Abstract
Purpose
The main objective of this work is to compare the standard bioequivalence tests based on individual estimates of the area under the curve and the maximal concentration obtained by non-compartmental analysis (NCA) to those based on individual empirical Bayes estimates (EBE) obtained by nonlinear mixed effects models.
Methods
We evaluate by simulation the precision of sample means estimates and the type I error of bioequivalence tests for both approaches. Crossover trials are simulated under H 0 using different numbers of subjects (N) and of samples per subject (n). We simulate concentration-time profiles with different variability settings for the between-subject and within-subject variabilities and for the variance of the residual error.
Results
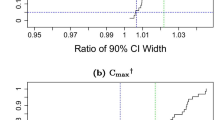
Bioequivalence tests based on NCA show satisfactory properties with low and high variabilities, except when the residual error is high, which leads to a very poor type I error, or when n is small, which leads to biased estimates. Tests based on EBE lead to an increase of the type I error, when the shrinkage is above 20%, which occurs notably when NCA fails.
Conclusions
For small n or data with high residual error, tests based on a global data analysis should be considered instead of those based on individual estimates.





Similar content being viewed by others
REFERENCES
FDA. Guidance for industry-statistical approaches to establishing bioequivalence. Technical report, FDA; 2001.
EMEA. Note for guidance on the investigation of bioavailability and bioequivalence. Technical report, EMEA; 2001.
Gabrielson J, Weiner D. Pharmacokinetic and pharmacodynamic data analysis: concepts and applications. Stockholm: Apotekarsocieteten; 2006.
Jusko WJ, Koup JR, Alván G. Nonlinear assessment of phenytoin bioavailability. J Pharmacokinet Biopharm. 1976;4:327–36.
Hayashi N, Aso H, Higashida M, Kinoshita H, Ohdo S, Yukawa E, et al. Estimation of rhG-CSF absorption kinetics after subcutaneous administration using a modified Wagner-Nelson method with a nonlinear elimination model. Eur J Pharm Sci. 2001;13:151–8.
EMEA. Guideline on similar biological medicinal products containing biotechnology-derived proteins as active substance: non-clinical and clinical issues. Technical report, EMEA; 2006.
Kaniwa N, Aoyagi N, Ogata H, Ishii M. Application of the NONMEM method to evaluation of the bioavailability of drug products. J Pharm Sci. 1990;79:1116–20.
Pentikis H, Henderson J, Tran N, Ludden T. Bioequivalence: individual and population compartmental modeling compared to noncompartmental approach. Pharm Res. 1996;13:1116–21.
Combrink M, McFadyen M-L, Miller R. A comparison of standard approach and the NONMEM approach in the estimation of bioavailability in man. J Pharm Pharmacol. 1997;49:731–3.
Maier GA, Lockwood GF, Oppermann JA, Wei G, Bauer P, Fedler-Kelly J, et al. Characterization of the highly variable bioavailability of tiludronate in normal volunteers using population pharmacokinetic methodologies. Eur J Drug Metab Pharmacokinet. 1999;24:249–54.
Hu C, Moore K, Kim Y, Sale M. Statistical issues in a modeling approach to assessing bioequivalence or PK similarity with presence of sparsely sampled subjects. J Pharmacokinet Pharmacodyn. 2003;31:312–39.
Zhou H, Mayer P, Wajdula J, Fatenejad S. Unaltered etanercept pharmacokinetics with concurrent methotrexate in patients with rheumatoid arthritis. J Clin Pharmacol. 2004;44:1235–43.
Fradette C, Lavigne J, Waters D, Ducharme M. The utility of the population approach applied to bioequivalence in patients. Ther Drug Monit. 2005;27:592–600.
Panhard X, Mentré F. Evaluation by simulation of tests based on non-linear mixed-effects models in pharmacokinetic interaction and bioequivalence cross-over trials. Stat Med. 2005;24:1509–24.
Hauschke D, Steinijans V, Pigeot I. Bioequivalence studies in drug development. Chichester: Wiley; 2007.
Lindstrom M, Bates D. Nonlinear mixed effects models for repeated measures data. Biometrics. 1990;46:673–87.
Pinheiro JC, Bates DM. Mixed-effects models in S and Splus. New York: Springer; 2000.
Delyon B, Lavielle M, Moulines E. Convergence of a stochastic approximation version of EM algorithm. Ann Stat. 1999;27:94–128.
Kuhn E, Lavielle M. Coupling a stochastic approximation version of EM with a MCMC procedure. ESAIM P & S. 2004;8:115–31.
Samson A, Lavielle M, Mentré F. The SAEM algorithm for group comparison tests in longitudinal data analysis based on non-linear mixed-effects model. Stat Med. 2007;26:4860–75.
The MONOLIX software http://software.monolix.org/ (accessed 05/07/09).
Lavielle M, Mentré F. Estimation of population pharmacokinetic of saquinavir in HIV patients and covariate analysis with the SAEM algorithm. J Pharmacokinet Pharmacodyn. 2007;34:229–49.
Comets E, Verstuyft C, Lavielle M, Jaillon P, Becquemont L, Mentré F. Modelling the influence of MDR1 polymorphism on digoxin pharmacokinetic parameters. Eur J Clin Pharmacol. 2007;63:437–49.
Bertrand J, Treluyer JM, Panhard X, Tran A, Auleley S, Rey E, et al. Influence of pharmacogenetics on indinavir disposition and short-term response in HIV patients initiating HAART. Eur J Clin Pharmacol. 2009;65:667–78. doi:667.
Panhard X, Taburet AM, Piketti C, Mentré F. Impact of modelling intra-subject variability on tests based on non-linear mixed-effects models in cross-over pharmacokinetic trials with application to the interaction of tenofovir on atazanavir in HIV patients. Stat Med. 2007;26:1268–84.
Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15:657–80.
Chow SC, Liu JP. Design and analysis of bioavailability and bioequivalence studies. New-York: Marcel Dekker; 2000.
Verbeke G, Molenberghs G. Linear mixed models for longitudinal data. New York: Springer; 2001.
Brown H, Prescott R. Applied mixed models in medicine. 2nd ed. Chichester: Wiley; 2006.
Berger R, Hsu J. Bioequivalence trials, intersection-union tests and equivalence confidence sets. Stat Sci. 1996;11:283–319.
Liu JP, Weng CS. Bias of two one-sided tests procedures in assessment of bioequivalence. Stat Med. 1995;14:853–61.
Savi R, Karlsson M. Shrinkage in empirical Bayes estimates for diagnosatics and estimation, 2007. PAGE 16 Abstr 1087 available at http://www.page-meeting.org/pdf_assets/9436-EBE_PAGE07_1_web.pdf (accessed 05/07/09).
Oehlert GW. A note on the delta method. Am Stat. 1992;46:27–9.
Girard P, Mentré F. A comparison of estimation methods in nonlinear mixed effects models using a blind analysis, 2005. PAGE 14 Abstr 834 available at http://www.page-meeting.org/page/page2005/PAGE2005O08.pdf (accessed 05/07/09).
Bauer R, Guzy S, Ng C. Survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. The AAPS Journal. 2007;9:60–83.
Rescigno A, Powers J, Herderick EE. Bioequivalent or nonbioequivalent? Pharmacol Res. 2001;43:543–6.
Bertrand J, Comets E, Laffont C, Chenel M, Mentré F. Pharmacogenetics and population pharmacokinetics: impact of the design on three tests using the SAEM algorithm. J Pharmacokinet Pharmacodyn. 2009;36:317–39.
Brown LD, Hwang JTG, Munk A. An unbiased test for the bioequivalence problem. Ann Stat. 1997;25:2345–67.
Cao L, Mathew T. A simple numerical approach toward improving the two-one sided test for average bioequivalence. Biom J. 2008;50:205–11.
Panhard X, Samson A. Extension of the SAEM algorithm for nonlinear mixed models with two levels of random effects. Biostat. 2009;10:121–35.
ACKNOWLEDGMENTS
We would like to thank the Modeling and Simulations group at Novartis Pharma AG, Basel, which supports by a grant Anne Dubois during this work.
Author information
Authors and Affiliations
Corresponding author
APPENDIX
APPENDIX
Approximation of the Variance of log(C max ) by the Delta Method
For a one-compartment model with first-order absorption and first-order elimination, C max is defined in Eq. 6 as a function of the three PK parameters: k a , CL/F and V/F. The variance of log(C max ), \( {{\omega }}_{{{\text{C}}_{{ \max }}}}^{\text{2}} \), is approximated by the delta method (33) as follows:
where log(μ) = (log(μ ka ), log(μ CL/F), log(μ V/F ))′. After computing the derivatives, \( {{\omega }}_{{{\text{C}}_{{ \max }}}}^{\text{2}} \) can be approximated by the following:
In this simulation study, the general formula above is applied to approximate the variance of log(C max ) for both treatment groups (Ref and Test). Given the treatment effect we simulate for the treatment Test, both approximations, \( \omega_{C_{\max }^{\left( {{\rm Re} f} \right)}}^2 \) and \( \omega_{C_{\max }^{\left( {Test} \right)}}^2 \), are equal.
To approximate the variance of log(C max ) by the delta method, we use the true simulated values of μ(Ref) and Ω(Ref) described in “Estimation Based on Nonlinear Mixed Effects Model.” To evaluate the delta method, we also estimate the variance of log(C max ), using the simulated parameter values of the rich design (N = 40, n = 10) for the reference treatment, under S l,l and S h,l . For both variability settings, \( \omega_{C_{\max }^{\left( {Ref} \right)}}^2 \) is estimated as the empirical variance of the 40000 true simulated values of log\( \left( {{C_{\max }}_i^{\left( {{\rm Re} f} \right)}} \right) \). For S l,l , the standard deviation of log(C max ) for the reference treatment expressed in percent is 10.5% both by simulation and the delta method. For S h,l , it is 46.3% and 46.7% by simulation and the delta method, respectively.
These results on the true simulated values validate the approximation of the variance of log(C max ) by the delta method. Consequently, we apply it to the data of each treatment group for each simulated trial of the simulation study to approximate \( \widehat\omega_{C_{_{\max }}^{\left( {{\rm Re} f} \right)}}^2 \) (\( \widehat\omega_{C_{_{\max }}^{\left( {Test} \right)}\;}^2 \)respectively) using \( {\mu^{\left( {{\rm Re} f} \right)}} \) (\( {\widehat\mu^{\left( {Test} \right)}} \) respectively) and \( {\widehat\Omega^{\left( {{\rm Re} f} \right)}} \) (\( {\widehat\Omega^{\left( {Test} \right)}} \) respectively).
Rights and permissions
About this article
Cite this article
Dubois, A., Gsteiger, S., Pigeolet, E. et al. Bioequivalence Tests Based on Individual Estimates Using Non-compartmental or Model-Based Analyses: Evaluation of Estimates of Sample Means and Type I Error for Different Designs. Pharm Res 27, 92–104 (2010). https://doi.org/10.1007/s11095-009-9980-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-009-9980-5