Abstract
Purpose
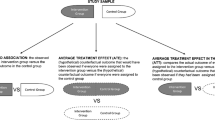
In this paper, we propose a robust Bayesian method for the assessment of average bioequivalence based on data from conventional crossover studies. We evaluate and motivate empirically the need for robust methods in bioequivalence studies by comparing the results of robust and conventional statistical methods in a large data pool of bioequivalence studies.
Methods
Robustness of the statistical methodology is achieved by replacing the normal distributions for residuals in the linear mixed model with skew-t distributions. In this way, the statistical model can accommodate skew and heavy-tailed data, particularly outliers, yielding robust statistical inference without the need for excluding outliers from the analysis. We performed a simulation study to investigate and compare the performance of the robust and conventional models.
Results
Our study shows that in some trials, the distribution of residuals is skew and heavy-tailed. In the presence of outliers, the 90% confidence intervals for the ratio of geometric means tend to be narrower for the robust methods than for the conventional method. Our simulation study shows that the robust method has suitable frequentist properties and yields more precise confidence intervals and higher statistical power than the conventional maximum likelihood method when outliers are present in the data.
Conclusions
As a sensitivity analysis, we recommend the fit of robust models for handling outliers that are occasionally encountered in crossover design bioequivalence data.





Similar content being viewed by others
Abbreviations
- θ :
-
GMR of bioavailability characteristic
- ABE:
-
Average bioequivalence
- ANOVA:
-
Analysis of variance
- BayesN :
-
Non-robust Bayesian model
- BayesT :
-
Robust Bayesian model
- CI:
-
Confidence interval
- CL:
-
Confidence limit
- eCDF:
-
Empirical cumulative distribution function
- GMR:
-
Geometric means ratio
- HPD:
-
Highest posterior density
- LML:
-
Log-marginal likelihood
- PK:
-
Pharmacokinetic
- REML:
-
Restricted maximum likelihood
- RMSE:
-
Root mean square error
- TOST:
-
Two one-sided test
References
FDA. Statistical approaches to establishing bioequivalence; 2001. Available at: https://www.fda.gov/ [accessed on 8 March 2020].
CHMP. Guideline on the investigation of bioequivalence; 2010. Available at: https://www.ema.europa.eu/en/ [accessed on 8 March 2020].
Meyners M. Equivalence tests–a review. Food Qual Prefer. 2012;26(2):231–45.
Schall R, Endrenyi L, Ring A. Residuals and outliers in replicate design crossover studies. J Biopharm Stat. 2010;20(4):835–49.
Schall R. The empirical coverage of confidence intervals: point estimates and confidence intervals for confidence levels. Biom J. 2012;54(4):537–51.
Chen ML, Blume H, Beuerle G, Mehta M, Potthast H, Brandt A, et al. Summary report of second EU-FEPS/AAPS conference on global harmonization in bioequivalence. Eur J Pharm Sci. 2019;127:24–8.
Chow SC, Tse SK. Outlier detection in bioavailability/bioequivalence studies. Stat Med. 1990;9(5):549–58.
Liu JP, Weng CS. Detection of outlying data in bioavailability/bioequivalence studies. Stat Med. 1991;10(9):1375–89.
Wang W, Chow SC. Examining outlying subjects and outlying records in bioequivalence trials. J Biopharm Stat. 2003;13(1):43–56.
Ramsay T, Elkum N. A comparison of four different methods for outlier detection in bioequivalence studies. J Biopharm Stat. 2004;15(1):43–52.
Zellner A. Bayesian and non-Bayesian analysis of the regression model with multivariate Student-t error terms. J Am Stat Assoc. 1976;71(354):400–5.
Lange KL, Little RJA, Taylor JMG. Robust statistical modeling using the t distribution. J Am Stat Assoc. 1989;84(408):881–96.
Pinheiro JC, Liu C, Wu YN. Efficient algorithms for robust estimation in linear mixed-effects models using the multivariate t distribution. J Comput Graph Stat. 2001;10(2):249–76.
Wang WL, Fan TH. Estimation in multivariate t linear mixed models for multiple longitudinal data. Stat Sin. 2011;21(4):1857–80.
Matos LA, Prates MO, Chen MH, Lachos VH. Likelihood-based inference for mixed-effects models with censored response using the multivariate-t distribution. Stat Sin. 2013;23(3):1323–45.
Azzalini A, Capitanio A. Distributions generated by perturbation of symmetry with emphasis on a multivariate skew t-distribution. J Roy Stat Soc B. 2003;65(2):367–89.
Azzalini A, Genton MG. Robust likelihood methods based on the skew-t and related distributions. Int Stat Rev. 2008;76(1):106–29.
Fernández C, Steel MFJ. On Bayesian modeling of fat tails and skewness. J Am Stat Assoc. 1998;93(441):359–71.
Yellowlees A, Bursa F, Fleetwood KJ, Charlton S, Hirst KJ, Sun R, et al. The appropriateness of robust regression in addressing outliers in an anthrax vaccine potency test. Bioscience. 2016;66(1):63–72.
Castro LM, Wang WL, Lachos VH, Inácio de Carvalho V, Bayes CL. Bayesian semiparametric modeling for HIV longitudinal data with censoring and skewness. Stat Methods Med Res. 2019;28(5):1457–76.
De Souza RM, Achcar JA, Martinez EZ, Mazucheli J. The use of asymmetric distributions in average bioequivalence. Stat Med. 2016;35(15):2525–42.
Ghosh P, Ntzoufras I. Testing population and individual bioequivalence: a hierarchical Bayesian approach. Department of Mathematics and Statistics, Georgia State University, Atlanta, GA & Department of Statistics, Athens University of Economics and Business, Greece; 2005.
Chow SC. Bioavailability and bioequivalence in drug development. Wiley Interdiscip Rev Comput Stat. 2014;6(4):304–12.
Arnold BC, Groeneveld RA. Measuring skewness with respect to the mode. Am Stat. 1995;49(1):34–8.
SAS Institute. SAS/IML user’s guide, Version 9.4. SAS Institute Cary, NC; 2013.
Robert CP. The Bayesian choice. 2nd ed. New York: Springer-Verlag; 2007.
Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 2006;1(3):515–33. https://doi.org/10.1214/06-BA117A.
Lunn D, Jackson C, Best N, Thomas A, Spiegelhalter D. The BUGS Book: A Practical Introduction to Bayesian Analysis. CRC Press; 2012.
Juárez MA, Steel MFJ. Model-based clustering of non-Gaussian panel data based on skew-t distributions. J Bus Econ Stat. 2010;28(1):52–66.
Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, et al. Stan: A probabilistic programming language. J Stat Softw 2017;76(1).
Stan Development Team. rstan: R interface to Stan; 2020. R package Version 2.19.3. Available from: http://CRAN.R-project.org/package=rstan
R Core Team. R: A language and environment for statistical computing. Vienna, Austria; 2018. Available from: https://www.R-project.org/
Ntzoufras I. Bayesian modeling using WinBUGS. Hoboken: John Wiley & Sons, Inc.; 2009.
Gronau QF, Singmann H, Wagenmakers EJ. bridgesampling: an R package for estimating normalizing constants. J Stat Softw. 2020;92(10):1–29.
Labes D, Schütz H. Inflation of type I error in the evaluation of scaled average bioequivalence, and a method for its control. Pharm Res. 2016;33(11):2805–14.
Tothfalusi L, Endrenyi L, Arieta AG. Evaluation of bioequivalence for highly variable drugs with scaled average bioequivalence. Clin Pharmacokinet. 2009;48(11):725–43.
ACKNOWLEDGMENTS AND DISCLOSURES
The datasets supporting this study’s findings are available on reasonable request from the corresponding author. The data are not publicly available due to privacy and ethics restrictions. The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work is based upon research supported by the South Africa National Research Foundation and South Africa Medical Research Council (South Africa DST-NRF-SAMRC SARChI Research Chair in Biostatistics, grant number 114613); and the Research Development Programme 219/2018 at the University of Pretoria, South Africa.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Burger, D.A., Schall, R. & van der Merwe, S. A robust method for the assessment of average bioequivalence in the presence of outliers and skewness. Pharm Res 38, 1697–1709 (2021). https://doi.org/10.1007/s11095-021-03110-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-021-03110-z