plants. In both the tested vineyards, bacteria-promotion effects largely rely in the formation of an extended epigeal system endowed of longer shoots with larger diameters and more nodes than non-inoculated plants.
Conclusions
PGP bacteria isolated in the laboratory can be successfully used to promote growth of grapevines in the field. The resulting larger canopy potentially increased the photosynthetic surface of the grapevine, promoting growth.
Similar content being viewed by others
Introduction
Plant Growth Promoting (PGP) bacteria can impart characteristics that improve plant growth either directly by providing nutrients to the roots or indirectly by contributing to plant hormone homeostasis and resistance to pathogens (Berg 2009; Lugtenberg and Kamilova 2009; Philippot et al. 2013; Panke-Buisse et al. 2015). These powerful properties have been shown at laboratory scale using in vitro assays (Zamioudis et al. 2013; Salomon et al. 2014) , soil microcosm tests (Sabir et al. 2012; Suarez et al. 2015), and field-like conditions (Shishido 1996; Chanway 1997; Aslantas et al. 2007; Magnin-Robert et al. 2013; Rolli et al. 2015). However, the role these symbionts play in field-scale agricultural environments remains poorly understood (Bashan et al. 2014 and reference therein; Berger et al. 2015; Garcia-Seco et al. 2015; Garima and Nath 2015). The establishment of a successful interaction with field-grown plants implies the ability of the amended bacteria to colonize both the root tissues - as endophytic bacteria - and the rhizoplane and to perform the beneficial effect on plant growth and development. They are thus called root-associated PGP bacteria (Schlaeppi and Bulgarelli 2015). Knowledge of detailed interactions between bacteria and plant is limited, even for many commercially available PGP bioformulations (Calvo et al. 2014). Most studies have been conducted on annual crop plants, such as radish, lettuce and sunflower (Berger et al. 2015; Sahin et al. 2015; Shahid et al. 2015), leaving the interaction between PGP bacteria and perennial arboreal plants poorly explored (Magnin-Robert et al. 2007; Magnin-Robert et al. 2013; Bashan et al. 2014; Gómez-Lama Cabanás et al. 2014; Aziz et al. 2015).
Grapevine is an economically important fruit crop cultivated over a large latitudinal range in Europe and the Middle East: in the northern hemisphere from 20 to 50°, from Saudi Arabia (Taif region) to Belgium (Wallonia region) (FAOSTAT n.d.). Grape productivity and fruit quality are threatened at all the latitudes (Chaves et al. 2010; Edwards and Clingeleffer 2013). Grapevine is a long-lasting cultivation: three years are required before new cuttings become productive, and, once established, adult plants can be maintained in vineyards for more than thirty years (Champagnol 1984; Vercesi 2010). The rhizosphere and root tissues, which comprise the grape root system, contain a subset of soil microorganisms that provide ecological services to the plant (Zarraonaindia et al. 2015). The grapevine core microbiome imparts similar PGP characteristics independent of geographical location, suggesting the persistence of a core set of PGP traits that influence plant development (Marasco et al. 2013). These bacteria can migrate from the root to the shoot of grapevine (Compant et al. 2010; Compant et al. 2011) surviving in the fruit (Bokulich N et al. 2014; Gilbert J et al. 2014). Moreover, recent works demonstrated how grape tissues can be colonized by both autochthonous and allochthonous bacteria (Compant et al. 2013; Rolli et al. 2015).
In this work, we assess whether bacteria of different geographical origins derived from different crop plants were able to effectively colonize grapevine roots, leading to a beneficial interaction that could result in a promoted growth in the field (Carvalho et al. 2016). We sorted 15 strains from a larger isolate collection for in vitro PGP characteristics and their ability to colonise roots. We assessed the ability of two subsets of the 15 selected strains to promote grapevine growth in two field-scale experiments in north and central Italy over two years. The aim of the work was to assess the potential of plant growth promoting bacteria to support the growth of grapevine under field conditions. We were not aimed to compare a set of bacteria on different cultivars but rather to test the largest number of strains as possible to demonstrate that bacteria are a useful biotechnological resource for grapevine crop management.
Materials and methods
Vineyards, plant material and experimental design
To maximize the number of observations, the field experiments were performed in two vineyards: Franciacorta (Lombardy region, Brescia province, northern Italy, 45°56′56″N; 9°97′53″E) owned by the “Castello Bonomi” farm (CB) and Montefalco (Umbria region, Perugia province, central Italy, 42°53′32″N; 12°38′53″E) owned by the “Arnaldo Caprai” farm (AC). Supplementary Fig. S1 and Supplementary Table S1 report the climate conditions and the chemical analyses of the soils at these farms. The field experiments were designed with, approved by and run by the technical staff of the two farms. Permission to conduct these experiments was granted by the owners of the two farms. The field study did not involve endangered or protected species. The field plots were chosen by the technical staff of the farms as the most suitable for the cultivars in the respective areas.
Field experiments were conducted on both grape plantlets (one-year-old Syrah plantlets grafted onto 1103P rootstock and one-year-old Sauvignon plantlets grafted on SO4 rootstock) and adult grape plants (17-year-old Syrah plants grafted onto Fercal rootstock) in 2010 and 2011. The two cultivars used in this study have been chosen mainly on the base of their distribution and importance in the two studied farms. We tested the effect of different PGP bacteria on plant growth by measuring vegetative (shoot diameter, shoot length, node number) and productive (number of grape bunches per plant and total fruit yield per plant) parameters. Six treatments (non-treated and treatment with strains B1, B2, B3, B4 or B5) were applied to 12 Syrah plantlets each distributed along three blocks that included four plants, randomly distributed in the CB vineyard in both 2010 and 2011. Eleven treatments (plants non-treated, and plants treated with strains B7, B8, B9, B10, B11, B12, B13, B14, B15 or B16) were applied to 50 Sauvignon plantlets each in two plots that included 25 plants, randomly distributed in the AC vineyard in 2010. In 2011, this experiment was repeated using only the best three treatments (plants treated with strains B7, B8 and B10 and the non-treated control plants) on the previously treated plantlets. During 2011, adult plants in the CB vineyard were also treated with the B1, B2, B3, B4 or B5 strains in order to test the potential of PGP bacteria to support and promote growth in adult plants under field conditions. Each treatment was on 12 random plants in three blocks that included four plants each. The control plants were untreated.
Origin and PGP potential of the selected bacteria
The 15 bacterial strains used in the study were a subset of a larger collection of bacteria isolated from the rhizosphere and the root endosphere of several Mediterranean plants, including grapevines, olive trees and pepper plants, according to the protocol described by Marasco et al. (2012). To span a large phylogenetic range, we included different bacterial genera and species among the fifteen selected strains. The strains, species affiliations and characteristics are reported in Table 1. The partial 16S rRNA gene sequences of the selected strains were deposited in the NCBI database (accession numbers reported in Table 1). Bacteria with multiple in vitro PGP activities, such as auxin (Indole-3-acetic acid, IAA) production, 1-Aminocyclopropane-1-Carboxylate (ACC) deaminase activity, exopolysaccharide production, phosphate solubilisation, siderophore production, potential nitrogen fixation, protease and ammonia production, were selected for screening assays. The screenings were performed as follows. Briefly, IAA production was estimated as described by Bric et al. (1991). The presence of IAA in the culture supernatant was determined spectrophotometrically at 530 nm. Pure IAA (Sigma-Aldrich, Italy) was used as the standard and uninoculated medium served as the control. ACC-deaminase activity was evaluated as described by Penrose and Glick (2003). Bacteria were streaked on DF-ACC medium prepared by adding, just prior to use, the ACC solution (Sigma-Aldrich, Italy) to autoclaved DF minimal medium. DF medium (without a nitrogen source) was used as the control. Solid DF and DF-ACC media were supplemented with 1.5 % ultrapure agar. DF-ACC and DF plates were incubated for 96 h at 30 °C. Bacteria able to grow only in the DF-ACC medium were identified as positive for ACC deaminase activity. Exopolysaccharide production (Ali et al. 2014) was estimated by growing bacteria on a RCV-sucrose agar medium as described by Santaella et al. (2008). Phosphate solubilization was assessed as described by Ahmad et al. (2008). The solubilization halo formed after one week of incubation at 30 °C was evaluated. Siderophore production was detected according to the protocol described by Milagres et al. (1999) using chrome azurol sulphonate (CAS) agar plates. The formation of orange halos on CAS agar plates after incubation for one week at 30 °C was evaluated. Potential nitrogen fixation activity was evaluated on a nitrogen-free (NFB) medium (Döbereiner 1980). Protease activity was determined by growing isolates on skimmed milk agar (Nielsen and Sørensen 1997). Ammonia production was tested in peptone water according to the protocol designed by Cappuccino and Sherman (1992).
Bacterial recolonization of plant roots
Recolonization assays were performed on grapevines and the model plant Arabidopsis thaliana using fluorescent-labelled strain derivatives. We aimed to study the early step of the colonization process of grapevine root by the beneficial bacteria. We also used Arabidopsis because of its little root that being thinner that the grapevine one, it allows a higher magnification of the whole root at the microscope. We are aware of the large biological differences between Arabidopsis and grapevine and that establishing a direct correlation between the patterns observed in the two plants is difficult. However, we considered Arabidopsis because it can provide more details in the bacterial colonization on the root, since the whole root tip can be easily observed at a relatively higher magnification than for grapevine at the confocal microscope. Arabidopsis is widely used as a tool to study plant-microbe interaction by overcoming the inherent problematics to perform research on woody long-living plants (Poupin et al. 2013; Baldan et al. 2015; Maldonado-Gonzalez et al. 2015). Following the protocol described by Marasco et al. (2012), we transformed two strains, Pseudomonas fluorescens gfp-B5 and Pseudomonas rhodesiae dsRED-B7, labelled with Gfp and DsRED fluorescent proteins, respectively.
Experiments with Arabidopsis
We evaluated the ability of the bacteria to adhere to the roots and recolonise one-week-old Arabidopsis seedlings. The Arabidopsis seeds were soaked for 20 min in 5 % household bleach and rinsed five times with sterile distilled water in microcentrifuge tubes. After this surface sterilisation step, the seeds were transferred to plates containing half-strength MS medium (2.15 g/L Murashige and Skoog salt (Sigma), 15 g/L saccharose, pH = 5.7), vernalised for 72 h at 4 °C and transferred to the growth chamber (150 μmol/m2 sec, 12 h/12 h light/dark, 22 °C). After one week, the plantlets were removed from the plates and the roots were dipped in 108 cells/mL bacterial suspensions for 48 h. Bacteria were inoculated in TSB (trypto soy broth) medium and incubated at 30 °C overnight in a shaking incubator (200 rpm). Bacterial cells were counted under the microscope and the different inoculum preparations were obtained by resuspending 108 cells in 1 ml of sterile water. Arabidopsis roots dipped in sterilized water were used as negative controls. After incubation, bacteria loosely attached to the surfaces of the roots were removed by washing the plants in sterile water. The colonized and non-treated roots were observed under a laser-scanning confocal microscope (Leica TCSNT) using GFP (excitation at 488 nm) and dsRED filters (excitation at 558 nm). The fluorescence in the acquired images was analysed by using the MBF-ImageJ software to quantify the colonisation of the bacterial cells on the surface of the roots.
Experiments with grapevine
To evaluate the ability of fluorescent-labelled bacterial strains to colonize a grapevine root system, the bacterial cultures were prepared as described above. Two-year-old ‘black magic’ grapevine plants were planted in pots containing soil from the CB or the AC farms and placed in the greenhouse for 7 days. The plants were appropriately irrigated throughout the experiment. The roots inoculated with the B5-gfp and B7-dsRED strains were gently removed from the soil and analysed under the laser-scanning confocal microscope as described for the experiments with Arabidopsis. Finally, the bacteria in the grapevine rhizosphere was analysed using denaturing gradient gel elecrophoresis (DGGE) as described by Rolli et al. (2015). The DGGE bands were excised from the gel using a sterile cutter and eluted in 50 μl of water at 37 °C for 5 h. The reamplification of DNA eluted from the DGGE bands was performed using 907R and 357F primers without GC-clamps according to the following protocol: 95 °C for 5 min, 30 cycles at 95 °C for 1 min, 61 °C for 1 min, 72 °C for 1 min and a final extension at 72 °C for 7 min. PCR products were checked by electrophoresis in 1 % agarose gel. The sequencing was performed at Macrogen Inc. (Korea). The gel band patterns were analysed using the BioRad GELDOC software.
Preparation of the bacterial cells and treatment of the grape plants
Since the aim of this study was to explore the use of PGP bacteria in the field, we chose the highest number of isolates to be tested in the two available field locations. Following this objective, five bacterial strains (B1-B5) were assayed for experiments at the CB farm, while the remaining ten strains were used in experiments at the AC farm (strains B7-B16). Following Bashan et al. (2016), we describe the strain preparation and application methods. The strains were streaked on tryptic soy agar (TSA) plates to verify their purity and then inoculated in 300 mL of TSB in a 1000 mL Erlenmeyer flask and incubated at 30 °C on a shaker (150 rpm) for 48 h. Five to ten flasks, depending on the strain and its biomass yield, were prepared to obtain sufficient biomass for plant bacterization according to the bacterial cell concentrations reported below. Bacteria were collected by centrifugation (4000 rpm, 15 min) and washed twice in 9 g/L NaCl. We used a simple water-based liquid formulation (Bashan et al. 2014) to prepare the bacterial inoculants. Each of the washed bacterial pellets was resuspended in sterile distilled water (3 L per each plant) and used to inoculate grapevine plantlets when planted or during irrigation of the soil when the plants were already established in the field.
Before planting in the soils collected from the farms, one-year old plantlets were rehydrated by placing the root system in water for 8 h and then soaking them in a 108 CFU/mL suspension of the tested bacterial strains for 48 h. Non-treated controls were created by dipping the plantlets’ roots in distilled sterile water. During the second year (2011), plantlets were retreated with the selected bacteria by irrigating the soil around the plant with a bacterial suspension to provide 2 x 1011 cells/plant per portion of soil (20 cm diameter). The treatment was performed in spring at the vegetative stage. The treatment performed in the second year at the AC farm included only the three best performing strains (B7, B8 and B10) from the first year trial. At the CB farm, adult plants were treated with the B1-B5 strains.
Measurement of vegetative parameters and statistical analysis of the data
We measured various vegetative parameters, including shoot length, shoot diameter and number of nodes, on all treated and control plants every 20 days after bacterization (dab) for a period ranging from two to four months, depending on the experiment. The measurements of vegetative parameters of the adult plant treatments were performed only in 2011 every 20 days until harvest time. Productive parameters of the adult plants (number of grape bunches per plant and total fruit yield per plant) were measured only in 2011 at harvest.
We considered the vegetative and productive parameters to be response variables to test the null hypothesis of no difference among all the treatments using the Statistical Package for the Social Sciences (SPSS) software. We used ANOVA with Duncan’s test, consisting of a series of pairwise comparisons between means, to determine statistical significance of the applied treatments: the vegetative and productive parameters (considered as response variables) among the different treatments were the fixed factors and plants treated with strains B1, B2, B3, B4, B5, B7, B8, B9 and B10 and non-treated plants were the orthogonal levels, across the experimental time (depending on the experimental procedures). Significant differences between the treatments were considered at p ≤ 0.05; these differences are indicated in the graphs with a star (*).
Results
In vitro evaluation of PGP of selected bacteria
The studied strains (species affiliations reported in Table 1) comprise fifteen bacterial families, Alcaligenaceae, Bacillaceae, Brucellaceae, Comomonadaceae, Enterobacteriaceae, Xanthomonadaceae, Microbacteriaceae, Moraxellaceae, Nocardiaceae, Paenibacillaceae, Promicromonosporaceae, Pseudomonadaceae, Sphingobacteriaceae, Streptomycetaceae and Rhizobiaceae. The strains isolated from both root tissues (seven strains) and rhizospheric soil (eight strains) exhibited a variety of PGP traits in vitro. These traits included the ability to solubilise insoluble phosphorous forms (12 strains), produce the auxin 1-indolacetic acid (11 strains) and release siderophores (10 strains), suggesting that a range of potential mechanisms supports plant growth (Table 1). The feasibility of the application of beneficial bacteria was further studied by assessing the bacterial rhizocompetence on grapevine roots. To better assess the details of the colonization process on whole roots, Arabidopsis plants were also used because of the thinner roots that the grapevine’s ones, allowing higher magnification of the whole root at the microscope. Two strains transformed with fluorescent-protein-coding genes were used for the colonization assays on germ-free Arabidopsis plantlets and non-sterilized grapevine roots.
Experiments with Arabidopsis
After 24 h of exposure to the B5-gfp bacterial suspension, confocal microscopy analysis showed that the Arabidopsis rhizoplane was massively colonized by gfp-tagged cells and the root hairs were completely enwrapped by bacterial cells (Fig. 1a). Epifluorescence microscopy provided adherence profile of the B5-gfp and B7-dsRED strains, which confirmed the ability of bacterial cells to colonize the root surfaces of Arabidopsis roots (Fig. 1b and c). Quantification of the number of fluorescent cells on the surfaces of the roots was used to estimate the colonization ability of the bacteria. The B5-gfp strain had coverage of 0.803 ± 0.383 cell/1000 μm2 of root surface after 24 h of exposure to the bacterial suspension (Fig. 1b). A similar number of cells was counted with the B7-dsRED strain (0.747 ± 0.337 cell/1000 μm2 of root surface, Fig. 1c).
Rhizocompetence of bacteria on Arabidopsis thaliana and grapevine roots and impact of bacteria treatment on rhizosphere communities associated with grape plants grown under field-like conditions. The plant root adhesion assay was performed using two fluorescent-labelled strains, marked respectively with gfp and dsRED and two model plants: Arabidopsis (a-c) and grape (d and e). a Confocal microscopy analysis of the adhesion profile of B5-gfp on the Arabidopsis rhizoplane after 24 h. b c Details of the adherence profile of strains B5-gfp and B7-dsRED on Arabidopsis root system under the epifluorescence microscope after 24 h. For each image set, the first panel refers to the fluorescence images to visualize the adherence profile of the gfp/dsRED-labelled cells, the second panel shows the phase contrast microscopy of Arabidopsis roots and the third one results from the merge of the phase contrast and the fluorescent images. Scale bars correspond to 100 μm in A, to 50 μm in B and C, respectively. d Confocal microscopy analysis of B5-gfp strain colonizing the grapevine root surface 7 days after biofertilization with the selected strain. The red channel was used to acquire the grape root autofluorescence, providing information about its structure. Arrows indicate gfp fluorescent bacteria along the root surface or root hair. Scale bars correspond to 100 μm in D. (E) DGGE profiles based on the 16S rRNA gene of the rhizosphere bacterial communities associated with grapevine cultivated outdoors. DGGE profiles of B5 and B7 are included. Stars and circles were used to mark in the DGGE gel the specific bands corresponding to strains B5 and B7 in the DGGE profiles of the 16S rRNA gene of the rhizospheric communities associated to the grape plants grown in greenhouse
Experiments with grapevine
Only B5-gfp bacterial cells were detected by confocal microscopy on non-sterile grapevine roots due to the strong auto-fluorescence of grapevine roots in the red channel. The B5-gfp cells were mainly detected in the rhizoplane and only a few cells were found on the root hairs, for a total of 0.567 ± 0.278 cells/1000 μm2 of root surface (Fig. 1d). The presence of inoculated strains was evaluated using molecular fingerprinting techniques. DGGE gel bands with the migration profiles similar to those of the bacterial strains were present only in plants treated with the bacteria. Given that PCR-DGGE cannot detect bacteria in a community below a 1 % cut-off (Muyzer et al. 1993), the relevant DGGE bands confirmed that the selected bacteria can effectively and stably colonize grapevine root systems (Fig. 1e).
Effects of the bacterial inoculation on grapevine plantlets during the first growing season
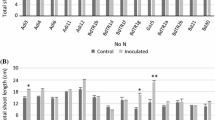
It is impossible to control the environmental conditions in the field. During the two years of the field experiment, the two farms experienced various climate events (Supplementary Fig. S1). Measurements were made every 20 days from March through May in 2010 in both vineyards (Supplementary Tables S2 and S3). The shoots of plantlets exposed to B2 and B3 at CB were significantly longer (109.5 ± 17.9 cm and 110.6 ± 34.5 cm, respectively) than the shoots of the untreated control plants (75.5 ± 29.1 cm; Fig. 2a and Supplementary Table S2). Data collected at 80 dab showed that strains B7, B8 and B10 had significantly higher node numbers, shoot lengths and shoot diameters compared with the non-treated control, suggesting that these isolates promote the formation of more robust epigeal systems under field conditions (Fig. 2b and Supplementary Table S3). While the shoot length in the control reached 32.8 ± 18.4 cm at 80 dab, the shoot length of the plantlets exposed to B7 reached 56.4 ± 26.5 cm, a 65 % increase in shoot length over the control plants. Plants exposed to B7 had statistically significant thicker shoot diameters (5.7 ± 1.6 mm vs. 3.3 ± 1.2 mm) and higher numbers of nodes (17.4 ± 7 vs. 12.6 ± 5.8) than the control plants, corresponding to 72 % and 38 % increases, respectively (Supplementary Table S3).
Effects of PGP bacteria on growth promotion of Syrah and Sauvignon grapevine plantlets by PGP bacteria during the first year experiment in the field. The selected bacteria promoted the formation of a larger shoot system by improving shoot length in Syrah grapevine plantlets in CB vineyard (a) and Sauvignon grapevine plantlets in AC vineyard (b). For each bacterial strain overlapping histograms are reported, representing the different times of shoot length measuring at 20, 40, 60 and 80 dab. The Duncan’s test statistical significance of the differences among treatments is indicated only for the last time of measuring. For the values at the other vegetative parameters, the data and the statistical significance are reported in Supplementary Tables S2 and S3. Dash-lines in the graphs mark the values for the untreated plants, in order to better evaluate the promotion effect mediated by the PGP bacteria
Effects of bacterial inoculation on grapevine plantlets and adults during the second growing season
At CB, plants treated with B2 and B3 exhibited increased shoot length at 120 dab (Fig. 3a and Supplementary Table S4), whereas an increase in shoot diameter was not observed in these plants (Fig. 3b). Longer shoots and increased numbers of nodes were observed in the treated plants at 40 dab and this trend continued during the season (Supplementary Table S4). For instance, plants treated with B2 had shoot lengths of 103.7 ± 56 cm with 23.11 ± 10.4 nodes compared to 68.94 ± 64.78 cm and 15.11 ± 11.38, respectively in the control plants. Plants exposed to B4 presented longer shoots and larger shoot diameters compared to the non-treated control (Fig. 3b and Supplementary Table S4).
Effects of PGP bacteria on growth promotion of Syrah and Sauvignon grapevine plantlets by PGP bacteria during the second year experiment in the field. The selected bacteria confirmed the formation of a larger shoot system by improving a, c shoot length and b, d shoot diameter in Syrah (CB vineyard) and Sauvignon (AC vineyard) plantlets, respectively. In panels b, c and d for each bacterial strain overlapping histograms are reported, representing the different times of shoot parameters measuring from 20 to 100 dab (each 20 days). The Duncan’s test statistical significance of the differences among treatments is indicated only for the last time of measuring. For the values of the others vegetative parameters, the data and the statistical significance are reported in Supplementary Tables S4 and S5. Dash-lines in the graphs mark the values for the untreated plants, in order to better evaluate the promotion effect mediated by the PGP bacteria
At AC, the increased growth induced by B7, B8 and B10, the three best performing strains during the first year, was maintained in the second year (Fig. 3c and d and Supplementary Table S5). Plants treated with these strains exhibited extended epigeal systems with longer shoots (B7 and B8) or increased shoot diameters (B7 and B10) and a higher number of nodes (B7 and B10) compared to untreated controls. The increases in the plants treated with B7 were observed at the first observation point. The increases in the plants treated with B8 and B10 were observed at the second observation point, while the two other strains determined a promotion effect only observable after the second time point of measure (Supplementary Table S5).
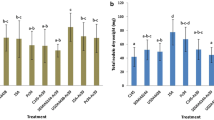
During the vegetative season, we observed an increase in shoot length in plants treated with B3, (Fig. 4a and Supplementary Table S6) in the 17-y-old productive grapevines. B2- and B3-treated adult plants exhibited increased fruit yield (Fig. 4c). B2-treated plants also displayed increased bunch numbers per plant in comparison with the untreated grapevine plants (Fig. 4d and Supplementary Table S6).
Effects of PGP bacteria on growth promotion of Syrah grapevine adult productive plants by PGP bacteria under field conditions. The selected bacteria promoted the formation of a larger canopy by increasing a shoot length, b the number of nodes, c bunch production and d number of grape bunches, per plant. In panels A and B for each bacterial strain overlapping histograms are reported, representing measurements at 20 and 40 dab. The Duncan’s test statistical significance of the differences among treatments is indicated only for the last time of measuring. For the values at of the others vegetative parameters, the data and the statistical significance are reported in Supplementary Tables S6. Dashed lines in the graphs mark the values for the untreated plants, in order to better evaluate the promotion effect mediated by the PGP bacteria
Discussion
There is overwhelming evidence that PGP bacteria increase plant production (Schlaeppi and Bulgarelli 2015). Several studies have demonstrated the beneficial effects exerted by PGP bacteria on the laboratory scale (Bhardwaj et al. 2014; Gontia-Mishra et al. 2014; Daffonchio et al. 2015; Haney et al. 2015), but relatively less data are available under on the field scale (Bashan et al. 2014 and references therein; Nelissen et al. 2014). Translating laboratory results to field trials is important for the future development of crop productivity (Tuberosa 2012; Bashan et al. 2014).
Grape is the second largest fruit crop in the world: 67 million tons of grapes are produced annually worldwide and viticulture is an important sector of the economy (FAOSTAT). Information on the involvement of the grape-associated microbiome in vineyard productivity and the contribution of this microbiome to the quality of the final product is emerging (Marasco et al. 2013; Bokulich N et al. 2014; Gilbert J et al. 2014). While data are available on the effects of PGP bacteria on grapevines at the laboratory and greenhouse scales (Rolli et al. 2015), no such data have been reported on the field scale, although field studies of the biocontrol of pathogens have been conducted (Magnin-Robert et al. 2007; Magnin-Robert et al. 2013; Aziz et al. 2015). Clearly, the study of the promotion of plant growth in the field is less advanced than that of biocontrol of pathogens in the field. Here, we assess the effects of PGP bacteria in the field. We found that several bacterial strains consistently increased one or more plant growth parameter in the two successive years of the experiments. These effects were achieved in the two climatic regions of north and central Italy. We observed the effects of PGP bacteria on young vine cuttings over two growing seasons and on productive adult vines over one growing season. The observed effect on the shoot growth parameters (length, diameter and number of nodes) and on fruit production parameters (number of grape bunches and weight of grapes) were statistically significant.
Despite the important economic and agricultural roles played by arboreal crops, little work has been done on the application of PGP bacteria in the field level. Only apple, desert legume and mango trees have been studied (Shishido 1996; Aslantas et al. 2007; Bashan et al. 2009; Bashan et al. 2012; Jaos Frederico et al. 2014). The effects of PGP bacteria on annual crops have also been the focus of research (Bashan et al. 2014 and references therein). For instance, phosphate-solubilizing bacteria were shown to be effective in enhancing maize yields (Khan and Khan 2015). Our data extend understanding of the role played by PGP bacteria at the field level in arboreal plants, such as grape.
Our data show that in the second year of the experiments, strains B2, B3 and B7 had a more pronounced effect on shoot length than on shoot diameter (Figs. 2 and 3 and Supplementary Tables S2–5). This could be linked to the effects of the strains on the plant hormone homeostasis, such as the auxin balance (Dimkpa et al. 2009; Kurepin et al. 2014).
Previous studies demonstrated that bacterial strains promote plant growth at the laboratory and greenhouse scales (Rojas-Tapias et al. 2012; Cherif et al. 2015). The PGP effects that we observed in the field could be attributed to beneficial interactions between the isolates we studied and the natural microbiomes of the grapevine roots, including beneficial bacteria and/or fungi. Although such interactions were beyond the scope of our study, this possibility is an important research topic that requires further investigation.
Before the field experiments we did not test the selected strains through in planta in the laboratory or greenhouse. Our selection of bacterial strains was based on in vitro laboratory screening of multiple PGP traits. It is commonly considered that functional screening on the laboratory scale is required to identify, among large isolate collections, the most promising PGP candidates (Bashan et al. 2014; Schlaeppi and Bulgarelli 2015). However, screenings based on pure culture assays may not identify the most powerful strains (Cardinale et al. 2015). Even though in vitro laboratory screening may not identify the most effective PGP isolates, our field experiment data confirmed that pre-screening is indeed helpful for the selection of PGP candidates (Garima and Nath 2015) that will perform under field conditions.
By evaluating the colonization potential of fluorescent-labelled bacteria, we introduced an intermediate step between the laboratory and the field scale. Indeed, root colonization is a crucial event in the establishment of beneficial interactions between symbiotic and non-symbiotic (mutualist) bacteria and the plant host (Drogue et al. 2012). By advanced microscopy analysis, it was previously demonstrated that the desert soil isolate Saccharothrix algeriensis is able to establish within 10 days on the root surface of grapevine plants (Compant et al. 2013). Similarly, Vitis vinifera cv. Glera host beneficial microbes that induce pleiotropic effect on Arabidopsis root architecture, finally resulting in a potential enhancement of water and nutrients (Baldan et al. 2015). In our study grapevine and Arabidopsis were used to assay whether the beneficial bacteria have broad rhizocompetence ability, in the vision of field application in vineyard. We included Arabidopsis in the experiments, because it has thinner roots than grapevine that can be observed as a whole root at a higher magnification than grapevine, offering the possibility to observe more details of the bacterial colonization. Moreover, we found that the autofluorescence in the red channel of the grapevine root does not allow a proper visualization of one of the strains we have labelled. By using Arabidopsis we could overcome the study of the root colonization by this strain. We recognize that the colonization patterns observed in Arabidopsis may not reflect the one occurring in grapevine, but provide hints of the general process of root colonization. Our fluorescent screens on plant roots confirmed that the assayed bacteria were able to colonise the root systems of the plants as previously observed by Rolli et al. (2015). Moreover, bacteria may became endophytic and moving from the interior of the roots to other plant tissues (Compant et al. 2010; Compant et al. 2011; Lareen et al. 2016).
We did not design our experiments to compare different bacterial strains on different grapevine cultivars in two climatic regions. We therefore cannot suggest that the strains will have similar effects on different cultivars or in different soils. Several factors contribute in shaping the grapevine associated microbiome, including the plant selective pressure, the soil structure (in particular the pH and C:N ratio), the vineyard management practises and biogeographic related parameters with a local heterogeneity even across small distances, homogenous climatic parameters and similar soil properties (Zarraonaindia et al. 2015). However, recent studies have shown that core microbiomes inhabit the same plant species whatever the cultivar or wherever it is grown (Bulgarelli et al. 2012; Lundberg et al. 2012; Peiffer J et al. 2013). Similarly, ecological or biocontrol services performed by bacteria have been shown to occur in root-associated microbiomes, under mild or more challenging environmental conditions (Marasco et al. 2013) or in different crop plants (Berg et al. 2002). The strains studied here should be further tested in specifically conceived experiments.
There is debate on how the origin of the plant host affects the bacteria’s ability to induce beneficial effects in other plant species (Drogue et al. 2012). PsJN, a Burkholderia phytofirmans strain and a well-assessed PGP grapevine endophyte, efficiently ameliorated the effects of drought on growth, physiology and yields of maize in a field trial (Naveed et al. 2014). Contrasting results in the literature on the cross effects of PGP bacteria on different plants suggest that that they should be considered case by case. We recognize that the number of strains tested in our experiments was not large enough to infer any role that plant provenance could play in PGP capacity. However, we found that isolates obtained from grapevine root systems had the most pronounced effects on growth parameters. Such an observation suggests that the origin of the bacteria is an important factor and that co-evolution with the plant host in the wild may play a role (Rodriguez et al. 2008).
Conclusion
Prospecting root-associated microbiomes for natural products that increase agricultural production without damaging the environment is a current biotechnological trend (Schlaeppi and Bulgarelli 2015). Although larger and longer field trials under different environmental conditions, on different cultivars and in different soils are needed, our results show that PGP bacteria reared in the laboratory can: i) rapidly establish in the root system of grapevine suggesting a versatile rhizocompetent profile; ii) improve grapevine growth in the field in a statistically significant way respect to non bacterized plants even if they originate from different grapevine cultivars and iii) induce promotion effect in the growth of grapevine plantlets in various biogeographical and vineyard microscale contexts. However, to further establish microorganism-based field-scale viticulture, the persistence and turnover of the applied bacteria in association with the plants over time and their interactions with the indigenous microbiome should be investigated.
References
Ahmad F, Ahmad I, Khan MS (2008) Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 163:173–181. doi: 10.1016/j.micres.2006.04.001
Ali SZ, Sandhya V, Rao LV (2014) Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann Microbiol 64:493–502. doi:10.1007/s13213-013-0680-3
Aslantas R, Cakmakci R, Sahin F (2007) Effect of plant growth promoting rhizobacteria on young apple tree growth and fruit yield under orchard conditions. Sci Hortic (Amsterdam) 111:371-377
Aziz A, Verhagen B, Magnin-Robert M, et al. (2015) Effectiveness of beneficial bacteria to promote systemic resistance of grapevine to gray mold as related to phytoalexin production in vineyards. Plant Soil:1-13. doi:10.1007/s11104-015-2783-z
Baldan E, Nigris S, Romualdi C, et al. (2015) Beneficial bacteria isolated from grapevine inner tissues shape Arabidopsis thaliana roots. PLoS One 10:1-18. doi:10.1371/journal.pone.0140252
Bashan Y, Salazar B, Puente ME (2009) Responses of native legume desert trees used for reforestation in the Sonoran Desert to plant growth-promoting microorganisms in screen house. Biol Fertil Soils 45:655-662. doi:10.1007/s00374-009-0368-9
Bashan Y, Salazar BG, Moreno M, et al. (2012) Restoration of eroded soil in the Sonoran Desert with native leguminous trees using plant growth-promoting microorganisms and limited amounts of compost and water. J Environ Manag 102:26-36. doi:10.1016/j.jenvman.2011.12.032
Bashan Y, de-Bashan LE, Prabhuz SR, Hernandez JP (2014) Advances in plant growth-romoting bacterial inoculant technology: Formulations and practical perspectives (1998-2013). Plant Soil 378:1-33. doi:10.1007/s11104-013-1956-x
Bashan Y, Kloepper JW, LE d-B, Nannipieri P (2016) A need for disclosure of the identity of microorganisms, constituents, and application methods when reporting tests with microbe-based or pesticide-based products. Biol Fertil Soils. doi:10.1007/s00374-016-1091-y
Berg G (2009) Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11-18. doi:10.1007/s00253-009-2092-7
Berg G, Roskot N, Steidle A, et al. (2002) Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host Pl. Appl Environ Microbiol 68:3328-3338. doi:10.1128/AEM.68.7.3328
Berger B, Wiesner M, Brock AK, Schreiner M (2015) K. Radicincitans, a beneficial bacteria that promotes radish growth under field conditions. Agron Sustain Dev:1521-1528. doi:10.1007/s13593-015-0324-z
Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Factories 13:66. doi:10.1186/1475-2859-13-66
Bokulich N a, Thorngate JH, Richardson PM, Mills D a (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:E139-E148. doi:10.1073/pnas.1317377110
Bric JM, Bostock RM, Silverstone SE, et al (1991) Rapid In Situ Assay for Indoleacetic Acid Production by Bacteria Immobilized on a Nitrocellulose Membrane Rapid In Situ Assay for Indoleacetic Acid Production by Bacteria Immobilized on a Nitrocellulose Membrane. 57:535-538.
Bulgarelli D, Rott M, Schlaeppi K, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91-95. doi:10.1038/nature11336
Calvo P, Nelson L, Kloepper JW (2014) Agricultural uses of plant biostimulants. Plant Soil 383:3-41. doi:10.1007/s11104-014-2131-8
Cappuccino JC, Sherman N (1992) Microbiology: A Laboratory Manual. Third, New York
Cardinale M, Ratering S, Suarez C, et al. (2015) Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol Res 181:22-32. doi:10.1016/j.micres.2015.08.002
Carvalho TLG, Ballesteros HGF, Thiebaut F, et al. (2016) Nice to meet you: genetic, epigenetic and metabolic controls of plant perception of beneficial associative and endophytic diazotrophic bacteria in non-leguminous plants. Plant Mol Biol 90:561-574. doi:10.1007/s11103-016-0435-1
Champagnol F (1984) Éléments de physiologie de la vigne et de viticulture générale. INRA Station de Recherches Viticoles Ecole Nationale Superieure Agronomique, Montpellier Cedex (FRA)
Chanway CP (1997) Inoculation of tree roots with plant growth promoting soil bacteria: an emerging Technology for Reforestation. For Sci 43:99-112
Chaves MM, Zarrouk O, Francisco R, et al. (2010) Grapevine under deficit irrigation: hints from physiological and molecular data. Ann Bot 105:661-676. doi:10.1093/aob/mcq030
Cherif H, Marasco R, Rolli E, et al. (2015) Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ Microbiol Rep 7:668-678. doi:10.1111/1758-2229.12304
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669-678. doi:10.1016/j.soilbio.2009.11.024
Compant S, Mitter B, Colli-Mull JG, et al. (2011) Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb Ecol 62:188-197. doi:10.1007/s00248-011-9883-y
Compant S, Muzammil S, Lebrihi A, Mathieu F (2013) Visualization of grapevine root colonization by the Saharan soil isolate Saccharothrix algeriensis NRRL B-24137 using DOPE-FISH microscopy. Plant Soil 370:583-591. doi:10.1007/s11104-013-1648-6
Daffonchio D, Hirt H, Berg G (2015) Principles of Plant-Microbe Interactions. Springer Int Publ Switz:265-276. doi:10.1007/978-3-319-08575-3
Dimkpa C, Weinand T, Asch F (2009) Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ 32:1682-1694. doi:10.1111/j.1365-3040.2009.02028.x
Döbereiner J (1980) Forage grasses and grain crops. In: Bergensen FJ (ed) Methods for evaluating biological nitrogen. Wiley, NY, pp 535–555
Drogue B, Doré H, Borland S, et al. (2012) Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res Microbiol 163:500-510. doi:10.1016/j.resmic.2012.08.006
Edwards EJ, Clingeleffer PR (2013) Interseasonal effects of regulated deficit irrigation on growth, yield, water use, berry composition and wine attributes of cabernet sauvignon grapevines. Aust J Grape Wine Res 19:261-276. doi:10.1111/ajgw.12027
FAOSTAT (n.d.) http://faostat.fao.org/.
Garcia-seco D, Zhang Y, Gutierrez-ma FJ, et al. (2015) Application of Pseudomonas fluorescens to blackberry under field conditions improves fruit quality by modifying flavonoid metabolism. PLoS One:1-23. doi:10.1371/journal.pone.0142639
Garima G, Nath JP (2015) Screening of potential PGPR candidates as future biofertilizers-a strategic approach from lab to field. Res J Biotechnol 10:48-62
Gilbert J a, der Lelie D v, Zarraonaindia I (2014) Microbial terroir for wine grapes. Proc Natl Acad Sci U S A 111:5-6. doi:10.1073/pnas.1320471110
Gómez-Lama Cabanás C, Schilirò E, Valverde-Corredor A, Mercado-Blanco J (2014) The biocontrol endophytic bacterium Pseudomonas fluorescens PICF7 induces systemic defense responses in aerial tissues upon colonization of olive roots. Front Microbiol 5:427. doi:10.3389/fmicb.2014.00427
Gontia-Mishra I, Sasidharan S, Tiwari S (2014) Recent developments in use of 1-aminocyclopropane-1-carboxylate (ACC) deaminase for conferring tolerance to biotic and abiotic stress. Biotechnol Lett 36:889-898. doi:10.1007/s10529-014-1458-9
Haney CH, Samuel BS, Bush J, Ausubel FM (2015) Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1:15051. doi:10.1038/nplants.2015.51
Jaos Frederico MDP, Costa PB, Costa MD, et al. (2014) Cultivable bacteria isolated from apple trees cultivated under different crop systems : diversity and antagonistic activity against Colletotrichum gloeosporioides. Genet Mol Biol 37:560-572. doi:10.1590/S1415-47572014000400013
Khan A, Khan A (2015) Phosphorus and compost management influence maize (Zea mays ) productivity under semiarid condition with and without phosphate solubilizing bacteria. Front Plant Sci 6:1-8. doi:10.3389/fpls.2015.01083
Kurepin LV, Park JM, Lazarovits G, Bernards M a (2014) Burkholderia phytofirmans-induced shoot and root growth promotion is associated with endogenous changes in plant growth hormone levels. Plant Growth Regul 75:199-207. doi:10.1007/s10725-014-9944-6
Lareen A, Burton F, Schäfer P, Scha P (2016) Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol:1-13. doi:10.1007/s11103-015-0417-8
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541-556. doi:10.1146/annurev.micro.62.081307.162918
Lundberg DS, Lebeis SL, Paredes SH, et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86-90. doi:10.1038/nature11237
Magnin-Robert M, Trotel-Aziz P, Quantinet D, et al. (2007) Biological control of Botrytis Cinerea by selected grapevine-associated bacteria and stimulation of chitinase and ??-1,3 glucanase activities under field conditions. Eur J Plant Pathol 118:43-57. doi:10.1007/s10658-007-9111-2
Magnin-Robert M, Quantinet D, Couderchet M, et al. (2013) Differential induction of grapevine resistance and defense reactions against Botrytis cinerea by bacterial mixtures in vineyards. BioControl 58:117-131. doi:10.1007/s10526-012-9474-y
Maldonado-Gonzalez MM, Bakker P a HM, Prieto P, Mercado-Blanco J (2015) Arabidopsis thaliana as a tool to identify traits involved in Verticillium dahliae biocontrol by the olive root endophyte Pseudomonas fluorescens PICF7. Front Microbiol 6:1-12. doi:10.3389/fmicb.2015.00266
Marasco R, Rolli E, Ettoumi B, et al. (2012) A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7:e48479. doi:10.1371/journal.pone.0048479
Marasco R, Rolli E, Fusi M, et al. (2013) Plant growth promotion potential is equally represented in diverse grapevine root-associated bacterial communities from different biopedoclimatic environments. Biomed Res Int 2013:491091. doi:10.1155/2013/491091
Milagres a MF, Machuca A, Napoleão D (1999) Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J Microbiol Methods 37:1-6. doi:10.1016/S0167-7012(99)00028-7
Muyzer G Uitterlinden AG, de Waal EC, Uitterlinden AG DWEC (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695-700. doi:0099-2240/93/030695-06$02.00/0
Naveed M, Mitter B, Reichenauer TG, et al. (2014) Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ Exp Bot 97:30-39. doi:10.1016/j.envexpbot.2013.09.014
Nelissen H, Moloney M, Inzé D (2014) Translational research: from pot to plot. Plant Biotechnol J 12:277-285. doi:10.1111/pbi.12176
Nielsen P, Sørensen J (1997) Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere.
Panke-buisse K, Poole AC, Goodrich JK, et al. (2015) Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J 9:980-989. doi:10.1038/ismej.2014.196
Peiffer J a, Spor A, Koren O, et al. (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548-6553. doi:10.1073/pnas.1302837110
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10-15. doi:10.1034/j.1399-3054.2003.00086.x
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789-799. doi:10.1038/nrmicro3109
Poupin MJ, Timmermann T, Vega A, et al. (2013) Effects of the plant growth-promoting bacterium Burkholderia phytofirmans PsJN throughout the life cycle of Arabidopsis thaliana. PLoS One 8:22-24. doi:10.1371/journal.pone.0069435
Rodriguez RJ, Henson J, Van Volkenburgh E, et al. (2008) Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404-416. doi:10.1038/ismej.2007.106
Rojas-Tapias D, Moreno-Galvan A, Pardo-Diaz S, et al. (2012) Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl Soil Ecol 61:264-272. doi:10.1016/j.apsoil.2012.01.006
Rolli E, Marasco R, Vigani G, et al. (2015) Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17:316-331. doi:10.1111/1462-2920.12439
Sabir A, Yazici MA, Kara Z, Sahin F (2012) Growth and mineral acquisition response of grapevine rootstocks (Vitis spp.) to inoculation with different strains of plant growth-promoting rhizobacteria (PGPR). J Sci Food Agric 92:2148-2153. doi:10.1002/jsfa.5600
Sahin U, Ekinci M, Kiziloglu FM, et al. (2015) Ameliorative effects of plant growth promoting bacteria on water-yield relationships, growth, and nutrient uptake of lettuce plants under different irrigation levels. Hortscience 50:1379-1386
Salomon MV, Bottini R, de Souza Filho GA, et al. (2014) Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol Plant 151:359-374. doi:10.1111/ppl.12117
Santaella C, Schue M, Berge O, et al. (2008) The exopolysaccharide of rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ Microbiol 10:2150-2163. doi:10.1111/j.1462-2920.2008.01650.x
Schlaeppi K, Bulgarelli D (2015) The plant microbiome at work. Mol Plant-Microbe Interact 28:212-217
Shahid M, Hameed S, Tariq M (2015) Characterization of mineral phosphate-solubilizing bacteria for enhanced sunflower growth and yield-attributing traits. Ann Microbiol:1525-1536. doi:10.1007/s13213-014-0991-z
Shishido M (1996) Effect of plant growth PromotingBacillusStrains on pine and spruce seedling growth and Mycorrhizal infection. Ann Bot 77:433-442. doi:10.1006/anbo.1996.0053
Suarez C, Cardinale M, Ratering S, et al. (2015) Plant growth-promoting effects of Hartmannibacter diazotrophicus on summer barley ( Hordeum vulgare L .) under salt stress. Appl Soil Ecol 95:23-30. doi:10.1016/j.apsoil.2015.04.017
Tuberosa R (2012) Phenotyping for drought tolerance of crops in the genomics era. Front Physiol 3:1-26. doi:10.3389/fphys.2012.00347
Vercesi A (2010) Linee guida per una corretta gestione dei versanti a vigneto nell’Oltrepò Pavese. Edizioni TCP, Pavia
Zamioudis C, Mastranesti P, Dhonukshe P, et al. (2013) Unraveling root developmental programs initiated by beneficial pseudomonas spp. bacteria. Plant Physiol 162:304-318. doi:10.1104/pp.112.212597
Zarraonaindia I, Owens SM, Weisenhorn P, et al. (2015) The soil microbiome influences grapevine-associated microbiota. MBio 6:1-10. doi:10.1128/mBio.02527-14.Editor
Acknowledgments
This research was supported by the EU project BIODESERT (European Community’s Seventh Framework Programme CSA-SA REGPOT-2008-2 under grant agreement no. 245746) and King Abdullah University of Science and Technology (KAUST). ER and FM acknowledge support by Università degli Studi di Milano, DeFENS, the European Social Fund (FSE) and Regione Lombardia (contract ‘Dote Ricerca’). The authors would like to thank the “Arnaldo Caprai” and “Castello Bonomi” farms for their kind availability and interest in performing the field trials in their vineyards. The authors would like to thank Usama A. El-Behairy and Ayman F. Abou-Hadid (Ain Shams University, Cairo, Egypt) and Roberto Gerbino (Fondazione Bussolera Branca, Casteggio, Italy) for support in grapevine sampling, Umberto Fascio (CIMA, Centro Interdipartimentale di Microscopia Avanzata, University of Milan, Milan, Italy) for technical support with the confocal microscope and the “Vitis Rauscedo” grapevine nursery for kindly providing the grapevine plantlets. The authors dedicate this work to Nicola Rubaga.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Stéphane Compant.
Eleonora Rolli and Ramona Marasco contributed equally to the work
Electronic supplementary material
ESM 1
(DOC 325 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rolli, E., Marasco, R., Saderi, S. et al. Root-associated bacteria promote grapevine growth: from the laboratory to the field. Plant Soil 410, 369–382 (2017). https://doi.org/10.1007/s11104-016-3019-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-3019-6