Abstract
The interaction between plants and plant pathogens can have significant effects on ecosystem performance. For their growth and development, both bionts rely on amino acids. While amino acids are key transport forms of nitrogen and can be directly absorbed from the soil through specific root amino acid transporters, various pathogenic microbes can invade plant tissues to feed on different plant amino acid pools. In parallel, plants may initiate an immune response program to restrict this invasion, employing various amino acid transporters to modify the amino acid pool at the site of pathogen attack. The interaction between pathogens and plants is sophisticated and responses are dynamic. Both avail themselves of multiple tools to increase their chance of survival. In this review, we highlight the role of amino acid transporters during pathogen infection. Having control over the expression of those transporters can be decisive for the fate of both bionts but the underlying mechanism that regulates the expression of amino acid transporters is not understood to date. We provide an overview of the regulation of a variety of amino acid transporters, depending on interaction with biotrophic, hemibiotrophic or necrotrophic pathogens. In addition, we aim to highlight the interplay of different physiological processes on amino acid transporter regulation during pathogen attack and chose the LYSINE HISTIDINE TRANSPORTER1 (LHT1) as an example.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Availability of nitrogen orchestrates plant pathogen resistance
A proper plant nitrogen (N) nutrition is warranted by the uptake of inorganic and organic N sources. Organic N such as proteins, peptides or amino acids (AAs) are taken up via specific root transporters (Paungfoo-Lonhienne et al. 2008; Nasholm et al. 2009; Tegeder and Rentsch 2010; Inselsbacher and Näsholm 2012; Tegeder and Masclaux-Daubresse 2018; Gratz et al. 2021) that have multiple functions within a plant (Yang et al. 2020; Yao et al. 2020). AAs represent an important storage and transport form of organic N and are precursors for protein synthesis. AAs are especially important for the development of roots, leaves, and seeds (Rentsch et al. 2007; Tegeder and Masclaux-Daubresse 2018), which makes AA transport systems a key component for plant development. Not only plants but also the microbial community relies on the availability of AAs, and it is not surprising that both compete for this N source (Roberts and Jones 2012; Kuzyakov and Xu 2013; Wilkinson et al. 2014). We identified the need of a concise survey highlighting the role of AA transporters (AATs) during pathogen infection due to the fact that literature mostly focusses on the influence of inorganic N on plant resistance (Ballini et al. 2013; Huang et al. 2017; Farjad et al. 2018; Sun et al. 2020).
Amino acid pools and fluxes are, however, dependent on N supply and the absolute majority of studies reporting on N effects on pathogen resistance have focused on comparisons of the inorganic N sources nitrate (NO3−) and ammonium (NH4+). In addition to reviewing the links between AATs and pathogen resistance, we therefore also performed a literature search aiming to compare effects of nitrate and ammonium addition on the plant’s ability to resist pathogens that differ in their nutrition strategy (Table 1, Supplementary Tables 1–3). Especially the different nutrient acquisition strategies by different pathogens such as biotrophic, hemibiotrophic as well as necrotrophic pathogens are important in this context. Biotrophic pathogens exhibit specialized feeding structures that allow nutrient retrieval from living cells. Hemibiotrophic microbes, however, first colonize the living cell but then transition into a necrotrophic phase. Necrotrophs obtain their nutrients from killed cells (Spanu and Panstruga 2017). Within biotrophic pathogens, the presence of different inorganic N sources led to strong and opposing effects: addition of NO3− reduced plant resistance in the majority of analyzed cases (11 out of 15 cases). Interestingly, not only the presence but also the rate of NO3− addition influenced defense responses of plants (Ding et al. 2021). Tomato plants infected with the biotroph Ralstonia solanacearum, for instance, demonstrated less disease lesions when grown on 1 mM compared to 7 mM NO3− (Ding et al. 2021). Interestingly, the presence of NH4+ as N source, though, demonstrated an opposing trend: in 10 out of 14 cases elevated plant resistance was found (Table 1, Supplementary Table 1). Plant resistance against hemibiotrophic pathogens seems not to display any clear response to different inorganic N sources and both positive and negative effects of NO3− and NH4+ addition have been reported (Table 1, Supplementary Table 2). Concentration-related effects such as reduced disease lesions were observed for tomato plants after infection with Pseudomonas syringae (P. syringae), when plants were grown on 1 mM compared to 7 mM NO3− (Ding et al. 2021). In 9 out of 15 cases NO3− led to a positive immune response such as increased resistance or hypersensitive response during necrotrophic attack (Table 1, Supplementary Table 3). Similar to biotrophic and hemibiotrophic infection, plant responses after necrotrophic interaction seem to dependent on the N rate (Farjad et al. 2018). Measurements of bacterial cell numbers of the necrotroph Erwinia amylovora in infected Arabidopsis thaliana (Arabidopsis) revealed lower numbers when grown on low NO3− (0.5 mM) compared to high NO3− (5 mM). This was associated with transcriptional reprograming of defense genes, e.g., PATHOGENESIS-RELATED GENE2 and 5 (PR2 and PR5) or salicylic acid (SA)-related genes (Farjad et al. 2018). Addition of NH4+, though, led to increased cases of elevated plant susceptibility, when infected with a necrotroph (7 out of 11 cases) (Table 1). Overall, we found that a plant’s ability to withstand biotrophic attacks tends to be more successful when NH4+ is accessible, the opposite of what was shown for necrotrophs. The overall N addition rate might serve as a proxy for plant N status, which influences susceptibility additionally.
As shown above, plant N sources play critical roles for plant resistance. This observation motivates a further analysis of N transporters during pathogen attack. Camanes et al. (2012) investigated the response of NO3− transporters AtNRT2.1 and AtNRT2.2 to infection by the hemibiotrophic bacteria P. syringae. The nrt2 mutant exhibited an increased immune response along with a reduced susceptibility and significant alterations in the transcriptome. The expression of SA marker genes was strongly increased compared to the wild type, and it was suggested that members of the AtNRT2 family might be important for the plant-pathogen interaction (Camanes et al. 2012). More recently it was shown that the nrt2.5 mutant displayed similar responses (du Toit et al. 2020). Similarly, also NH4+ transporters such as AtAMT1.1 seem to play an important role for plant resistance (Pastor et al. 2014). amt1.1 plants infected with P. syringae and Plectosphaerella cucumerina, a hemibiotrophic and a necrotrophic organism respectively, exhibited increased resistance, an effect that was enhanced by N depletion (Pastor et al. 2014). These findings lead to the hypothesis, that N transporters play a role in plant immune responses, by acting as regulators in N supply. We therefore ask the question whether other transporters that are involved in N uptake and N translocation and in particular the AATs could potentially also play a role in plant resistance.
A dual utilization of amino acids
It is well established that pathogens can feed on plant N reserves, mainly AAs, which makes them crucial players in the plant-pathogen interaction (Struck et al. 2004; Zeier 2013; Sonawala et al. 2018; Yang et al. 2020; Sharma 2020). It is energetically more beneficial for pathogens to directly acquire and metabolize plant AAs which is why a range of pathogens can directly target the induction of genes needed for AAT (Sonawala et al. 2018; Li et al. 2020). Having control over a plant’s AA uptake and transport system can, therefore, be decisive for the survival of either the plant or the pathogen.
Li et al. found substantial reprogramming of N and C metabolic pathways in kiwifruit tissues upon infection with P. syringae, i.e., an accumulation of specific AAs (Li et al. 2020). While the accumulation of some AAs can be beneficial for the pathogen, others can play important roles in plant resistance. Tryptophan and methionine, for instance, are known precursors for the synthesis of secondary metabolites with antimicrobial effects (Ahuja et al. 2012). Depending on the microbe, these metabolites accumulate in individual root cell layers and can contribute to increased resistance (Froschel et al. 2021). A similar response of citrus plants was described upon infection with the phloem-feeding biotroph Candidatus liberibacter, as the phloem sap of tolerant plants exhibited high amounts of tryptophan, tyrosine or phenylalanine; well-studied precursors for secondary metabolites and phenolics (Killiny and Hijaz 2016). Proline, a known radical scavenger, contributes to the regulation of cellular redox homeostasis (Smirnoff and Cumbes 1989). Gupta et al. (2020) recently corroborated the positive properties of proline during infection and analyzed upstream components. They identified miRNA involved in the regulation of proline biosynthesis, which is not only important for the plant immune response but is also involved in regulation of abiotic stresses (Gupta et al. 2020).
The above suggests that it is crucial to understand the molecular regulation of AA transport and accumulation because AAs can be used as N sources for the pathogen but also as protective agents for the plant. This leads to the question whether AATs are differently expressed during plant-pathogen interaction and if so, who the driver of this regulation is. Having control over the expression can, thus, decide over the fate of both, plants or pathogens (Hammes et al. 2006; Liu et al. 2010; Elashry et al. 2013; Pariyar et al. 2018; Sonawala et al. 2018; Froschel et al. 2021).
Responses of plant amino acid transporters to pathogen infection
The products of about 100 genes are known to facilitate AA transport in Arabidopsis and similar AATs have additionally been identified in many crop and tree species (Tegeder and Ward 2012; Pratelli and Pilot 2014; Yang et al. 2020). ATF (amino acid transporter family), APC (amino acid-polyamine-choline transporter family) and UMAMIT (usually multiple acids move in and out transporter family) represent the three main AAT families (Rentsch et al. 2007; Pratelli and Pilot 2014; Dinkeloo et al. 2018; Yang et al. 2020). ATFs can be divided into several subfamilies such as, e.g., AAPs (amino acid permeases) or LHTs (lysine histidine transporters) (Rentsch et al. 2007). CATs (cationic amino acid transporters) represent a subfamily within the APCs (Tegeder and Rentsch 2010).
Amino acid permeases (AAPs)
AAPs, a group of one-directional transporters, are involved in root AA uptake, phloem loading, xylem-phloem transfer, and seed loading (Fischer et al. 1995; Okumoto et al. 2002, 2004; Lee et al. 2007; Svennerstam et al. 2008; Zhang et al. 2010; Santiago and Tegeder 2016). It is well known that AAPs are highly conserved between various species (Benedito et al. 2010; Zhao et al. 2012, 2017; Limpens et al. 2013; Garneau et al. 2018; Duan et al. 2020; Llebrés et al. 2021; Omari Alzahrani 2021).
Several members of the AAP family were found to be differentially regulated upon biotrophic interactions. AtAAPs demonstrated enhanced gene expression after plant-parasitic nematode infection and increased resistance in respective knockout mutants (Hammes et al. 2005; Elashry et al. 2013; Marella et al. 2013). Analysis of aap1, aap2 and aap6 knockout mutants displayed decreased reproduction of cyst nematodes (Elashry et al. 2013). Similarly, aap3 and aap6 exhibited reduced reproduction of root-knot nematodes (Marella et al. 2013). Recently, the role of CsAAP2A in cucumber became evident as knockout plants displayed resistance to downy mildew (Berg et al. 2021). A functional analysis of AAPs in tomato plants, when challenged with the hemibiotrophic Phytophthora infestans (P. infestans), displayed that mutations in the tomato homologues SIAAP5A and SIAAP5B led to similar effects (Berg et al. 2021). It is reasonable that an infection causes a differential regulation of local AATs in specific cell types. It would also be conceivable that a transporter is being regulated in opposing directions upon infection of the same pathogen, however, in different cells. A recent study zoomed in on these questions and compared expression patterns in four specific root cell layers (rhizodermis, cortex, endodermis, and stele), when Arabidopsis was challenged with, in their nutrition strategy varying, microbes (Froschel et al. 2021). When looking at the cell layer-specific transcript abundance after hemibiotrophic P. parasitica infection, it was found that AtAAP3, AtAAP5 and AtAAP6 were induced in the stele, however, AtAAP6 was additionally upregulated in the cortex (Froschel et al. 2021). Responses to hemibiotrophic, vascular Verticillium longisporum (V. longisporum) varied within the AtAAP family: AtAAP4 was the only representative that was upregulated and only in the cortex. AtAAP1, in the cortex, and AtAAP2, in the rhizodermis, were found to be downregulated after infection (Froschel et al. 2021).
Based on the above publications, it can be suggested that AAPs are negative regulators in plant defense against (hemi-) biotrophic pathogens. An increase in AAT transcript abundance might reduce plant defense reactions which would be beneficial for the pathogen. Alternatively, these transporters might be exploited by pathogens to steer plant AA transport, elevating the amount of accessible AAs in infected leaves and creating an artificial sink that pathogens can feed on (Berg et al. 2021).
Cationic amino acid transporters (CATs)
Some AATs affect the plant immune system in a positive way, like AtCAT1 (Yang et al. 2014). The infection with hemibiotrophic P. syringae caused elevated transcript levels of AtCAT1 and increased resistance. Overexpression of AtCAT1 led to the constitutive expression of SA related and PR1 genes, as well as an increase in SA levels. Since AtCAT1 expression responded quickly to the infection it seems that it is involved in the systemic resistance of the plant (Yang et al. 2014).
Usually multiple acids move in and out transporter family (UMAMITs)
Most AATs operate as one-directional symporter, transporting AAs along a proton gradient (Bush 1993; Frommer et al. 1993; Hsu et al. 1993), however, UMAMITs are an exception. Driven by an electrochemical gradient, UMAMITs transport AAs in both directions (Ladwig et al. 2012; Muller et al. 2015). Due to their bi-directional activity, AtUMAMITs are involved in multiple physiological roles ranging from phloem loading/unloading, over xylem-phloem transport, to transport to sink tissues (Ladwig et al. 2012; Muller et al. 2015; Besnard et al. 2016). When looking at the cell layer-specific transcript abundance, all differentially regulated AtUMAMIT genes found upon presence of the hemibiotroph P. parasitica were downregulated: AtUMAMIT11/38/41 were differentially regulated in the rhizodermis and the cortex. Besides, AtUMAMIT11 was additionally downregulated in the stele. AtUMAMIT33 was regulated in the cortex and AtUMAMIT5 in the rhizodermis as well as the stele (Froschel et al. 2021). AtUMAMIT18 expression in the rhizodermis and stele, AtUMAMIT5 in the stele, and AtUMAMIT34 expression in the cortex were downregulated upon hemibiotrophic V. longisporum infection. The opposite effect, an increase in transcripts, was seen for AtUMAMIT5/31 (cortex), AtUMAMIT38 (endodermis) and AtUMAMIT14 (stele) (Froschel et al. 2021). Based on the analysis of transgenic Arabidopsis lines, Besnard et al. (2021) suggested that AtUMAMIT14 is a positive regulator in plant pathogen resistance. When challenged with the biotrophic oomycota Hyaloperonospora arabidopsidis, AtUMAMIT14 overexpression lines displayed enhanced expression of SA marker genes as well as SA levels, leading to increased resistance (Besnard et al. 2021). The example of UMAMITs visualizes a diverse set of responses, where individual genes can be regulated opposingly depending on the cell type, and genes within the transporter family are regulated inconsistently. It might be that their bi-directional transport ability causes different responses, which is why the individual role of each transporter during plant-pathogen interaction needs to be carefully evaluated.
Lysine histidine transporters (LHTs)
In Arabidopsis, 10 AtLHT paralogs (Rentsch et al. 2007) exist with different specificity and cellular location. AtLHT1, the first identified transporter of this family (Chen and Bush 1997; Hirner et al. 2006; Svennerstam et al. 2007) is involved in leaf mesophyll import as well as root uptake of acidic and neutral AAs, both at naturally occurring concentrations (Svennerstam et al. 2011), and from agricultural soil (Ganeteg et al. 2017). AtLHT1 also transports non-proteinogenic AAs, like 1-aminocyclopropane-1-carboxylic acid (ACC), just as its paralog AtLHT2 (Shin et al. 2015; Choi et al. 2019). ACC serves as a precursor of the phytohormone ethylene (ET) and as a signaling molecule on its own (Van de Poel and Van Der Straeten 2014; Vanderstraeten et al. 2019). AtLHT1 can be exploited to shuttle novel AA-coupled pesticides inside a plant (Jiang et al. 2018; Chen et al. 2018). Homologs of AtLHT1 were also identified and studied in, e.g., rice, poplar, lotus, tea and ginseng (Guether et al. 2011; Zhang et al. 2013; Wang et al. 2019; Guo et al. 2020; Gratz et al. 2021; Li et al. 2021). The Arabidopsis knockout mutant lht1-1 displayed an early senescence phenotype (Hirner et al. 2006; Svennerstam et al. 2007).
The role of AtLHT1 during pathogen infection has been investigated in several studies: AtLHT1 transcript levels were elevated when the host was infected with the biotrophic powdery mildew fungus Erysiphe cichoracearum (E. cichoracearum) (Liu et al. 2010) or the biotrophic nematode Heterodera schachtii (Elashry et al. 2013). Also, upon infection with the hemibiotrophic bacteria P. syringae, the fungi Colletotrichum higginsianum (C. higginsianum) (Liu et al. 2010) and V. longisporum (Froschel et al. 2021) as well as the oomycete P. parasitica (Froschel et al. 2021), AtLHT1 was upregulated. Most biotrophs feed on the apoplast or apoplast-like compartments and assimilate nutrients directly from their living host (Szabo and Bushnell 2001; Fatima and Senthil-Kumar 2015; Wang et al. 2020). It has been shown, that pathogens can reprogram plant transport proteins for their benefit, in order to, e.g., gain nutrients (Delmotte et al. 2009; Spanu and Panstruga 2017). This opens for the possibility that the pathogen, rather than the host plant, may steer the expression of AtLHT1.
From a plant’s perspective, it would be beneficial to increase the uptake of AAs from the apoplast to lower AAs accessibility for biotrophic pathogens and to secure its AA resources away from the infected area. This means an increased remobilization of AAs would require increased expression of AATs as part of a slash-and-burn defense strategy (Masclaux-Daubresse et al. 2010) (Fig. 1a). The increased expression of AtLHT1 could be seen as a defense strategy caused by the plant to drain a maximum of AAs out of the apoplast in order to starve the pathogen.
Response of the plant amino acid transporter AtLHT1 to pathogen attack. Upon attack by biotrophic pathogens (orange P), the transcript abundance of AtLHT1 is increased (a). An increased gene expression leads to an increased AtLHT1 protein abundance at the plasma membrane, which causes an active import of AAs (purple dots) into the cytosol. As a consequence, a depletion of apoplastic- and an increase of cytosolic AA concentrations occurs. This might be a direct response by the plant to apoplastic-feeding pathogens, in order to empty the apoplast and shuttle AAs into the cytosol. From there, AAs can be exported to healthy plant tissues. Due to the fact that lht1-1 mutants display increased pathogen resistence due to the accumulation of reactive oxygen species (ROS), salicylic acid (SA) production and pathogenesis-related (PR) gene expression, the upregulation of AtLHT1 might by steered by the biotrophic pathogen itself (dotted arrow). This action might avoid SA defense responses and might increase chances for the pathogen to survive. Upon attack by a necrotrophic pathogen, AtLHT1 is also elevated (b). This might, however, be an exclusive response by the plant. AtLHT1 transports the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (yellow dots). Mostly ET/JA-mediated responses contribute to the defense against necrotrophic pathogens. Additionally, an upregulation of the transporter might contribute to the shuttling of AAs to healthy, more distal plant tissues. Hence, the observed upregulation of AtLHT1 might be mostly a protective measure, steered by the plant
However, and in contrast to the predictions from this hypothesis, lht1-1 knockout mutants displayed increased resistance to P. syringae, C. higginsianum and E. cichoracearum, highlighting that AtLHT1 is a negative regulator in plant defenses (Liu et al. 2010). Disruption of AtLHT1 displayed different defense responses such as increased callose deposition, hypersensitive cell death and the constitutive expression of genes belonging to the SA defense pathway such as PR1 (Liu et al. 2010). The response is very similar to what was described for mutants of different AAPs (Elashry et al. 2013; Marella et al. 2013; Berg et al. 2021; Froschel et al. 2021). Liu et al. (2010) hypothesized that AtLHT1’s role in plant resistance was linked to its ability to transport glutamine. The absence of AtLHT1 causes a lack of glutamine within the cell, which leads to an altered redox status and enhanced immunity due to an accumulation of reactive oxygen species (ROS) and induced programmed cell death (PCD) (Liu et al. 2010). This suggests that the increased expression of AtLHT1 observed during the infection may be caused by the biotrophic pathogens, in order to inhibit the activation of the SA defense and, hence, an increase in plant resistance (Fig. 1a).
On the contrary, necrotrophic pathogens break plasma membranes and induce PCD in the host prior to nutrient uptake. While the SA pathway plays little role, the ethylene/jasmonic acid (ET/JA)-mediated response contributes to defense against necrotrophic pathogens (Glazebrook 2005; Pieterse et al. 2012; Huang et al. 2020). Furthermore, it has been shown that plants react in an analogous way to nematodes as to necrotrophic pathogens by activating the ET/JA pathway (Przybylska and Obrępalska-Stęplowska 2020). Similar to what has been observed for biotrophic pathogens, increased LHT1 transcript levels were also found upon interaction with necrotrophic pathogens Botrytis cinerea (Xiong et al. 2018) and Erwinia amylovora (Farjad et al. 2018). Farjad et al. confirmed the involvement of AtLHT1 during pathogen attack: AtLHT1 resembled the expression profile of other defense associated genes by being induced during infection, behaving opposing to other N metabolism related genes. Potentially this serves an increased transport of ACC, supporting ET-based plant defense, as AtLHT1 and AtLHT2 were found to transport the ET precursor (Shin et al. 2015; Choi et al. 2019). This hypothesis is in line with the finding, that lht1-1 mutants displayed no increased resistance to necrotrophic pathogen infection such as Sclerotinia sclerotiorum (Liu et al. 2010) or the nematode H. schachtii (Elashry et al. 2013). Necrotrophic pathogens would not benefit from increasing the transcript abundance of AtLHT1, which therefore might display a plant response in order to transport ACC as defense mechanism as well as to transport AAs away from the invaded tissue (Fig. 1b).
Regulation of amino acid transporters through additional physiological processes
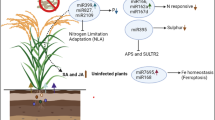
The dominant players in plant defense are the antagonistic phytohormones SA and ET/JA (Huang et al. 2020; Zhang et al. 2020). The involvement of other phytohormones and crosstalk among the different players is well studied (Pieterse et al. 2012; Huang et al. 2020; Zhang et al. 2020; Aerts et al. 2021). The SA-mediated defense seems to be more effective against biotrophs and hemibiotrophs whereat the ET/JA-mediated defense targets necrotrophic microbes (Glazebrook 2005; Huang et al. 2020; Zhang et al. 2020). The link between SA-mediated defense and AAT regulation has been studied (Liu et al. 2010; Yang et al. 2014; Besnard et al. 2021), whereas not much is known about ET/JA-regulated defense against necrotrophs in connection to AAT regulation. Recently, much work has been done on understanding the molecular underpinnings of leaf senescence. Due to the fact that the lht1-1 mutant displays an early senescence-like phenotype (Hirner et al. 2006; Svennerstam et al. 2007), we aimed to identify regulatory targets, that play a role in plant senescence and pathogen defense, and at the same time display a connection to the regulation of AATs (Fig. 2).
The molecular regulation of amino acid transporters is influenced by diverse regulatory pathways. Using the example of AtLHT1, the influence of individual key players important for ethylene (ET) signaling and senesence in the context of pathogen defense is depicted. AtLHT1 transports the signaling molecule and ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC). The ET receptor kinase ETHYLENE RESPONSE SENSOR2 (AtERS2) might interact with AtLHT1 and thus depicts a potential feedback loop in dependence of ET. ET presence in parallel represses the activity of miRNA164, through the action of the transcription factor (TF) EIN3. miRNA164 itself is a negative regulator of the TF ORESARA1 (AtORE1), a key player in plant senescence. AtEIN3 activates AtORE1 transcription directly whereas AtORE1 then activates the expression of ACC SYNTHASE2 (AtACS2), displaying a feed-forward loop. AtORE1 itself is directly regulated by the ubiquitin ligase NITROGEN LIMITATION ADAPTATION (AtNLA), which also regulates AtLHT1 through either direct or indirect action. Dashed lines indicate potential regulatory connections that remains to be tested
The transcription factor ORESARA1 (AtORE1) targets promoters of senescence-associated genes and directly mediates PCD (Oh et al. 1997; Kim et al. 2009; Balazadeh et al. 2010; Farage-Barhom et al. 2011; Al-Daoud and Cameron 2011; Matallana-Ramirez et al. 2013; Qiu et al. 2015; Durian et al. 2020). AtORE1 itself is targeted for degradation by the RING-type E3 ubiquitin ligase NITROGEN LIMITATION ADAPTATION (AtNLA) (Park et al. 2018). Deubiquitination events, however, stabilize AtORE1 and promote leaf senescence (Park et al. 2019). ET is involved in a positive regulation of AtORE1. More specifically, AtEIN3, a transcription factor acting downstream of EIN2, represses miR164, a negative regulator of AtORE1, and can in parallel bind to the AtORE1 promoter (Kim et al. 2009, 2014; Li et al. 2013). Together with AtEIN3, AtORE1 then activates transcription of chlorophyll catabolic genes in an ET dependent manner (Qiu et al. 2015). AtORE1 additionally activates ACC SYNTHASE2 (AtACS2) and AtACS6 expression, leading to enhanced ET production, displaying a coherent feed-forward loop for ET dependent leaf senescence (Qiu et al. 2015; Zhang et al. 2021). Interestingly, the action of AtORE1 and AtNLA are tightly connected to plant defense responses (Zhang et al. 2021). Arabidopsis infection with the hemibiotroph V. dahliae caused premature leaf senescence. It was shown that a microbial elicitor interfered with the interaction between AtORE1 and AtNLA, which, in turn, stabilized AtORE1, enhanced ET production and, thus, promoted senescence (Zhang et al. 2021). Recently, it was shown that AtORE1 is activated through protein phosphorylation via the calcium (Ca2+) kinase AtCPK1 (Durian et al. 2020). This kinase has previously been analyzed and it was shown that AtCPK1 is upregulated upon pathogen infection and was found to be a positive regulator in plant resistance due to activation of SA biosynthesis (Coca and San Segundo 2010). Interestingly, also plants infected with necrotrophs displayed increased resistance, although no ET derived defense responses were found (Coca and San Segundo 2010). In a preprinted study, it was suggested that AtNLA displays a negative regulator in plant defense against necrotrophs (Val-Torregrosa et al. 2021-preprint). nla mutants displayed increased callose deposition as well as increased resistance. Upon pathogen attack, transcript levels of AtNLA were reduced (Val-Torregrosa et al. 2021-preprint).
It was recently shown that AtORE1 and AtNLA additionally play a role in the regulation of AtLHT1 (Fig. 2). The ubiquitin ligase AtNLA targets pathways connected to organic N remobilization by targeting AATs during N deficiency (Liao et al. 2020). Transcript abundance of several AATs was found upregulated in the nla mutant and AtLHT1 displayed the highest regulation. A proteomic analysis confirmed the regulation of AtLHT1 by AtNLA (Liao et al. 2020), however, it remains to be tested whether this regulation is due to a direct interaction between AtLHT1 and the ligase. The authors additionally speculated whether AtORE1 is controlling transcription of AtLHT1 (Liao et al. 2020), however, an upregulation of AtLHT1 in AtORE1 overexpression lines has not been observed (Matallana-Ramirez et al. 2013). It remains unclear whether AtORE1 serves as TF regulating AtLHT1.
Given this complex regulatory crosstalk between different physiological processes, it can be speculated whether AtLHT1 is subject to additional molecular regulation. Due to the fact that miR164 is an important player at the interface between ET signaling and senescence (Kim et al. 2009, 2014; Li et al. 2013), and miRNAs in general play important roles in plant immunity (Val-Torregrosa et al. 2021), future studies should evaluate whether AtLHT1 may also be regulated through the action of miRNAs. As mentioned above, the signaling compound and ET precursor ACC is transported by members of the AtLHT family (Van de Poel and Van Der Straeten 2014; Shin et al. 2015; Choi et al. 2019; Vanderstraeten et al. 2019), which provides a direct link between the ET signaling- and AA uptake pathways. In addition, Chen et al 2012 found the ER-localized ETHYLENE RESPONSE SENSOR2 (ERS2) (Hua et al. 1998), to interact with AtLHT1 in yeast (Chen et al. 2012). Novel findings about the poplar homolog PtrLHT1.2 being not exclusively localized at the PM but also at the ER (Gratz et al. 2021), raise the question about a potential functional importance of this potential interaction, that remains to be tested in planta. Given the fact that ERS2 is a receptor kinase (Moussatche and Klee 2004) whose activity is not needed for ET signaling, it raises the question whether the kinase targets substrates outside the ET pathway and, thus, could be involved in additional responses (Chen et al. 2009; Lacey and Binder 2014). This opens up for the hypothesis that AtLHT1 could be post-translationally modified in an ET-dependent way; a speculation that remains to be tested. The strong connection between AtLHT1 and ET leads to the question if unknown defense responses against necrotrophs exist, that involve the action of AtLHT1. Pathogen attack triggers Ca2+ influx into the cell (Nishad et al. 2020), which can then lead to phosphorylation and activation of AtORE1 (Coca and San Segundo 2010; Durian et al. 2020). Overexpression of AtCPK1 leads to increased resistance of plants upon necrotrophic attack, the molecular regulation for this is, however, so far unknown (Coca and San Segundo 2010). The suggested downregulation of AtNLA upon necrotrophic interaction (Val-Torregrosa et al. 2021-preprint) would lead to a potential reduction in AtORE1 degradation. Overall, this would increase AtORE1 activity and PCD as well as senescence (Oh et al. 1997; Kim et al. 2009; Balazadeh et al. 2010; Farage-Barhom et al. 2011; Al-Daoud and Cameron 2011; Matallana-Ramirez et al. 2013; Qiu et al. 2015; Durian et al. 2020). This, a beneficial outcome for necrotrophs, would stand in contrast to the fact that a high accumulation of AtORE1 would increase ACC production via ACS2/6, and thus, ET accumulation (Qiu et al. 2015; Zhang et al. 2021). Reduced transcript accumulation of AtNLA would additionally lead to an increase in AtLHT1 (Liao et al. 2020). AtLHT1 could then contribute to the production of ET by transport of ACC (Shin et al. 2015; Choi et al. 2019) and, potentially, ET triggered resistance to necrotrophic microbes. It becomes evident that many common players in the regulation of pathogen resistance, leaf senescence and AAT regulation have overlapping functions. In future experiments, it has to be carefully determined, in which way the crosstalk between those players has an influence on plant microbes and plant resistance.
The complex network behind plant pathogen defense depends on several factors such as soil N availability and composition of the soil N pool which would affect both the internal N status of the plant and its energy status. Both plants and pathogens possess toolboxes, containing different signaling molecules such as ROS or hormones, but also transcription factors to concur the respective other. These responses are deeply interwoven with a machinery of cell-type specific regulation of AATs and, hence, the accumulation or depletion of specific AAs. The unique response signatures that are being formed upon association of a pathogen then contributes to the susceptibility of the plant.
Data availability
Not applicable.
Code availability
Not applicable.
Material availability
Not applicable.
References
Aerts N, Pereira Mendes M, Van Wees SCM (2021) Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J 105(2):489–504. https://doi.org/10.1111/tpj.15124
Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17(2):73–90. https://doi.org/10.1016/j.tplants.2011.11.002
Al-Daoud F, Cameron RK (2011) ANAC055 and ANAC092 contribute non-redundantly in an EIN2-dependent manner to age-related resistance in Arabidopsis. Physiol Mol Plant Pathol 76(3):212–222. https://doi.org/10.1016/j.pmpp.2011.09.003
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62(2):250–264. https://doi.org/10.1111/j.1365-313X.2010.04151.x
Ballini E, Nguyen TT, Morel JB (2013) Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice (N Y) 6(1):32. https://doi.org/10.1186/1939-8433-6-32
Benedito VA, Li H, Dai X, Wandrey M, He J, Kaundal R, Torres-Jerez I, Gomez SK, Harrison MJ, Tang Y, Zhao PX, Udvardi MK (2010) Genomic inventory and transcriptional analysis of Medicago truncatula transporters. Plant Physiol 152(3):1716–1730. https://doi.org/10.1104/pp.109.148684
Berg JA, Hermans FWK, Beenders F, Abedinpour H, Vriezen WH, Visser RGF, Bai Y, Schouten HJ (2021) The amino acid permease (AAP) genes CsAAP2A and SlAAP5A/B are required for oomycete susceptibility in cucumber and tomato. Mol Plant Pathol 22(6):658–672. https://doi.org/10.1111/mpp.13052
Besnard J, Pratelli R, Zhao C, Sonawala U, Collakova E, Pilot G, Okumoto S (2016) UMAMIT14 is an amino acid exporter involved in phloem unloading in Arabidopsis roots. J Exp Bot 67(22):6385–6397. https://doi.org/10.1093/jxb/erw412
Besnard J, Sonawala U, Maharjan B, Collakova E, Finlayson SA, Pilot G, McDowell J, Okumoto S (2021) Increased expression of UMAMIT amino acid transporters results in activation of salicylic acid dependent stress response. Front Plant Sci. https://doi.org/10.3389/fpls.2020.606386
Bush DR (1993) Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol 44(1):513–542. https://doi.org/10.1146/annurev.pp.44.060193.002501
Camanes G, Pastor V, Cerezo M, Garcia-Andrade J, Vicedo B, Garcia-Agustin P, Flors V (2012) A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol 158(2):1054–1066. https://doi.org/10.1104/pp.111.184424
Chen L, Bush DR (1997) LHT1, a lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiol 115(3):1127–1134. https://doi.org/10.1104/pp.115.3.1127
Chen T, Liu J, Lei G, Liu Y-F, Li Z-G, Tao J-J, Hao Y-J, Cao Y-R, Lin Q, Zhang W-K, Ma B, Chen S-Y, Zhang J-S (2009) Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol 50(9):1636–1650. https://doi.org/10.1093/pcp/pcp107
Chen J, Lalonde S, Obrdlik P, Noorani Vatani A, Parsa SA, Vilarino C, Revuelta JL, Frommer WB, Rhee SY (2012) Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front Plant Sci 3:124. https://doi.org/10.3389/fpls.2012.00124
Chen Y, Yan Y, Ren Z-F, Ganeteg U, Yao G-K, Li Z-L, Huang T, Li J-H, Tian Y-Q, Lin F, Xu H-H (2018) AtLHT1 transporter can facilitate the uptake and translocation of a glycinergic-chlorantraniliprole conjugate in Arabidopsis thaliana. J Agric Food Chem 66(47):12527–12535. https://doi.org/10.1021/acs.jafc.8b03591
Choi J, Eom S, Shin K, Lee R-A, Choi S, Lee J-H, Lee S, Soh M-S (2019) Identification of lysine histidine transporter 2 as an 1-aminocyclopropane carboxylic acid transporter in Arabidopsis thaliana by transgenic complementation approach. Front Plant Sci. https://doi.org/10.3389/fpls.2019.01092
Coca M, San Segundo B (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63(3):526–540. https://doi.org/10.1111/j.1365-313X.2010.04255.x
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA 106(38):16428–16433
Ding S, Shao X, Li J, Ahammed GJ, Yao Y, Ding J, Hu Z, Yu J, Shi K (2021) Nitrogen forms and metabolism affect plant defence to foliar and root pathogens in tomato. Plant Cell Environ 44(5):1596–1610. https://doi.org/10.1111/pce.14019
Dinkeloo K, Boyd S, Pilot G (2018) Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin Cell Dev Biol 74:105–113. https://doi.org/10.1016/j.semcdb.2017.07.010
Duan Y, Zhu X, Shen J, Xing H, Zou Z, Ma Y, Wang Y, Fang W (2020) Genome-wide identification, characterization and expression analysis of the amino acid permease gene family in tea plants (Camellia sinensis). Genomics 112(4):2866–2874. https://doi.org/10.1016/j.ygeno.2020.03.026
Durian G, Sedaghatmehr M, Matallana-Ramirez LP, Schilling SM, Schaepe S, Guerra T, Herde M, Witte C-P, Mueller-Roeber B, Schulze WX, Balazadeh S, Romeis T (2020) Calcium-dependent protein kinase CPK1 controls cell death by in vivo phosphorylation of senescence master regulator ORE1[OPEN]. Plant Cell 32(5):1610–1625. https://doi.org/10.1105/tpc.19.00810
du Toit Y, Coles DW, Mewalal R, Christie N, Naidoo S (2020) eCALIBRATOR: a comparative tool to identify key genes and pathways for eucalyptus defense against biotic stressors. Front Microbiol 11:216. https://doi.org/10.3389/fmicb.2020.00216
Elashry A, Okumoto S, Siddique S, Koch W, Kreil DP, Bohlmann H (2013) The AAP gene family for amino acid permeases contributes to development of the cyst nematode Heterodera schachtii in roots of Arabidopsis. Plant Physiol Biochem 70:379–386. https://doi.org/10.1016/j.plaphy.2013.05.016
Farage-Barhom S, Burd S, Sonego L, Mett A, Belausov E, Gidoni D, Lers A (2011) Localization of the Arabidopsis senescence-and cell death-associated BFN1 nuclease: from the ER to fragmented nuclei. Mol Plant 4(6):1062–1073. https://doi.org/10.1093/mp/ssr045
Farjad M, Rigault M, Pateyron S, Martin-Magniette ML, Krapp A, Meyer C, Fagard M (2018) Nitrogen limitation alters the response of specific genes to biotic stress. Int J Mol Sci 19(11):3364. https://doi.org/10.3390/ijms19113364
Fatima U, Senthil-Kumar M (2015) Plant and pathogen nutrient acquisition strategies. Front Plant Sci 6:750. https://doi.org/10.3389/fpls.2015.00750
Fischer WN, Kwart M, Hummel S, Frommer WB (1995) Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem 270(27):16315–16320. https://doi.org/10.1074/jbc.270.27.16315
Frommer WB, Hummel S, Riesmeier JW (1993) Expression cloning in yeast of a cDNA encoding a broad specificity amino acid permease from Arabidopsis thaliana. Proc Natl Acad Sci USA 90(13):5944–5948. https://doi.org/10.1073/pnas.90.13.5944
Froschel C, Komorek J, Attard A, Marsell A, Lopez-Arboleda WA, Le Berre J, Wolf E, Geldner N, Waller F, Korte A, Droge-Laser W (2021) Plant roots employ cell-layer-specific programs to respond to pathogenic and beneficial microbes. Cell Host Microbe 29(2):299-310.e297. https://doi.org/10.1016/j.chom.2020.11.014
Ganeteg U, Ahmad I, Jämtgård S, Aguetoni-Cambui C, Inselsbacher E, Svennerstam H, Schmidt S, Näsholm TJP (2017) Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant Cell Environ 40(3):413–423
Garneau MG, Tan Q, Tegeder M (2018) Function of pea amino acid permease AAP6 in nodule nitrogen metabolism and export, and plant nutrition. J Exp Bot 69(21):5205–5219. https://doi.org/10.1093/jxb/ery289
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Gratz R, Ahmad I, Svennerstam H, Jämtgård S, Love J, Holmlund M, Ivanov R, Ganeteg U (2021) Organic nitrogen nutrition: LHT1.2 protein from hybrid aspen (Populus tremula L. x tremuloides Michx) is a functional amino acid transporter and a homolog of Arabidopsis LHT1. Tree Physiol. https://doi.org/10.1093/treephys/tpab029
Guether M, Volpe V, Balestrini R, Requena N, Wipf D, Bonfante P (2011) LjLHT1.2—a mycorrhiza-inducible plant amino acid transporter from Lotus japonicus. Biol Fertil Soils 47(8):925–936. https://doi.org/10.1007/s00374-011-0596-7
Guo N, Hu J, Yan M, Qu H, Luo L, Tegeder M, Xu G (2020) Oryza sativa lysine-histidine-type transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J 103(1):395–411. https://doi.org/10.1111/tpj.14742
Gupta A, Patil M, Qamar A, Senthil-Kumar M (2020) ath-miR164c influences plant responses to the combined stress of drought and bacterial infection by regulating proline metabolism. Environ Exp Bot 172:103998. https://doi.org/10.1016/j.envexpbot.2020.103998
Hammes UZ, Schachtman DP, Berg RH, Nielsen E, Koch W, McIntyre LM, Taylor CG (2005) Nematode-induced changes of transporter gene expression in Arabidopsis roots. Mol Plant Microbe Interact 18(12):1247–1257. https://doi.org/10.1094/MPMI-18-1247
Hammes UZ, Nielsen E, Honaas LA, Taylor CG, Schachtman DP (2006) AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J 48(3):414–426. https://doi.org/10.1111/j.1365-313X.2006.02880.x
Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W (2006) Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18(8):1931–1946. https://doi.org/10.1105/tpc.106.041012
Hsu LC, Chiou TJ, Chen L, Bush DR (1993) Cloning a plant amino acid transporter by functional complementation of a yeast amino acid transport mutant. Proc Natl Acad Sci USA 90(16):7441–7445. https://doi.org/10.1073/pnas.90.16.7441
Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10(8):1321–1332. https://doi.org/10.1105/tpc.10.8.1321
Huang H, Nguyen Thi Thu T, He X, Gravot A, Bernillon S, Ballini E, Morel JB (2017) Increase of fungal pathogenicity and role of plant glutamine in nitrogen-induced susceptibility (NIS) to rice blast. Front Plant Sci 8:265. https://doi.org/10.3389/fpls.2017.00265
Huang S, Zhang X, Fernando WD (2020) Directing trophic divergence in plant-pathogen interactions: antagonistic phytohormones with no doubt? Front Plant Sci. https://doi.org/10.3389/fpls.2020.600063
Inselsbacher E, Näsholm TJNP (2012) The below-ground perspective of forest plants: soil provides mainly organic nitrogen for plants and mycorrhizal fungi. New Phytol 195(2):329–334
Jiang X, Xie Y, Ren Z, Ganeteg U, Lin F, Zhao C, Xu H (2018) Design of a new glutamine-fipronil conjugate with alpha-amino acid function and its uptake by A. thaliana lysine histidine transporter 1 ( AtLHT1). J Agric Food Chem 66(29):7597–7605. https://doi.org/10.1021/acs.jafc.8b02287
Killiny N, Hijaz F (2016) Amino acids implicated in plant defense are higher in Candidatus Liberibacter asiaticus-tolerant citrus varieties. Plant Signal Behav 11(4):e1171449. https://doi.org/10.1080/15592324.2016.1171449
Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science (New York, NY) 323(5917):1053–1057. https://doi.org/10.1126/science.1166386
Kim HJ, Hong SH, Kim YW, Lee IH, Jun JH, Phee BK, Rupak T, Jeong H, Lee Y, Hong BS, Nam HG, Woo HR, Lim PO (2014) Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot 65(14):4023–4036. https://doi.org/10.1093/jxb/eru112
Kuzyakov Y, Xu X (2013) Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol 198(3):656–669. https://doi.org/10.1111/nph.12235
Lacey RF, Binder BM (2014) How plants sense ethylene gas—the ethylene receptors. J Inorg Biochem 133:58–62. https://doi.org/10.1016/j.jinorgbio.2014.01.006
Ladwig F, Stahl M, Ludewig U, Hirner AA, Hammes UZ, Stadler R, Harter K, Koch W (2012) Siliques are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol 158(4):1643–1655. https://doi.org/10.1104/pp.111.192583
Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M (2007) AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J 50(2):305–319. https://doi.org/10.1111/j.1365-313X.2007.03045.x
Li Z, Peng J, Wen X, Guo H (2013) Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25(9):3311–3328. https://doi.org/10.1105/tpc.113.113340
Li Y, Wang X, Zeng Y, Liu P (2020) Metabolic profiling reveals local and systemic responses of kiwifruit to Pseudomonas syringae pv. actinidiae. Plant Direct 4(12):e00297. https://doi.org/10.1002/pld3.297
Li F, Dong C, Yang T, Bao S, Fang W, Lucas WJ, Zhang Z (2021) The tea plant CsLHT1 and CsLHT6 transporters take up amino acids, as a nitrogen source, from the soil of organic tea plantations. Hortic Res 8(1):178. https://doi.org/10.1038/s41438-021-00615-x
Liao Q, Tang T-J, Zhou T, Song H-X, Hua Y-P, Zhang Z-H (2020) Integrated transcriptional and proteomic profiling reveals potential amino acid transporters targeted by nitrogen limitation adaptation. Int J Mol Sci 21(6):2171. https://doi.org/10.3390/ijms21062171
Limpens E, Moling S, Hooiveld G, Pereira PA, Bisseling T, Becker JD, Kuster H (2013) Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE 8(5):e64377. https://doi.org/10.1371/journal.pone.0064377
Liu G, Ji Y, Bhuiyan NH, Pilot G, Selvaraj G, Zou J, Wei Y (2010) Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22(11):3845–3863. https://doi.org/10.1105/tpc.110.079392
Llebrés MT, Castro-Rodríguez V, Pascual MB, Avila C, Cánovas FM (2021) The amino acid permease PpAAP1 mediates arginine transport in maritime pine. Tree Physiol. https://doi.org/10.1093/treephys/tpab089
Marella HH, Nielsen E, Schachtman DP, Taylor CG (2013) The amino acid permeases AAP3 and AAP6 are involved in root-knot nematode parasitism of Arabidopsis. Mol Plant Microbe Interact 26(1):44–54. https://doi.org/10.1094/MPMI-05-12-0123-FI
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105(7):1141–1157
Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue G-P, Dröge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant 6(5):1438–1452. https://doi.org/10.1093/mp/sst012
Moussatche P, Klee HJ (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279(47):48734–48741. https://doi.org/10.1074/jbc.M403100200
Muller B, Fastner A, Karmann J, Mansch V, Hoffmann T, Schwab W, Suter-Grotemeyer M, Rentsch D, Truernit E, Ladwig F, Bleckmann A, Dresselhaus T, Hammes UZ (2015) Amino acid export in developing Arabidopsis seeds depends on UmamiT facilitators. Curr Biol 25(23):3126–3131. https://doi.org/10.1016/j.cub.2015.10.038
Nasholm T, Kielland K, Ganeteg U (2009) Uptake of organic nitrogen by plants. New Phytol 182(1):31–48. https://doi.org/10.1111/j.1469-8137.2008.02751.x
Nishad R, Ahmed T, Rahman VJ, Kareem A (2020) Modulation of plant defense system in response to microbial interactions. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01298
Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12(3):527–535. https://doi.org/10.1046/j.1365-313x.1997.00527.x
Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277(47):45338–45346. https://doi.org/10.1074/jbc.M207730200
Okumoto S, Koch W, Tegeder M, Fischer WN, Biehl A, Leister D, Stierhof YD, Frommer WB (2004) Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J Exp Bot 55(406):2155–2168. https://doi.org/10.1093/jxb/erh233
Omari Alzahrani F (2021) Genome wide analysis of amino acid transporter superfamily in Solanum lycopersicum. Plants (Basel) 10(2):289. https://doi.org/10.3390/plants10020289
Pariyar SR, Nakarmi J, Anwer MA, Siddique S, Ilyas M, Elashry A, Dababat AA, Leon J, Grundler FM (2018) Amino acid permease 6 modulates host response to cyst nematodes in wheat and Arabidopsis. Nematology 20(8):737–750. https://doi.org/10.1163/15685411-00003172
Park BS, Yao T, Seo JS, Wong ECC, Mitsuda N, Huang C-H, Chua N-H (2018) Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat Plant 4(11):898–903. https://doi.org/10.1038/s41477-018-0269-8
Park SH, Jeong JS, Seo JS, Park BS, Chua NH (2019) Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol 223(3):1447–1460. https://doi.org/10.1111/nph.15879
Pastor V, Gamir J, Camanes G, Cerezo M, Sanchez-Bel P, Flors V (2014) Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front Plant Sci 5:231. https://doi.org/10.3389/fpls.2014.00231
Paungfoo-Lonhienne C, Lonhienne TG, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S (2008) Plants can use protein as a nitrogen source without assistance from other organisms. Proc Natl Acad Sci USA 105(11):4524–4529. https://doi.org/10.1073/pnas.0712078105
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521. https://doi.org/10.1146/annurev-cellbio-092910-154055
Pratelli R, Pilot G (2014) Regulation of amino acid metabolic enzymes and transporters in plants. J Exp Bot 65(19):5535–5556. https://doi.org/10.1093/jxb/eru320
Przybylska A, Obrępalska-Stęplowska A (2020) Plant defense responses in monocotyledonous and dicotyledonous host plants during root-knot nematode infection. Plant Soil. https://doi.org/10.1007/s11104-020-04533-0
Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, Zhou X (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11(7):e1005399. https://doi.org/10.1371/journal.pgen.1005399
Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581(12):2281–2289. https://doi.org/10.1016/j.febslet.2007.04.013
Roberts P, Jones DL (2012) Microbial and plant uptake of free amino sugars in grassland soils. Soil Biol Biochem 49:139–149. https://doi.org/10.1016/j.soilbio.2012.02.014
Santiago JP, Tegeder M (2016) Connecting source with sink: the role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol 171(1):508–521. https://doi.org/10.1104/pp.16.00244
Sharma I (2020) Chapter 7—phytopathogenic fungi and their biocontrol applications. In: Sharma VK, Shah MP, Parmar S, Kumar A (eds) Fungi bio-prospects in sustainable agriculture, environment and nano-technology. Academic Press, Cambridge, pp 155–188. https://doi.org/10.1016/C2019-0-04952-7
Shin K, Lee S, Song W-Y, Lee R-A, Lee I, Ha K, Koo J-C, Park S-K, Nam H-G, Lee Y, Soh M-S (2015) Genetic identification of ACC-RESISTANT2 reveals involvement of LYSINE HISTIDINE TRANSPORTER1 in the uptake of 1-aminocyclopropane-1-carboxylic acid in Arabidopsis thaliana. Plant Cell Physiol 56(3):572–582. https://doi.org/10.1093/pcp/pcu201
Smirnoff N, Cumbes QJ (1989) Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 28(4):1057–1060. https://doi.org/10.1016/0031-9422(89)80182-7
Sonawala U, Dinkeloo K, Danna CH, McDowell JM, Pilot G (2018) Functional linkages between amino acid transporters and plant responses to pathogens. Plant Sci 277:79–88. https://doi.org/10.1016/j.plantsci.2018.09.009
Spanu PD, Panstruga R (2017) Editorial: biotrophic plant-microbe interactions. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00192
Struck C, Mueller E, Martin H, Lohaus G (2004) The Uromyces fabae UfAAT3 gene encodes a general amino acid permease that prefers uptake of in planta scarce amino acids. Mol Plant Pathol 5(3):183–189. https://doi.org/10.1111/j.1364-3703.2004.00222.x
Sun Y, Wang M, Mur LAJ, Shen Q, Guo S (2020) Unravelling the roles of nitrogen nutrition in plant disease defences. Int J Mol Sci 21(2):572. https://doi.org/10.3390/ijms21020572
Svennerstam H, Ganeteg U, Bellini C, Näsholm T (2007) Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol 143(4):1853–1860. https://doi.org/10.1104/pp.106.092205
Svennerstam H, Ganeteg U, Nasholm T (2008) Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol 180(3):620–630. https://doi.org/10.1111/j.1469-8137.2008.02589.x
Svennerstam H, Jamtgard S, Ahmad I, Huss-Danell K, Nasholm T, Ganeteg U (2011) Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. New Phytol 191(2):459–467. https://doi.org/10.1111/j.1469-8137.2011.03699.x
Szabo LJ, Bushnell WR (2001) Hidden robbers: the role of fungal haustoria in parasitism of plants. Proc Natl Acad Sci USA 98(14):7654–7655. https://doi.org/10.1073/pnas.151262398
Tegeder M, Masclaux-Daubresse C (2018) Source and sink mechanisms of nitrogen transport and use. New Phytol 217(1):35–53. https://doi.org/10.1111/nph.14876
Tegeder M, Rentsch D (2010) Uptake and partitioning of amino acids and peptides. Mol Plant 3(6):997–1011. https://doi.org/10.1093/mp/ssq047
Tegeder M, Ward JM (2012) Molecular evolution of plant AAP and LHT amino acid transporters. Front Plant Sci 3:21. https://doi.org/10.3389/fpls.2012.00021
Val-Torregrosa B, Bundó M, Chiou T-J, Flors V, Segundo BS (2021-preprint) NITROGEN LIMITATION ADAPTATION functions as a negative regulator of Arabidopsis immunity. bioRxiv. https://doi.org/10.1101/2021.12.09.471910
Val-Torregrosa B, Bundó M, San Segundo B (2021) Crosstalk between nutrient signalling pathways and immune responses in rice. Agriculture 11(8):747
Van de Poel B, Van Der Straeten D (2014) 1-Aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front Plant Sci. https://doi.org/10.3389/fpls.2014.00640
Vanderstraeten L, Depaepe T, Bertrand S, Van Der Straeten D (2019) The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front Plant Sci. https://doi.org/10.3389/fpls.2019.01591
Wang X, Yang G, Shi M, Hao D, Wei Q, Wang Z, Fu S, Su Y, Xia J (2019) Disruption of an amino acid transporter LHT1 leads to growth inhibition and low yields in rice. BMC Plant Biol 19(1):268. https://doi.org/10.1186/s12870-019-1885-9
Wang Y, Wang Y, Wang Y (2020) Apoplastic proteases: powerful weapons against pathogen infection in plants. Plant Commun 1(4):100085. https://doi.org/10.1016/j.xplc.2020.100085
Wilkinson A, Hill PW, Farrar JF, Jones DL, Bardgett RD (2014) Rapid microbial uptake and mineralization of amino acids and peptides along a grassland productivity gradient. Soil Biol Biochem 72:75–83. https://doi.org/10.1016/j.soilbio.2014.01.026
Xiong J-S, Zhu H-Y, Bai Y-B, Liu H, Cheng Z-M (2018) RNA sequencing-based transcriptome analysis of mature strawberry fruit infected by necrotrophic fungal pathogen Botrytis cinerea. Physiol Mol Plant Pathol 104:77–85. https://doi.org/10.1016/j.pmpp.2018.08.005
Yang H, Postel S, Kemmerling B, Ludewig U (2014) Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ 37(6):1404–1414. https://doi.org/10.1111/pce.12244
Yang G, Wei Q, Huang H, Xia J (2020) Amino acid transporters in plant cells: a brief review. Plants (Basel) 9(8):967. https://doi.org/10.3390/plants9080967
Yao X, Nie J, Bai R, Sui X (2020) Amino acid transporters in plants: identification and function. Plants (Basel) 9(8):972. https://doi.org/10.3390/plants9080972
Zeier J (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36(12):2085–2103. https://doi.org/10.1111/pce.12122
Zhang L, Tan Q, Lee R, Trethewy A, Lee YH, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22(11):3603–3620. https://doi.org/10.1105/tpc.110.073833
Zhang R, Zhu J, Cao HZ, Xie XL, Huang JJ, Chen XH, Luo ZY (2013) Isolation and characterization of LHT-type plant amino acid transporter gene from Panax ginseng Meyer. J Ginseng Res 37(3):361–370. https://doi.org/10.5142/jgr.2013.37.361
Zhang Y, Wang H-L, Li Z, Guo H (2020) Genetic network between leaf senescence and plant immunity: crucial regulatory nodes and new insights. Plants (Basel) 9(4):495. https://doi.org/10.3390/plants9040495
Zhang Y, Gao Y, Wang HL, Kan C, Li Z, Yang X, Yin W, Xia X, Nam HG, Li Z, Guo H (2021) Verticillium dahliae secretory effector PevD1 induces leaf senescence by promoting ORE1-mediated ethylene biosynthesis. Mol Plant 14(11):1901–1917. https://doi.org/10.1016/j.molp.2021.07.014
Zhao H, Ma H, Yu L, Wang X, Zhao J (2012) Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 7(11):e49210. https://doi.org/10.1371/journal.pone.0049210
Zhao Y, Xu Y, Wang Z, Zhang J, Chen X, Li Z, Li Z, Jin L, Wei P, Zhang L (2017) Genome-wide identification and characterization of an amino acid permease gene family in Nicotiana tabacum. RSC Adv 7(60):38081–38090. https://doi.org/10.1039/C7RA05610A
Acknowledgements
We thank the Kempe Foundations and the Knut and Alice Wallenberg Foundation for financial support. In addition, we are grateful to Dr. Rumen Ivanov for his valuable comments on this manuscript. Sincere apologies to all colleagues whose scientific contributions could not be included in this review due to space limitations.
Funding
Open access funding provided by Swedish University of Agricultural Sciences. This review was supported by the Kempe Foundations (Grant Numbers JCK-2015.1 and JCK-2122 awarded to Torgny Näsholm and Regina Gratz) and the Knut and Alice Wallenberg Foundation (Grant Number 2016.0352 awarded to Torgny Näsholm).
Author information
Authors and Affiliations
Contributions
Conceptualization: RG; Literature search: LT, JC, RG; Writing—original draft preparation: LT, RG; Writing—review and editing: LT, JC, TN, RG; Funding acquisition: TN, RG; Supervision: TN, RG.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tünnermann, L., Colou, J., Näsholm, T. et al. To have or not to have: expression of amino acid transporters during pathogen infection. Plant Mol Biol 109, 413–425 (2022). https://doi.org/10.1007/s11103-022-01244-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-022-01244-1