Abstract
Background and aims
In the symbiotic bacterium Rhizobium leguminosarum bv. trifolii, a majority of proteins involved in exopolysaccharide (EPS) synthesis are encoded by genes located in a large polysaccharide synthesis cluster (Pss). The aim of this study was genetic characterization of the Pss region in the Rt24.2 strain in the context of EPS production and symbiosis with red clover (Trifolium pratense).
Methods
The expression of genes located in the Pss cluster was determined using constructed pss-lacZ transcriptional fusions. The role of transcriptional regulator RosR in pss transcription was confirmed using a rosR mutant and the Rt24.2(pBR1) strain carrying multiple rosR copies. An EPS-deficient mutant, Rt770 was obtained using a random mutagenesis and mTn5SSgusA40 transposon. Symbiotic properties of the Rt770 strain in interaction with clover were characterized in inoculation experiments. Infection of host roots and nodule occupancy by this mutant were investigated using both light and electron microscopy.
Results
Transcriptional levels of particular pss genes differed significantly; the genes encoding glycosyltransferases and enzymes modifying EPS have promoters of weak activities, whereas those encoding proteins involved in EPS polymerization and export possess stronger promoters. Furthermore, RosR affected expression of some pss genes. A mutation in Rt24.2 pssS encoding glucosyltransferase totally abolished EPS synthesis, decreased motility, and increased sensitivity to some stressors. The pssS mutant Rt770 induced formation of nodules on clover roots, which were ineffective in nitrogen fixation.
Conclusion
EPS secreted by Rt24.2 is required for both adaptation to soil conditions and the establishment of effective symbiosis with clover plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extracellular polysaccharides (EPS) are produced by a wide range of microorganisms. Various functions are ascribed to these polymers, among them protection against stress factors and plant antimicrobial compounds, nutrient gathering, attachment to both abiotic and biotic surfaces, and biofilm formation, which ensures adaptation of bacterial cells to changing environmental conditions (Fujishige et al. 2006; Flemming and Wingender 2010; Jaszek et al. 2014; Janczarek et al. 2015a). In the case of soil nitrogen-fixing bacteria, which are able to establish symbiotic interactions with legumes, EPS is also indispensable for effective infection and nodulation of many host plants (e.g., Medicago, Trifolium, Pisum, Vicia spp.) (Downie 2010; Janczarek et al. 2015b). Strains defective in EPS production from several rhizobial species, such as S. meliloti, and Rhizobium leguminosarum bvs. trifolii and viciae, induce only small, partially infected or even empty nodule-like structures on their host plants that are ineffective in nitrogen fixation (Cheng and Walker 1998; van Workum et al. 1998; Rolfe et al. 1996; Janczarek and Rachwał 2013).
R. leguminosarum bv. trifolii induces formation of nodules on clover roots (Trifolium pratense). The structure of EPS synthesized by this bacterium has been established in detail. It is a heteropolymer composed of octasaccharide repeating units consisting of a backbone of two glucose and two glucuronic acid residues, and a side chain of three glucose and one galactose residues. These subunits are additionally modified by non-sugar substituents (one O-acetyl and two pyruvyl groups) (Robertsen et al. 1981; McNeil et al. 1986; Cremers et al. 1991; O’Neill et al. 1991; Breedveld et al. 1993). However, the biosynthesis pathway of this polysaccharide in R. leguminosarum is known only fragmentarily. So far, functions of enzymes engaged in the four steps of the EPS unit assembly have been experimentally confirmed. The initiation of EPS biosynthesis is conducted by a glucosyl-IP-transferase, which transfers glucose-1-phosphate from UDP-glucose to a C55-isoprenylphosphate (IP) carrier anchored at the cytoplasmic site of the inner membrane (Pollock et al. 1998). This enzyme is encoded by the pssA gene, which is located in the chromosome at a long distance from other EPS synthesis genes and transcribed as a monocistronic mRNA (Borthakur et al. 1988; Ivashina et al. 1994; van Workum et al. 1997). Glucuronosyl-(β1-4)-glucosyl transferase PssDE and glucuronosyl-(β1-4)-glucuronosyl transferase PssC are involved in the second and the third step of the unit synthesis, respectively (Pollock et al. 1998). These proteins are encoded by the pssC, pssD, and pssE genes located in a cluster named Pss-I (Król et al. 2007). Mutations in the pssA and pssD genes totally abolished EPS synthesis (Borthakur et al. 1988; Ivashina et al. 1994; Janczarek and Urbanik-Sypniewska 2013), whereas a disruption of the pssC gene resulted in a decrease in EPS synthesis to 40 % of the amount produced by the wild-type strain (van Workum et al. 1997). pssJ encodes a galactosyltransferase, which probably participates in the last step of the unit assembly (Breedveld et al. 1993). A strain exo344::Tn5 carrying a mutation in the pssJ gene produced only residual amounts of EPS, whose repeating units did not contain terminal galactose.
However, genes involved in the remaining steps of the unit synthesis have not yet been identified, except pssM, which is located in this cluster. PssM is a ketal pyruvate transferase responsible for one of the non-sugar modifications of EPS (pyruvylation of the subterminal glucose in the repeating unit) (Ivashina et al. 2010). A mutation in pssM resulted in failed symbiosis with the compatible host plant. Moreover, some genes involved in EPS polymerization and export were characterized (pssTNOP, pssL and pssP2), among them a majority are located in the polysaccharide synthesis region (Mazur et al. 2005; Marczak et al. 2013, 2014).
The amount of produced EPS and its biochemical properties seem to be very important for functions of this polysaccharide in both adaptation to soil conditions and symbiosis. However, the knowledge of EPS biosynthesis in R. leguminosarum and factors engaged in the regulation of this process is still limited. Up to now, functions of four proteins (PsiA, PsrA, ExoR, and RosR) have been confirmed. psrA and psiA, which are located on symbiotic megaplasmids of R. leguminosarum bv. phaseoli strains, were discovered as the first regulatory genes (Borthakur and Johnston 1987; Borthakur et al. 1988). Although a mutation in psiA did not affect the amounts of produced EPS, multiple copies of this gene prevented EPS synthesis. This effect was overcome in the presence of additional copies of psrA or pssA, indicating that balanced copy numbers of these genes are needed for a proper level of EPS production (Borthakur and Johnston 1987; Latchford et al. 1991; Mimmack et al. 1994). Another gene, exoR also negatively influenced EPS synthesis in R. leguminosarum, since an exoR mutant produced significantly more EPS than the wild-type strain (Reeve et al. 1997).
In contrast, a protein encoded by rosR positively affected EPS synthesis in R. leguminosarum. A mutation in this gene resulted in a 3-fold decrease, whereas multiple rosR copies increased the synthesis of this polymer nearly 2-fold (Janczarek and Skorupska 2007; Janczarek et al. 2009). RosR belongs to the family of Ros/MucR transcriptional regulators, which are involved in regulation of EPS synthesis in several rhizobial species (S. meliloti, Rhizobium etli, Agrobacterium tumefaciens, Agrobacterium radiobacter) (D’Souza-Ault et al. 1993; Keller et al. 1995; Bittinger et al. 1997; Chou et al. 1998). This protein contains a Cys2His2-type zinc-finger motif responsible for binding to a 22-bp-long sequence called the RosR-box. Previously, it was confirmed that RosR affected transcription of pssA and its own gene (Janczarek and Skorupska 2007; Janczarek and Urbanik-Sypniewska 2013).
Up to now, no comprehensive analysis of transcription of genes involved in R. leguminosarum EPS synthesis has been performed so far. In this work, genetic organization of the polysaccharide synthesis cluster (Pss) of the Rt24.2 strain was established and a detailed transcriptional analysis of the pss genes located in it was performed. In addition, a mutant strain in one of the pss genes of this cluster (pssS) was obtained and characterized in relation to EPS production and symbiosis with clover.
Materials and methods
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1, whereas oligonucleotide primers are mentioned in Supplementary Material (Online Resource 1). E. coli strains were routinely grown in Luria-Bertani (LB) medium at 37 °C (Sambrook et al. 1989). R. leguminosarum strains were cultured in 79CA medium with 1 % glycerol as a carbon source (Kowalczuk and Lorkiewicz 1979) and in M1 minimal medium (Sambrook et al. 1989) containing additionally 2 ml l−1 vitamin stock solution (Brown and Dilworth 1995) at 28 °C. When required, antibiotics were used at the following final concentrations: rifampicin, 40 μg ml−1; kanamycin, 40 μg ml−1; tetracycline, 10 μg ml−1; ampicillin, 100 μg ml−1; and spectinomycin, 40 μg ml−1.
DNA methods and sequence analysis
Standard techniques were used for genomic DNA and plasmid isolation, restriction enzyme digestion, cloning, transformation, hybridization, and sequencing (Sambrook et al. 1989). For PCR reaction, REDTaq Ready PCR Reaction Mix (Sigma) was used. In order to obtain the nucleotide sequence of the Pss cluster, genomic DNA of Rt24.2 and primer walking techniques were used. The resulting sequence of 28 613 bp was deposited in GenBank under the accession no. KP067320. PCR products and plasmid constructs were sequenced using the BigDye terminator cycle sequencing kit (Applied Biosystems) and the ABI Prism 310 sequencer. Database searches were done with the FASTA and BLAST programs available from the National Center for Biotechnology Information (Bethesda, MD, USA) and the European Bioinformatic Institute (Hinxton, UK). Promoter prediction in the pss regulatory regions was done using the BDGP Neural Network Promoter Prediction (http://www.fruitfly.org). The probability of occurrence of promoter sequences was determined by the use of Promoter prediction (P) score, which can have values from 0 to 1 (a value close to 1 means very high probability, whereas that below 0.7 suggests a lack of promoter sequences). Subsequently, the searches for identification of motifs in the pss upstream regions, for which P values were higher than 0.7, were performed with Malign and Fuzznuc programs using S. meliloti CTTGAC-N17-18-CTATAT and E. coli TTGACA-N17-18-TATAAT promoter consensuses (MacLellan et al. 2006) as query sequences (http://www.genebee.msu.su/services/malign, http://emboss.ch.embnet.org/Pise). RosR-box consensus TGAAATCTAGGGGTAGATTTCA was used as a query sequence for searching of RosR motifs in the pss upstream regions (http://www.genebee.msu.su/services/malign). Amino acid sequence analyses were performed using the BLASTP program (http://blast.ncbi.nlm.nih.gov/) and the CAZy database ( www.cazy.org/).
Construction of plasmids bearing transcriptional pss-lacZ fusions
To construct plasmids containing regulatory regions of the pss genes from the Rt24.2 Pss cluster, a broad-host-range plasmid pMP220 carrying a promoterless lacZ gene was used. Based on the sequence of this region, primers complementary to promoters of the pss genes were designed and used in PCR reactions with the genomic DNA of this strain (Supplementary Material, Online Resource 1). A set of promoter fragments was generated by PCR amplification using primer pairs: mgl2-27 and mgl2-28 primers for the pMgl16 plasmid fusion, pssV-69 and pssV-70 for pV90, pssW-29 and pssW-30 for pW74, pssM-33 and pssM-34 for pM65, pssL-101 and pssL-102 for pL52, pssK-31 and pssK-32 for pK48, pssI-35 and pssI-36 for pI90, pssF-37 and pssF-38 for pH65, pssC-39 and pssC-40 for pC55, pssD-41 and pssD-79 for pD35, pssP-42 and pssP-89 for pP85, pssO-43 and pssO-44 for pO66, pssN-45 and pssN-46 for pN76, pssT-47 and pssT-48 for pT80, and pssR-Eco1 and pssR-Pst1 for pR64. The PCR products obtained were digested with appropriate restriction enzymes, cloned into the corresponding sites in the pUC19 vector, and verified by sequencing. Then, plasmid-derived fragments were re-cloned between the respective sites of the pMP220 vector, yielding plasmids with the pss upstream regions fused to the promoterless lacZ gene. These plasmids were introduced into E. coli S17-1 by transformation and subsequently into Rt24.2 derivatives by biparental conjugation (Sambrook et al. 1989).
β-Galactosidase assay
R. leguminosarum derivatives harboring the pss-lacZ fusions were grown for 24 h in 79CA medium supplemented with tetracycline and, where necessary, kanamycin. The assay of β-galactosidase activity was carried out according to the protocol described by Miller (Miller 1972). The reported values are given in Miller units and are averages of at least five independent experiments.
Mutagenesis of the pssS gene
In order to obtain Rt24.2 derivatives defective in EPS synthesis, random mutagenesis of this strain was performed using the S17-1 strain containing the mTn5SSgusA40 transposon with a promoterless gusA gene (Wilson et al. 1995). As a result of the biparental conjugation experiments, in total 2000 strains carrying mutations were obtained, among them five clones had a non-mucoid phenotype. The insertion site of the mTn5SSgusA40 transposon in the genome of the Rt24.2 derivatives was established by restriction, hybridization, and sequencing analyses. Among these, only the one mutant strain, named Rt770 proved to have the transposon insertion located within the Pss region, in the pssS gene.
Construction of plasmids for complementation of a pssS mutation
To construct a set of plasmids containing different fragments of the pssWSR region, the pBBR1MCS-2 vector and amplicons obtained in PCR reactions with primers complementary to this region were used (Supplementary Material, Online Resource 1). Primers W-FW3 and W-RW5 were used for construction of a pW5 plasmid, primers W-FW3 and S-RW4 for construction of pWS4, and primers W-FW3 and R-RW3 for construction of pWSR2. The PCR products obtained were digested with BamHI and SacI enzymes, cloned into the corresponding sites in the pBBR1MCS-2 vector, and inserts of the resulting plasmids were verified by sequencing. Then, the pW5, pWS4, and pWSR2 plasmids were introduced into E. coli S17-1 by transformation and subsequently into Rt770 by biparental conjugation.
EPS isolation and quantification
For EPS isolation, 10-ml cultures of rhizobial strains were grown in 79CA medium supplemented with 1 % glycerol for 2 days at 28 °C. Then, the cultures were centrifuged for 20 min at 14,000 rpm. EPS was precipitated from obtained supernatants with 4 vol. of 96 % ethanol, collected by centrifugation, dissolved in 2 ml of water and analyzed for carbohydrates (Loewus 1952). The total sugar content was calculated as glucose equivalents.
Motility assay
The motility of the rhizobial strains was established using 0.3 % 79CA medium. 5-μl aliquots of bacterial suspensions (OD600 = 0.4) of these strains prepared in sterile water were stabbed into agar. Then, the plates were incubated at 28 °C for 48 h, and the migration distance of bacteria from the injection site was measured. The assay was repeated three times with three repetition for each strain analyzed.
Sensitivity assay to stress factors
In order to determine the sensitivity of the Rt770, Rt24.2 and Rt770(pWS4) strains to several stress factors (sodium dodecyl sulfate, sodium deoxycholate, and ethanol), the minimal inhibitory concentration of the individual stressor was determined. In this purpose, bacterial suspensions of these strains of OD600 = 0.2 were prepared into sterile water and 10-μl aliquots were placed on the plates with 79CA medium and defined concentrations of the tested compound (SDS - concentrations from 0.05 to 1 % w/v, DOC – from 0.05 to 1 % w/v, ethanol – from 0.05 to 5 % v/v). The bacterial growth on the individual media was determined after 48-h incubation at 28 °C.
Plant experiments
Seeds of red clover (Trifolium pratense cv. Diana) were surface sterilized as described previously (Jaszek et al. 2014). Next, the seeds were placed on plates with Fåhraeus agar (Vincent 1970), incubated for 2–3 days at 22 °C, and seedlings were subsequently placed onto Fåhraeus slants. After 4 days, the seedlings were inoculated with bacterial suspensions with OD600 = 0.2 (100-μl aliquot per plant). The plants were grown for 28 days under natural light supplemented with artificial light (14 h at 24 °C and 10 h at 18 °C) in a greenhouse. Nodules appearing on the roots were counted after each 7 days. 4-week plants were harvested, and their wet shoot masses were estimated. The experiment was repeated three times using 20 plants for each treatment.
Nodule analysis using light and electron microscopy
To establish nodule colonization by the Rt770 mutant, the enzymatic activity of β-glucuronidase encoded by gusA in the mTn5SSgusA40 transposon was used. In addition, Rt24.2 tagged with the pJBA21Tc plasmid carrying gusA was used as a control (Janczarek and Rachwał 2013). Clover seedlings were inoculated with these strains and grown up to 3 weeks. Next, the nodules were stained using 50 mM sodium phosphate buffer (pH 7.2) containing 50 μg ml−1 of 5-bromo-4-chloro-3-indolyl-β-D-glucuronide as described previously (Janczarek and Rachwał 2013) and analyzed under a Nikon light microscope (OPTIPHOT2). In order to characterize in detail the structures of the nodules induced by the Rt24.2 strain and the Rt770 mutant, plant material was prepared for the electron microscopy analysis as described earlier (Król et al. 1998).
Results
The sequence analysis of the Pss region in R. leguminosarum bv. trifolii 24.2
Rhizobia secrete large amounts of EPS into the environment (D’Haeze et al. 2004; Quelas et al. 2006; Janczarek et al. 2015a). Although some data concerning genetic characteristics of a few genes involved in EPS synthesis in R. leguminosarum have been published previously, no comprehensive studies on the transcriptional activity of genes grouped in the EPS synthesis region have been performed so far. Therefore, we decided to establish the genetic organization of this cluster in R. leguminosarum bv. trifolii strain 24.2 and transcriptional activity of the individual genes present in it. At first, sequencing of the Rt24.2 Pss cluster was performed and the 28.613-kb sequence obtained was deposited in the GenBank database under Accession No. KP067320. Based on the nucleotide sequence analysis, 23 open reading frames (ORFs) were identified in this region, which showed significant identity to genes from other rhizobial species involved in biosynthesis, export, and modification of extracellular polysaccharides. Comparative sequence analysis of this cluster with nucleotide sequences of R. leguminosarum bvs. trifolii TA1 and viciae 3841 strains as well as closely related species Rhizobium etli CFN42 indicated that this DNA region showed high synteny (Fig. 1). The pss genes of the Rt24.2 cluster showed the same genetic organization as that found in the RtTA1, Rv3841, and ReCFN42 bacteria. Only its fragment encompassing the plyA, rapA, and orf1 genes proved to be more divergent, and this difference was especially visible in the case of the RtTA1 strain. The entire Rt24.2 Pss cluster showed the highest nucleotide sequence identity to the homologous region of RtTA1 (95 % identity), and a slightly lower level to the sequences of Rv3841 (91 %) and ReCFN42 (83 %) strains, confirming that these genetic regions are highly conserved among these rhizobial strains. Depending on putative functions in EPS biosynthesis, the pss genes of the Rt24.2 Pss cluster were divided into 3 groups. Genes involved in the subunit synthesis were classified into the first group: pssJ encoding galactosyltransferase, pssS, pssI, and pssF coding for glycosyltrasferases, and pssC, pssD, and pssE genes coding for glucuronosyltransferases. The second group included genes involved in addition of non-sugar residues to the EPS subunits: pssR for acetyltrasferase and two pssM and pssK genes for ketal pyruvate transferases. The third group encompassed the genes involved in EPS polymerization and export: pssP encoding an EPS polymerization protein, pssO encoding an outer membrane protein, and the pssN, pssT and pssL genes encoding Wza-type, Wzy-type and Wzx-type polysaccharide export proteins, respectively. Moreover, other genes not directly involved in EPS biosynthesis were identified in the Rt24.2 Pss cluster. These were: pssW encoding a glycosyl hydrolase probably engaged in EPS processing, the prsD and prsE genes encoding components of the type-I protein secretion system and three genes, named plyA, rapA, and orf3, which encode proteins that are substrates for this transport system.
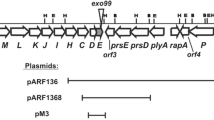
Genetic organization of exopolysaccharide synthesis clusters in different rhizobia. a Physical and genetic map of the Pss region of R. leguminosarum bv. trifolii 24.2. Arrows below the map indicate the direction of transcription. Promoter regions of the pss genes cloned into the pMP220 vector and present in the particular pss-lacZ transcriptional fusions are marked as rectangles: black – DNA regions characterized by very high probability of the promoter occurrence (P values) higher than 0.9, grey - regions with the P values higher than 0.8, white - regions with the P values higher than 0.7 (http://www.fruitfly.org). Selected restriction sites are marked: E- EcoRI, H- HindIII, B- BamHI. Location of the mTn5SSgusA40 insertion in the Pss region of R. leguminosarum bv. trifolii mutant Rt770 is marked by dark grey vertical arrow. b Genetic map of the Pss-I region of R. leguminosarum bv. trifolii TA1, c Genetic map of the R. leguminosarum bv. viciae 3841 EPS synthesis cluster, d Genetic map of the R. etli CFN42 EPS synthesis cluster
In conclusion, the Rt24.2 Pss cluster shows very similar genetic organization with those of other R. leguminosarum strains, although some minor differences were found.
Transcriptional activity of the pss genes located in the Rt24.2 Pss region
In order to identify putative promoters of the pss genes present in the Rt24.2 Pss cluster, in silico sequence analysis of upstream regions of these genes was performed using both S. meliloti (5′-CTTGAC-N17-18-CTATAT-3′) and canonical E. coli (5′-TTGACA-N17-TATAAT-3′) promoter consensus motifs recognized by RNA polymerase with ϭ70. It was established that a great majority of the analyzed genes possessed motifs in their regulatory regions of a high probability (P) of occurrence of promoter sequences (P score higher than 0.9 for mgl2, pssV, pssW, pssR, pssM, pssI, pssF, pssP, pssO, pssN, pssD and pssT). The values between 0.8 and 0.7 for pssL, pssK, pssD, and pssC suggested that these genes had motifs of the significantly lower probability of presence of promoter sequences. In the case of the upstream regions of the pssS, pssJ, and pssE genes, no motifs of the predicted promoter activity were identified. Next, the regulatory regions of these genes, for which the determined P scores were higher than 0.7, were amplified in PCR reactions and cloned upstream of a promoterless lacZ gene into the pMP220 vector. As a result, a set of 15 pss-lacZ transcriptional fusions containing the promoter regions of the different genes from the Rt24.2 Pss cluster were constructed. These plasmids were introduced into the wild-type Rt24.2 strain and β-galactosidase activity was assayed (Table 2).
In general, for almost all of the regulatory pss regions chosen for cloning, the presence of functional promoters in their sequences was confirmed. Moreover, high diversity concerning the levels of the promoter activity was observed among these genes. The highest transcription was observed for pssN, pssO, pssR, mgl2, and pssT, indicating that the upstream regions of these genes contain strong promoters (Table 2). Lower values of β-galactosidase activities were found for the pssI-lacZ, pssP-lacZ, pssV-lacZ, pssF-lacZ, and pssW-lacZ fusions (a range from 1106 to 661 Miller units). The lowest transcriptional activity was detected for the pssM, pssC, pssK, and pssD genes (from 325 to 157 Miller units), confirming the presence of weak promoters in their sequences. In the case of the pssL-lacZ fusion, the level of β-galactosidase activity was nearly the same as that determined for the control plasmid pMP220, indicating that no promoter sequence was present in the upstream region of this gene. Based on all these results, it was confirmed that the upstream regions of the pssV, pssW, pssR, pssM, pssK, pssI, pssF, pssC, pssD, pssT, pssN, pssO, pssP, and mgl2 genes contain functional promoters, although of different strengths of action. In addition, we conclude that the pssV, pssI, pssF, pssC, pssR, pssT, pssN, pssO, pssP, and mgl2 genes constitute single transcription units, whereas the transcription of other pss genes is linked (pssWS, pssML, pssKJ, and pssDE operons).
Promoter sequences of the pss genes
Up to now, the ability to recognize and predict non-ϭ54 promoters in alphaproteobacteria has not been well developed. Among rhizobial species, the data concerning this issue were established in detail only for S. meliloti, in which consensus 5′-CTTGAC-N17-18-CTATAT-3′ based on 25 experimentally verified promoter sequences was established (MacLellan et al. 2006). The structure of this consensus shares significant similarity with the E. coli ϭ70 consensus (5′-TTGACA-N17-18-TATAAT-3′), but is skewed by 1 nucleotide to the left.
In this study, a comparative analysis of the regulatory sequences of the pss genes was performed using both E. coli and S. meliloti ϭ70 promoter consensuses. These data are presented in Table 3. In general, a majority of the promoter sequences of the R. leguminosarum pss genes shared significant similarity with the S. meliloti ϭ70 consensus (pssV, pssW, pssF, pssC, pssD, pssT, pssO, pssR, pssN, and pssP). Moreover, it was established that the −35 motifs of the pss promoters had higher identity than the −10 motifs with the corresponding hexamers of the S. meliloti consensus. To find reasons for the extremely high promoter activity of pssN and pssO, we additionally searched the upstream regions of these genes for identification of other regulatory motifs affecting the level of transcription such as an upstream promoter element (UP) and extended −10 element. It is well known that these −10 elements significantly enhance the promoter strength (Ross et al. 1998; Estrem et al. 1999). The 30-bp-long A/T-rich UP sequence is located upstream of the −35 hexamer, whereas the 3-bp TGN motif upstream of the −10 hexamer. The A/T-rich sequences were found upstream of the pssN −35 motif (5′-ACAATCAATCTTTTCCGAAAATCTTATTAAAATCCATCCATTTAAACTTTAA-3′) and the pssO −35 motif (5′-AACTTGACCAATTCAACAGAGTCCGAATTGGAAATATAGATAATATCCTTGTCTT-3′). This indicated the significant role of these UP elements in enhancing transcription of these rhizobial genes and explained the highest activity of these promoters among all the studied pss promoters.
The role of RosR in regulation of expression of the pss genes
Previously, it was established that RosR positively affected EPS production in R. leguminosarum (Janczarek and Skorupska 2007; Janczarek et al. 2009). Therefore, we decided to determine whether RosR regulates EPS synthesis via modulation of transcription of the pss genes present in the Pss cluster. In order to verify this hypothesis, the set of the pss-lacZ fusions was introduced into the Rt2472rosR mutant, the Rt24.2(pBR1) strain containing additional rosR copies, and the Rt24.2(pBK1) strain containing the empty pBBR1MCS-2 vector, and β-galactosidase activity was measured (Table 2). In a majority of the studied pss-lacZ fusions, similar levels of β-galactosidase activity were observed in the four strains used: Rt24.2, Rt2472, Rt24.2(pBR1) and Rt24.2(pBK1) (which corresponded to values of nearly 1.0 for both Rt2472/Rt24.2 and Rt24.2(pBR1)/Rt24.2(pBK1) ratios). This indicated that the RosR protein did not influence the transcription of these genes. However, significant differences in the transcription levels of the pssO, pssN, pssP, and pssF genes were detected, when the β-galactosidase activities for these individual pss-lacZ fusions were compared between Rt24.2 and Rt2472, and between Rt24.2(pBR1) and Rt24.2(pBK1) strains, respectively (Table 2). Among these genes, the highest Rt2472/Rt24.2 ratio was found for the pssF-lacZ fusion (2.09). For the remaining genes, slightly lower Rt2472/Rt24.2 ratios were established. On the other hand, the Rt24.2(pBR1)/Rt24.2(pBK1) ratios were determined for these fusions to be significantly lower than 1.0, confirming that multiple rosR copies in the Rt24.2(pBR1) strain affected expression of these genes more strongly than the one rosR copy in the wild-type strain. These data indicated that the RosR protein was engaged in regulation of transcription of the pssO, pssN, pssP, and pssF genes.
In previous studies, it was confirmed that RosR recognized a sequence motif, called the RosR-box, located in the upstream regions of the rosR and pssA genes and affected their transcription (Janczarek and Skorupska 2007; Janczarek and Urbanik-Sypniewska 2013). The RosR-box is a 22-bp-long sequence containing 9-bp inverted repeats separated by a 4-bp spacer (Fig. 2). In order to establish whether the upstream regions of the pssO, pssN, pssP, and pssF genes possess RosR-box motifs, the promoter sequences of these genes were analyzed. We found motifs with significant sequence similarity to the RosR consensus in the pss upstream regions, which were located downstream of the promoters and upstream of translation start sites (with the exception of pssN) (Fig. 2). Among these, the motif identified in the pssF upstream region showed the highest sequence identity to the RosR consensus. Also, motifs found in the regulatory regions of pssO, pssN, and pssP showed significant similarity to the consensus sequence, suggesting that RosR influenced transcription of these genes via the RosR-boxes.
Phenotype of the Rt770 mutant strain and complementation of a pssS mutation
In order to establish mutations in the pss genes which totally abolish EPS synthesis, random mutagenesis of the Rt24.2 strain was performed using a mTn5SSgusA40 transposon (Wilson et al. 1995). As a result, about 2000 derivatives were obtained, among them only five strains were unable to produce EPS. Based on the PCR reaction, hybridization, and sequencing analyses, it was established that only one strain, named Rt770, had transposon insertion inside the Pss cluster (in the pssS gene) (Fig. 1). In addition, the presence of the one copy of the mTn5SSgusA40 transposon in the Rt770 genome was confirmed using hybridization. This mutant formed characteristic small nonmucoid colonies on agar plates, which significantly differed from those formed by wild-type bacteria (data not shown). Quantitative analysis indicated that the Rt770 strain did not produce any amounts of EPS (0 ± 0 mg L−1 in comparison with the 1281 ± 157 mg L−1 produced by Rt24.2). Moreover, although the Rt770 strain grew nearly as effectively as the parental strain in both the energy-rich media (79CA and TY), it exhibited approx. 3-fold slower growth in the minimal M1 medium (OD600 = 0.27 ± 0.03 for Rt770 and OD600 = 0.84 ± 0.06 for Rt24.2 in the 72-h cultures). Furthermore, the Rt770 strain was characterized by significantly reduced motility and increased sensitivity to several stress factors (Table 4). This mutant was more sensitive to such compounds as SDS, DOC, and ethanol, indicating that a lack of EPS resulted in a decrease of adaptation ability of this bacterium to stress conditions.
The complementation analysis of the mutation in the Rt770 strain performed by us using pBBR1MCS-2 derivatives containing different fragments of the pssWSR region indicated that a mucoid phenotype (Exo+) of this mutant was recovered after introduction of pWSR2 and pWS4 plasmids, but not by the pW5 plasmid (Fig. 3). The pWS4 plasmid containing the pssWS genes without pssR complemented the mutation from the Exo− to Exo+ phenotype. These data confirm that pssS plays a significant role in EPS synthesis and that the mutation in this gene does not have a polar effect on downstream-located pssR. The pssS gene encodes a 383-aa long protein, which according to homology search data was referred to the glycosyltransferases from GTB-type superfamily, GT family 1 (EC 2.4.1.-, CAZy database), which form glycosidic bonds with stereochemistry identical to that of the glycosyl donor. The Rt770 strain harboring pWS4 produced a similar amount of EPS (1397 ± 172 mg L−1) as Rt24.2 (1281 ± 157 mg L−1), and showed similar motility and sensitivity to the tested stress factors as the wild-type strain (Table 4).
Symbiotic properties of the Rt770 strain
In addition, symbiotic properties of the Rt770 strain in interaction with clover plants were characterized. It was found that this mutant had dramatically reduced infection effectiveness. After 14 days post infection, only 5 % of the clover plants were infected by this bacterium, and less than half of the plants were infected even after 4 weeks (Table 5). Besides the significant delay in nodule formation, also the total number of nodules induced by the pssS mutant was essentially lower than that induced by the wild-type strain (4-fold for the 28-day-old plants). Moreover, the nodules elicited by the mutant were small, white, often with the atypical shape and unable to fix nitrogen. This was confirmed by the very low fresh shoot mass of the clover plants, which was 2-fold lower than the mass of the plants infected by the wild-type bacteria, and nearly the same as that of uninfected plants (Table 5).
In order to compare colonization of nodules by Rt770 and Rt24.2, bacteria of these strains tagged with gusA encoding β-glucuronidase were used. It was confirmed in this experiment that the wild-type strain colonized the nodules effectively and all zones with the exception of the meristem were occupied by the bacteria (Fig. 4a and b). In contrast, nodule colonization by the Rt770 strain was drastically decreased. A great majority of the nodules were not occupied by the mutant bacteria, which were visible only on the nodule surface (Fig. 4c). In sporadically formed nodules, the bacteria were found inside only single nodule cells (Fig. 4d).
Light microscopy of nodules induced on clover roots (Trifolium pratense) by the R. leguminosarum bv. trifolii wild-type strain 24.2 and the pssS mutant Rt770 harboring the gusA reporter gene for β-glucuronidase. (a, b) Rt24.2 wild-type nodules at 7 and 14 days post infection, respectively; bars = 0.15 and 0.4 mm (c, d) Rt770 nodules at 6 and 14 days post infection, respectively; bars = 0.25 and 0.6 mm. The nodules were stained for GUS activity
The nodules induced by the wild-type strain Rt24.2 on clover roots exhibited a typical structure with all zones, including a large nitrogen-fixation zone (Fig. 5a). Infection threads had normally formed a thread wall with a large amount of thread matrix (Fig. 5b), and mature infected plant cells contained properly differentiated bacteroids (Fig. 5c).
Wild-type clover root nodule. a Semithin section of a 3-week-old nodule induced by the R. leguminosarum bv. trifolii wild-type strain 24.2: OC - outer cortex; IC - inner cortex; VB - nodule vascular bundle; RS - root stela; M - meristem; IZ - infection zone; II/III - interzone; NF - nitrogen fixation zone; arrow heads - nodule endodermis; asterisks - infected cells; bar = 100 μm. b Ultrastructure of the infection thread: TW - thread wall; TM - thread matrix; BA – bacteria; A - amyloplast; bar = 1 μm. c The mature infected plant cell with properly differentiated bacteroids (B); bar = 5 μm
In contrast, 3-week-old nodules induced by the pssS mutant were surrounded by cortex built of large, loosely arranged cells and by an endodermis. Semi-thin sectioning of these nodules revealed that a considerable part of central nodule tissue contained uninfected parenchymatous cells with starch grains and only a few cells were infected. The infected cells often formed clusters separated from one another by uninfected parenchyma. The nodules contained the meristem and vascular bundle connected with the root stele (Fig. 6a). The walls of root epidermal cells, being in contact with the mutant bacteria, were thickened and stained intensely with Azur A and Methylene Blue. The root cortical cells penetrated by infection threads were noticeably enlarged and their walls were also thicker in comparison to other cortex cells (Fig. 6b). The mutant-induced infection threads were wide and often branched. The thread walls were thick, irregularly formed, with knobs and protrusions, which was a consequence of incorporation of additional wall material (Fig. 6c). Another striking morphological feature of these infection threads was lack of typical thread matrix, in which rhizobia are usually embedded in the wild-type nodules. The bacterial cells were tightly packed inside these infection threads and showed high variability in their shape and size. The process of plant cell infection was abnormal, being characterized by “explosion-like” mass endocytosis of the bacteria into the plant cell cytoplasm (Fig. 6d). The symbiosomes formed in these cells contained a single bacteroid, and those with more than one bacteroid were observed only occasionally (Fig. 6e). Microscopic analysis revealed that some bacteroids differentiated, while others degenerated precociously. The morphologically pronounced bacteroid differentiation was different from that observed in wild-type nodules. The bacteroids were larger than normal, abnormally swollen and often deformed, and underwent rapid senescence. Their homogenous cytoplasm became electron-dense, marbled or completely dark (black) (Fig. 6f). The bacteroids degenerated inside the peribacteroid membranes or, when the peribacteroid membranes were fused, were aggregated in lytic compartments (Fig. 6g).
Semithin section of a 3-week-old clover root nodule induced by the R. leguminosarum bv. trifolii mutant Rt770 (a). M - meristem; OC - outer cortex; RS - root stela; VB - nodule vascular bundle; white asterisks - infected cells; arrow head - nodule endodermis; bar = 100 μm. b) The cortical region of the nodule induced by the mutant Rt770: M - meristem; long arrow - infection thread; short arrows - thickened cell walls of root epidermis and cortical cells; bar = 100 μm. c) Ultrastructure of the infection thread: BA – bacteria; TW - thread wall; arrows - material deposited between osmiophilic layers of the thread wall; bar = 2 μm. d) “Explosion-like” mass endocytosis of bacteria into the plant cytoplasm: ID - infection droplet; IT - infection thread; N - nucleus; double arrow heads - young bacteroids; bar = 10 μm. e) Mature infected cell: S – starch granules; rosette - abnormally differentiated bacteroids; triangle - degenerated bacteroids; bar = 5 μm. f) The infected cell with adult degrading bacteroids (stars); N - nucleus; S – starch granules, bar = 5 μm. g) Lytic compartments with degrading bacteroids (stars); bar = 5 μm
In conclusion, all these data indicate that symbiosis of the Rt770 strain with clover was impaired in the early stages of symbiosis, i.e., the infection process. The mutant bacteria infected only sporadic nodule cells, and bacteroids inside these plant cells degenerated early, which in consequence made the nodules unable to fix nitrogen.
Discussion
Rhizobia belong to a very important group of soil bacteria which possess a unique ability of establishing nitrogen-fixing symbiosis with legumes (Broughton et al. 2003). This type of the plant-microbe interaction is essential for biosphere functioning, since it provides nutrients to plants, increases soil fertility and restores deranged/damaged ecosystem (Kennedy et al. 1997; Yanni et al. 1997; Qin et al. 2011; Abreu et al. 2012; Angus et al. 2013). The process of biological nitrogen fixation is carried out by microsymbionts in specialized structures formed on legume roots, called nodules. However, rhizobia must survive often a long period of time in the soil as free-living organisms until they meet their symbiotic partner. During this period, they are exposed to the action of several environmental factors (e.g., drought, pH, temperature, salinity, nutrient limitation, heavy metals, oxidative stress) (Frey and Blum 1994; Rӓsӓnen et al. 2004; Soussou et al. 2013; Talbi et al. 2013; Suzuki et al. 2014; Cazenave et al. 2014; Pini et al. 2014; Burchill et al. 2014; Barthelemy-Delaux et al. 2014; Jaszek et al. 2014). Extracellular polysaccharide secreted in large amounts by rhizobial cells plays a significant protective role in their adaptation to stress conditions. Several environmental factors such as the type of carbon source, nitrogen and phosphorus availability, salinity, pH, and temperature, affect the level of EPS produced by rhizobia (Quelas et al. 2006; Janczarek and Skorupska 2009, 2011; Huang et al. 2012; Janczarek et al. 2015a). The synthesis of EPS is a multi-step process, which involves a coordinated action of many enzymatic proteins. In R. leguminosarum, a great majority of genes encoding these proteins are located in a large chromosomal cluster designated Pss (Polysaccharide synthesis). In this study, the Pss region of R. leguminosarum bv. trifolii strain 24.2 was sequenced and genetically characterized. It displayed high synteny with the homologous regions of other strains from R. leguminosarum species (Rv3841, RtTA1) and closely related bacterium R. etli CFN42 (Fig. 1) (Young et al. 2006; Król et al. 2007; González et al. 2006). In general, the organization and the genetic content of these regions are very similar, and the sequence identity is very high. The detailed sequence analysis of the Rt24.2 Pss cluster resulted in identification of promoters located upstream of the pss genes (Table 3), which indicated significant similarity with the S. meliloti or canonical E. coli ϭ70 promoters (MacLellan et al. 2006; Schlüter et al. 2013). Based on levels of the transcriptional activity of the pss-lacZ fusions, we conclude that the pss genes encoding enzymatic proteins involved in the synthesis and modification of EPS are transcribed at low levels (e.g., pssDE, pssWS, pssC, and pssKJ), whereas those encoding proteins engaged in polymerization and export of EPS have stronger promoters (pssN, pssO, pssT, and pssP). This was especially visible for the pssN and pssO genes, whose upstream regions contained the additional regulatory element UP (Estrem et al. 1999; Browning and Busby 2004). Up to now, UP elements increasing promoter activity were described in only a small number of bacterial promoters, among them the rosR promoter (Aiyar et al. 1998; Ross et al. 1998; Janczarek and Skorupska 2009).
Our previous studies indicated that RosR plays an essential role in positive regulation of EPS synthesis in R. leguminosarum, since a rosR mutation significantly decreases the production of this polysaccharide. RosR is a Rhizobium-specific transcriptional regulator, which binds to a sequence motif, called the RosR-box located in the upstream regions of the regulated genes. Homologs of the Rt24.2 RosR were found in other closely related bacteria such as R. etli (Bittinger et al. 1997), A. radiobacter (D’Souza-Ault et al. 1993), A. tumefaciens (Chou et al. 1998; Malgieri et al. 2007), and S. meliloti (Rüberg et al. 1999). Therefore, we decided to establish whether expression of some pss genes located in the Rt24.2 Pss cluster is regulated by RosR. For this purpose, transcriptional activities of the individual pss-lacZ fusions were compared between the wild-type and the Rt2472rosR mutant, and between Rt24.2(pBR1) containing multiple rosR copies and Rt24.2(pBK1) containing the empty vector pBBR1MCS-2, respectively. Statistically significant differences in expression levels were found for the pssF, pssP, pssO, and pssN genes (Rt2472/Rt24.2 and Rt24.2(pBR1)/Rt24.2(pBK1) ratios) (Table 2). In the upstream regions of these genes, motifs resembling the RosR-box were found, suggesting that these genes belong to the RosR regulon (Fig. 2). Similarly in S. meliloti, MucR, which is a RosR homolog, affected the synthesis of both exopolysaccharides: succinoglycan and galactoglucan via repression of some genes (wgaA, wgdA, wgeA, wggR, exoH, and exoX) and activation of other genes (exoYFQ and exoK) (Bertram-Drogatz et al. 1998; Quester and Becker 2004). Also in A. tumefaciens, the RosR homolog (Ros) negatively regulated expression of virC and virD genes, which positively affected EPS production in this bacterium (Chou et al. 1998).
The production of EPS is very important not only for free-living rhizobia, but also for establishment of effective symbiosis with many legumes (e.g., clover, alfalfa, vetch, and peas) (Ivashina et al. 1994; Cheng and Walker 1998; Janczarek and Rachwał 2013). In this study, we have characterized a strain Rt770 having a mutation in the pssS gene, which encodes glycosyltransferase belonging to the GTB-type superfamily (GT family 1, CAZy database). Glucose (Glc) is a dominant sugar component of EPS in R. leguminosarum, which is present as the first, fourth, fifth, sixth, and seventh residues in the repeating units (Robertsen et al. 1981). All sugar residues in the unit are bound by β-linkages (β-1,3, β-1,4, and β-1,6); the only exception is Glc at the fourth position, which is linked by an α-1,4 glycosidic bond. Based on amino acid sequence homology, the PssS protein might be involved in the fourth step of the unit synthesis. Similarly, Ivashina and Ksenzenko (2012) have recently proposed a model for the EPS repeating unit assembly in R. leguminosarum VF39 and TA1 strains, in which the fourth sugar residue (Glc) is linked by PssS, whereas addition of three further Glc residues is conducted by PssF, PssI/PssG, and PssI/PssH proteins, respectively, which belong to the GTA-type superfamily.
The phenotype of the Rt770 mutant obtained by us confirms that the PssS enzyme plays an essential role in EPS synthesis and symbiosis (Tables 4 and 5). In contrast to the wild-type strain, this EPS-non-producing mutant was unable to establish effective symbiosis with clover; the nodules induced on the host roots were very small, white, and contained only sporadically infected cells, in which bacteroids degenerated prematurely (Figs. 4, 5, and 6). The infection of plant cells was abnormal, visible as “explosion-like” mass endocytosis of the bacteria into the cell cytoplasm. This type of simultaneous endocytosis of a huge number of bacterial cells was previously reported for the pssD mutant of R. leguminosarum (Król et al. 1998) and the SGEFix−-1 mutant in pea with a mutation in sym40 controlling early nodule developmental stages (Tsyganov et al. 1998).
Previously, it was indicated for other EPS-deficient strains of R. leguminosarum and S. meliloti that absence of EPS led to similar strong disturbances in symbiosis (Rolfe et al. 1996; Guerreiro et al. 2000; Cheng and Walker 1998). Mutations in the pssA and pssD genes of R. leguminosarum bvs. trifolii and viciae also totally abolished EPS synthesis and resulted in inefficient symbiosis with the compatible host plants, which form indeterminate-type nodules (sporadically formed nodules were small and hardly occupied by bacteria) (Ivashina et al. 1994; Rolfe et al. 1996; van Workum et al. 1997; Król et al. 1998; Laus et al. 2005; Janczarek and Rachwał 2013). The formation of infection threads was only sporadically initiated and they were early aborted. Furthermore, Guerreiro et al. (2000) have reported that mutations in these genes had pleiotropic effects and they, among others, resulted in changed levels of synthesis of several proteins (over 20 proteins in the pssA mutant and 8 proteins in the pssD mutant). It was established that the symbiotic interaction of the pssD mutant with its host, clover was impaired in the step of endocytosis of the bacteria into the plant cell cytoplasm (Król et al. 1998). In contrast, an EPS-deficient pssA mutant of R. leguminosarum bv. phaseoli induced nitrogen-fixing nodules on its host, Phaseolus, which forms determinate-type nodules (Borthakur et al. 1988; Latchford et al. 1991). This indicates that the significance of EPS in symbiosis is mainly dependent on the kind of the host plant. Similarly, S. meliloti strains having mutations in genes encoding glycosyltransferases (exoY, exoA, exoL, exoM, exoO and exoW) were defective in production of succinoglycan and nodule invasion (Reuber et al. 1991; Becker et al. 1993a, b; Cheng and Walker 1998). Also, a lack of the PssV protein responsible for addition of pyruvyl residues to the units abolished production of this polysaccharide and negatively affected symbiosis with alfalfa (Glucksmann et al. 1993; Becker et al. 1993a). Furthermore, mutations in genes involved in the synthesis of nucleotide sugar precursors also caused strong negative effects on both EPS production and infection host plants. For example, a mutation in R. leguminosarum exo5 encoding UDP-glucose dehydrogenase responsible for oxidation of UDP-glucose to UDP-glucuronic acid, affected all glucuronic and galacturonic acid-containing polysaccharides including EPS and, in a consequence, successful colonization of the host roots (Laus et al. 2004). Also, mutations in S. meliloti exoC and exoB genes responsible for the synthesis of glucose-1-phosphate and UDP-galactose, respectively, abolished production of succinoglycan and other polysaccharides containing galactose (galactoglucan and LPS) (Glucksmann et al. 1993; Becker et al. 1993a, b). All these data indicate that EPS is indispensable for host root infection and effective nitrogen-fixation in symbiotic interactions with legumes forming indeterminate nodules. Mutations in genes encoding enzymes involved in the sugar precursor synthesis as well as those encoding glycosyltransferases result in similar effects, i.e., a lack of EPS synthesis and inefficient infection of the host plants.
Several functions are ascribed to rhizobial EPS in symbiosis such as facilitation of bacterial adhesion to and invasion of host roots, a structural function as a component of matrix inside infection threads, and protection against plant defense reactions (Downie 2010; Cheng and Walker 1998; Jaszek et al. 2014). D’Haeze et al. (2004) indicated for Azorhizobium caulinodans that a mutant strain producing significantly less EPS that the wild-type bacterium was more sensitive to oxidative stress and induced only ineffective pseudo-nodules on its host plant, Sesbania rostrata. In addition, these researchers showed a negative correlation between the amount of produced EPS and the level of H2O2 inside the bacterial cells. These data indicated that EPS produced by wild-type rhizobia forms a diffusion barrier protecting bacterial cells against huge amounts of hydrogen peroxide encountered during early stages of symbiosis (i.e., legume infection and nodule development). Also, EPS-deficient mutants of R. leguminosarum bv. trifolii showed decreased survival and adaptation to oxidative stress conditions (Jaszek et al. 2014).
References
Abreu I, Cerda ME, Pérez de Nanclares M, Baena I, Lloret J, Bonilla I, Bolaños L, Reguera M (2012) Boron deficiency affects rhizobia cell surface polysaccharides important for suppression of plant defense mechanisms during legume recognition and for development of nitrogen-fixing symbiosis. Plant Soil 361:385–395
Aiyar SE, Gourse RL, Ross W (1998) Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc Natl Acad Sci U S A 95:14652–1467
Angus AA, Lee A, Lum MR, Shehayeb M, Hessab R, Fujishige NA, Yerrapragada S, Kano S, Song N, Yang P, de los Santos PE, de Faria SM, Dakora FD, Weinstock G, Hirsch AM (2013) Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a nodulating and plant growth promoting beta-proteobacterium, are influenced by environmental factors. Plant Soil 369:543–562
Barthelemy-Delaux C, Marburger D, Delaux PM, Conley S, Ané JM (2014) Effect of drought on Bradyrhizobium japonicum populations in Midwest soils. Plant Soil 382:165–173
Becker A, Kleickmann A, Keller M, Arnold W, Pühler A (1993a) Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol Gen Genet 241:367–379
Becker A, Kleickmann A, Küster H, Keller M, Arnold W, Pühler A (1993b) Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol Plant Microbe Interact 6:735–744
Bertram-Drogatz PA, Quester I, Becker A, Pühler A (1998) The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol Gen Genet 257:433–441
Bittinger MA, Milner JL, Saville BJ, Handelsman J (1997) rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol Plant Microbe Interact 10:180–186
Borthakur D, Johnston AW (1987) Sequence of psi, a gene on the symbiotic plasmid of Rhizobium phaseoli which inhibits exopolysaccharide synthesis and nodulation, and demonstration that its transcription is inhibited by psr, another gene on the symbiotic plasmid. Mol Gen Genet 207:149–154
Borthakur D, Barker RF, Latchford JW, Rossen L, Johnston AW (1988) Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol Gen Genet 213:155–162
Breedveld MW, Zevenhuizen LPTM, Canter Cremers HCJ, Zehnder AJB (1993) Influence of growth conditions on production of capsular and extracellular polysaccharides by Rhizobium leguminosarum. Antonie Van Leeuwenhoek 64:1–8
Broughton WJ, Zhang F, Perret X, Staehelin C (2003) Signals exchanged between legumes and Rhizobium, agricultural uses and perspectives. Plant Soil 252:129–137
Brown CM, Dilworth MJ (1995) Ammonia assimilation by Rhizobium cultures and bacteroids. J Gen Microbiol 86:39–48
Browning DF, Busby SJ (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2:57–65
Burchill W, James E, Li D, Lanigan G, Williams M, Iannetta P, Humphreys J (2014) Comparisons of biological nitrogen fixation in association with white clover (Trifolium repens L.) under four fertiliser nitrogen inputs as measured using two 15N techniques. Plant Soil 385:287–302
Cazenave AB, Salon C, Jeudy C, Duc G, Voisin AS (2014) N2 fixation of pea hypernodulating mutants is more tolerant to root pruning than that of wild type. Plant Soil 378:397–412
Cheng HP, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180:5183–5191
Chou AY, Archdeacon J, Kado CI (1998) Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc Natl Acad Sci U S A 95:5293–5298
Cremers HC, Batley M, Redmond JW, Wijfjes AH, Lugtenberg BJ, Wijffelman CA (1991) Distribution of O-acetyl groups in the exopolysaccharide synthesized by Rhizobium leguminosarum strains is not determined by the Sym plasmid. J Biol Chem 266:9556–9564
D’Haeze W, Glushka J, De Rycke R, Holsters M, Carlson RW (2004) Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol Microbiol 52:485–500
Downie JA (2010) The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev 34:150–170
D’Souza-Ault MR, Cooley MB, Kado CI (1993) Analysis of the Ros repressor of Agrobacterium virC and virD operons: molecular intercommunication between plasmid and chromosomal genes. J Bacteriol 175:3486–3490
Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev 13:2134–2147
Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633
Frey SD, Blum LK (1994) Effect of pH on competition for nodule occupancy by type I and type II strains of Rhizobium leguminosarum bv. phaseoli. Plant Soil 163:157–164
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206
Glucksmann MA, Reuber TL, Walker GC (1993) Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol 175:7045–7055
González V, Santamaría RI, Bustos P, Hernández-González I, Medrano-Soto A, Moreno-Hagelsieb G, Janga SC, Ramírez MA, Jiménez-Jacinto V, Collado-Vides J, Dávila G (2006) The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci U S A 103:3834–3839
Guerreiro N, Ksenzenko VN, Djordjevic MA, Ivashina TV, Rolfe BG (2000) Elevated levels of synthesis of over 20 proteins results after mutation of the Rhizobium leguminosarum exopolysaccharide synthesis gene pssA. J Bacteriol 182:4521–4532
Huang KH, Chen BY, Shen FT, Young CC (2012) Optimization of exopolysaccharide production and diesel oil emulsifying properties in root nodulating bacteria. World J Microbiol Biotechnol 28:1367–1373
Ivashina TV, Ksenzenko VN (2012) Exopolysaccharide biosynthesis in Rhizobium leguminosarum: from genes to functions. In: The Complex World of Polysaccharides. InTech: Rijeka, Croatia, pp 99–127
Ivashina TV, Khmelnitsky MI, Shlyapnikov MG, Kanapin AA, Ksenzenko VN (1994) The pss4 gene from Rhizobium leguminosarum biovar viciae VF39: cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene 50:111–116
Ivashina TV, Fedorova EE, Ashina NP, Kalinchuk NA, Druzhinina TN, Shashkov AS, Shibaev VN, Ksenzenko VN (2010) Mutation in the pssM gene encoding ketal pyruvate transferase leads to disruption of Rhizobium leguminosarum bv. viciae - Pisum sativum symbiosis. J Appl Microbiol 109:731–742
Janczarek M, Rachwał K (2013) Mutation in the pssA gene involved in exopolysaccharide synthesis leads to several physiological and symbiotic defects in Rhizobium leguminosarum bv. trifolii. Int J Mol Sci 14:23711–23735
Janczarek M, Skorupska A (2007) The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol Plant Microbe Interact 20:867–881
Janczarek M, Skorupska A (2009) Rhizobium leguminosarum bv. trifolii rosR gene expression is regulated by catabolic repression. FEMS Microbiol Lett 291:112–119
Janczarek M, Skorupska A (2011) Modulation of rosR expression and exopolysaccharide production in Rhizobium leguminosarum bv. trifolii by phosphate and clover root exudates. Int J Mol Sci 12:4132–4155
Janczarek M, Urbanik-Sypniewska T (2013) Expression of the Rhizobium leguminosarum bv. trifolii pssA gene involved in exopolysaccharide synthesis is regulated by RosR, phosphate and the carbon source. J Bacteriol 195:3412–3423
Janczarek M, Jaroszuk-Ściseł J, Skorupska A (2009) Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Van Leeuwenhoek 96:471–486
Janczarek M, Rachwał K, Cieśla J, Ginalska G, Bieganowski A (2015a) Production of exopolysaccharide by Rhizobium leguminosarum bv. trifolii and its role for bacterial attachment and surface properties. Plant Soil 388:211–227
Janczarek M, Rachwał K, Marzec A, Grządziel J, Palusińska-Szysz M (2015b) Signal molecules and cell-surface components involved in early stages of the legume-rhizobium interactions. Appl Soil Ecol 85:94–113
Jaszek M, Janczarek M, Kuczyński K, Piersiak T, Grzywnowicz K (2014) The response of the Rhizobium leguminosarum bv. trifolii wild-type and exopolysaccharide-deficient mutants to oxidative stress. Plant Soil 376:75–94
Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Pühler A (1995) Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol Plant Microbe Interact 8:267–277
Kennedy IR, Pereg-Gerk LL, Wood C, Deaker R, Gilchrist K, Katupitiya S (1997) Biological nitrogen fixation in non-leguminous field crops: facilitating the evolution of an effective association between Azospirillum and wheat. Plant Soil 194:65–79
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kowalczuk E, Lorkiewicz Z (1979) Transfer of RP4 and R68.45 factors to Rhizobium. Acta Microbiol Pol 28:221–229
Król JE, Wielbo J, Mazur A, Kopcińska J, Łotocka B, Golinowski W, Skorupska A (1998) Molecular characterization of pssCDE genes of Rhizobium leguminosarum bv. trifolii strain TA1: pssD mutant is affected in exopolysaccharide synthesis and endocytosis of bacteria. Mol Plant Microbe Interact 11:1142–1148
Król JE, Mazur A, Marczak M, Skorupska A (2007) Syntenic arrangements of the surface polysaccharide biosynthesis genes in Rhizobium leguminosarum. Genomics 89:237–247
Latchford JW, Borthakur D, Johnston AW (1991) The products of Rhizobium genes, psi and pss, which affect exopolysaccharide production, are associated with the bacterial cell surface. Mol Microbiol 5:2107–2114
Laus MC, Logman TJ, Van Brussel AA, Carlson RW, Azadi P, Gao MY, Kijne JW (2004) Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 186:6617–6625
Laus MC, Van Brussel AA, Kijne JW (2005) Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol Plant Microbe Interact 18:533–538
Loewus FA (1952) Improvement in the anthrone method for determination of carbohydrates. Anal Chem 24:219
MacLellan SR, MacLean AM, Finan TM (2006) Promoter prediction in the rhizobia. Microbiology 152:1751–1763
Malgieri G, Russo L, Esposito S, Baglivo I, Zaccaro L, Pedone EM, Di Blasio B, Isernia C, Pedone PV, Fattorusso R (2007) The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain. PNAS USA 104:17341–17346
Marczak M, Dźwierzyńska M, Skorupska A (2013) Homo- and heterotypic interactions between Pss proteins involved in the exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii. Biol Chem 394:541–559
Marczak M, Matysiak P, Kutkowska J, Skorupska A (2014) PssP2 is a polysaccharide co-polymerase involved in exopolysaccharide chain-length determination in Rhizobium leguminosarum. PLoS One 9, e109106
Mazur A, Marczak M, Król JE, Skorupska A (2005) Topological and transcriptional analysis of pssL gene product: a putative Wzx-like exopolysaccharide translocase in Rhizobium leguminosarum bv. trifolii TA1. Arch Microbiol 184:1–10
McNeil M, Darvill J, Darvill AG, Albersheim P (1986) The discernible structural features of the acidic polysaccharides secreted by different Rhizobium species are the same. Carbohydr Res 146:307–326
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Mimmack ML, Hong GF, Johnston AW (1994) Sequence and regulation of psrA, a gene on the Sym plasmid of Rhizobium leguminosarum biover phaseoli which inhibits transcription of the psi genes. Microbiology 140:455–461
O’Neill MA, Darvill AG, Albersheim P (1991) The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J Biol Chem 266:9549–9555
Pini F, Spini G, Galardini M, Bazzicalupo M, Benedetti A, Chiancianesi M, Florio A, Lagomarsino A, Migliore M, Mocali S, Mengoni A (2014) Molecular phylogeny of the nickel-resistance gene nreB and functional role in the nickel sensitive symbiotic nitrogen fixing bacterium Sinorhizobium meliloti. Plant Soil 377:189–201
Pollock TJ, Workum WA, Thorne L, Mikolajczak MJ, Yamazaki M, Kijne JW, Armentrout RW (1998) Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol 180:586–593
Qin L, Jiang H, Tian J, Zhao J, Liao H (2011) Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant Soil 349:25–36
Quelas JI, López-García SL, Casabuono A, Althabegoiti MJ, Mongiardini EJ, Pérez-Giménez J, Couto A, Lodeiro AR (2006) Effects of N-starvation and C-source on Bradyrhizobium japonicum exopolysaccharide production and composition, and bacterial infectivity to soybean roots. Arch Microbiol 186:119–128
Quester I, Becker A (2004) Four promoters subject to regulation by ExoR and PhoB direct transcription of the Sinorhizobium meliloti exoYFQ operon involved in the biosynthesis of succinoglycan. J Mol Microbiol Biotechnol 7:115–132
Reeve WG, Dilworth MJ, Tiwari RP, Glenn AR (1997) Regulation of exopolysaccharide production in Rhizobium leguminosarum WSM710 involves exoR. Microbiology 143:1951–1958
Reuber TL, Reed J, Glazebrook J, Glucksmann MA, Ahmann D, Marra A, Walker GC (1991) Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance. Biochem Soc Trans 19:636–641
Robertsen BK, Aman P, Davill AG, McNeil M, Albersheim P (1981) Host-symbiont interactions. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol 67:389–400
Rolfe BG, Carlson RW, Ridge RW, Dazzo RW, Mateos FB, Pankhurst CE (1996) Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust J Plant Physiol 23:285–303
Ross W, Aiyar SE, Salomon J, Gourse RL (1998) Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol 180:5375–5383
Rüberg S, Pühler A, Becker A (1999) Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145:603–611
Rӓsӓnen LA, Saijets S, Jokinen K, Lindstrӧm K (2004) Evaluation of the roles of two compatible solutes, glycine betaine and trehalose, for the Acacia senegal- Sinorhizobium symbiosis exposed to drought stress. Plant Soil 260:237–251
Sambrook J, Fitsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schlüter JP, Reinkensmeier J, Barnett MJ, Lang C, Krol E, Giegerich R, Long SR, Becker A (2013) Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics 14:156
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784–791
Soussou S, Mahieu S, Brunel B, Escarré J, Lebrun M, Banni M, Boussetta H, Cleyet-Marel JC (2013) Zinc accumulation patterns in four Anthyllis vulneraria subspecies supplemented with mineral nitrogen or grown in the presence of their symbiotic bacteria. Plant Soil 371:423–434
Spaink HP, Okker RJH, Wijffelman CA, Pees E, Lugtenberg BJJ (1987) Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol 9:27–39
Suzuki Y, Adhikari D, Itoh K, Suyama K (2014) Effects of temperature on competition and relative dominance of Bradyrhizobium japonicum and Bradyrhizobium elkanii in the process of soybean nodulation. Plant Soil 374:915–924
Talbi C, Argandoña M, Salvador M, Alché JD, Vargas C, Bedmar EJ, Delgado MJ (2013) Burkholderia phymatum improves salt tolerance of symbiotic nitrogen fixation in Phaseolus vulgaris. Plant Soil 367:673–685
Tsyganov VE, Morzhina EV, Stefanov SY, Borisov AY, Lebsky VK, Tikhonovich IA (1998) The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function. Mol Gen Genet 259:491–503
van Workum WA, Canter Cremers HC, Wijfjes AH, van der Kolk C, Wijffelman CA, Kijne JW (1997) Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant Microbe Interact 10:290–301
van Workum WA, van Slageren S, van Brussel AAN, Kijne JW (1998) Role of exopolysaccharides of Rhizobium leguminosarum bv. viciae as host plant-specific molecules required for infection thread formation during nodulation of Vicia sativa. Mol Plant Microbe Interact 11:1233–1241
Vincent JM (1970) A manual for the practical study of root nodule bacteria. In: International biological program handbook no 15. Blackwell Scientific Publications Ltd., Oxford, UK
Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA (1995) β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691–1705
Yanni YG, Rizk RY, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, Orgambide G, De Bruijn F, Stoltzfus J, Buckley D, Schmidt TM, Mateos PF, Ladha JK, Dazzo FB (1997) Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 194:99–114
Young JP, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, Hull KH, Wexler M, Curson AR, Todd JD, Poole PS, Mauchline TH, East AK, Quail MA, Churcher C, Arrowsmith C, Cherevach I, Chillingworth T, Clarke K, Cronin A, Davis P, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmond M, Whitehead S, Parkhill J (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34
Acknowledgments
This work was supported by the grant of the National Science Centre no. 2012/07/B/NZ1/00099 and partially by the grant from the Ministry of Science and Higher Education no. N303 092234.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jeff R. Powell.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Janczarek, M., Rachwał, K. & Kopcińska, J. Genetic characterization of the Pss region and the role of PssS in exopolysaccharide production and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Plant Soil 396, 257–275 (2015). https://doi.org/10.1007/s11104-015-2567-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2567-5