Abstract
The origin of translation is critical for understanding the evolution of life, including the origins of life. The canonical genetic code is one of the most dominant aspects of life on this planet, while the origin of heredity is one of the key evolutionary transitions in living world. Why the translation apparatus evolved is one of the enduring mysteries of molecular biology. Assuming the hypothesis, that during the emergence of life evolution had to first involve autocatalytic systems which only subsequently acquired the capacity of genetic heredity, we propose and discuss possible mechanisms, basic aspects of the emergence and subsequent molecular evolution of translation and ribosomes, as well as enzymes as we know them today. It is possible, in this sense, to view the ribosome as a digital-to-analogue information converter. The proposed mechanism is based on the abilities and tendencies of short RNA and polypeptides to fold and to catalyse biochemical reactions. The proposed mechanism is in concordance with the hypothesis of a possible chemical co-evolution of RNA and proteins in the origin of the genetic code or even more generally at the early evolution of life on Earth. The possible abundance and availability of monomers at prebiotic conditions are considered in the mechanism. The hypothesis that early polypeptides were folding on the RNA scaffold is also considered and mutualism in molecular evolutionary development of RNA and peptides is favoured.
Similar content being viewed by others
“Nature! We obey her laws even when we rebel against them; we work with her even when we desire to work against her.”
(Johann Wolfgang von Goethe)
“What we name as an idea; that what is shown up and for this reason is approaching to us as a law of all phenomena.”
(Johann Wolfgang von Goethe)
Introduction
Explaining the Emergence of Life - Perhaps the most Central and Challenging Question in Modern Science
For thousands of years, philosophers and naturalists have been on a quest to find an answer to the intriguing question ‘How did life begin?’ The question on the origin of life is perhaps as old as mankind. For the Greek philosophers, life was inherent to matter. It is eternal and appears spontaneously whenever the conditions are favourable. These ideas were clearly stated by Thales, Democritus, Epicurus, Lucretius and Plato. Aristotle gathered the different claims into a real theory. However, Louis Pasteur and Charles Darwin were the first to formulate a modern approach to the chemical origin of life (Brack 2010). Modern technology offers various scientific approaches to this intriguing question (Brack 2010; Goodwin et al. 2014). In 1924, the Russian biochemist Aleksander Oparin introduced his thesis, which states that, in its initial phase, the development of living creatures should have been the subject of a purely chemical evolution. Under the influence of various natural circumstances, such as, for example, electrical discharges, the first organic substances (simple organic molecules (monomers) such as amino acids) were synthesised from various inorganic substances, which led to the formation of the first complex organic substances and creation of the so-called “primordial soup.” The latter was supposed to be composed of molecules which represented the basic building blocks for the creation of the first living cells (Dixon 1994; Luisi 2006; Brack 2010). On Earth, life probably appeared at least 3.95 billion years ago (Tashiro et al. 2017) with molecular systems organized in liquid water. Though biopolymers like peptides, proteins or nucleic acid are thermodynamically unstable in liquid water, they are kinetically stabilized by high activation energy. However, they are unstable at an extreme pH (Kaddour and Sahai 2014). Hydrolysis by complex biomolecules is not very plausible at prebiotic conditions, although amyloid peptides that could be present before sustainable life first appeared, possess certain hydrolytic activities (Friedmann et al. 2015). However, if the rate of formation of biopolymers is assumed to be faster than the rate of hydrolysis, then this dilemma can be avoided. Amide bonds are unstable at extreme acidic and basic pH and their hydrolysis or condensation can be further catalysed by metal ions. The instability of particular covalent bonds has long been considered disadvantageous to their potential utility in prebiotic networks but recent advances in systems chemistry show that an equilibrium between condensation and hydrolysis may create dynamic combinatorial networks from which the kinetic selection of amyloid-forming peptides occurs. Such networks may have played a foundational role in the early stages of life’s origins on Earth (Taran et al. 2017). Because primitive life developed several billion years ago, it was very likely different from that existing today, hence only its hypothetical descriptions can be proposed. By analogy with contemporary life, it is generally believed that primitive life originated from the processing of organic molecules made of carbon, hydrogen, oxygen, nitrogen and sulphur, which are often referred to as the CHONS (Brack 2010). Albeit a posteriori and without knowledge of the actual chemical steps that carried this evolution, the single assessment one can safely make about life’s origin on Earth is that it must have been an emergent process, through which biogenic atoms and molecules gained their complex associative and interactive states which we can observe in even the simplest forms of extant life (Pizzarello and Shock 2010). Three general approaches are currently used to understand the nature of primitive life: i) the first approach includes the reconstitution of artificial life in a laboratory, ii) the second approach consists of geological research which also includes the search for fossil traces of life in Archeaen sediments, an investigation of volcanism including the submarine volcanism of hydrothermal systems that may have been an important source of biomolecules on primitive Earth (Martin et al. 2008; Dalai et al. 2016) and iii) the third and final approach, a research which is based on the hypothesis that the prebiotic organic molecules had an extraterrestrial origin.
Concerning the first approach, Oparin’s idea was tested and confirmed plausible with Miller and Urey’s famous experiment in 1953 (Miller 1953). The experiment simulated the Earth’s atmosphere from a couple of billion years ago. They also simulated the electric discharges. The result was astonishing, several proteinogenic amino acids and other organic compounds were formed, which supposedly composed the “primordial soup,” as it existed a couple of billion years ago. Such abiotic chemical mechanisms supplied the monomers that could be used for the synthesis of polymer molecules needed for life to emerge (Miller 1953; Maynard Smith and Szathmáry 1995; Brack 2010; Dalai et al. 2016). Miller’s laboratory synthesis of amino acids occurs efficiently when a reducing gas mixture containing significant amounts of hydrogen is used. On the contrary, the dominant view in recent years is that the early Earth atmosphere consisted mainly of carbon dioxide, nitrogen, and water along with small amounts of carbon monoxide and hydrogen (Kasting 1993; Kasting and Catling 2003; Kasting and Howard 2006; Shaw 2008). Only small amounts of amino acids are formed in such a mixture (Miller 1998). Recent studies show that the low yields previously reported appear to be the outcome of oxidation of the organic compounds during the hydrolytic workup by nitrite and nitrate produced in the reactions. The amount of amino acids is greatly increased when oxidation inhibitors, such as ferrous iron, are added, suggesting that endogenous synthesis from neutral atmospheres may be more important than previously thought (Cleaves et al. 2008; Brack 2010). However, despite the experimental successes of the prebiotic synthesis, there are several drawbacks like the time limitations to reproduce the emergence of life in the laboratory and the concentration problem. High concentrations of reactants are typically required; otherwise it is very unlikely that the reactants will react. Therefore, it is reasonable to infer, that it is also not yet possible to predict the complexity of life that can be created in a test tube. Moreover, the issue is not to synthesize specific molecules but the need for the spontaneous emergence of non-equilibrium self-organizing systems with an evolvable capacity (Tessera 2011; Vitas and Dobovišek 2014, 2017). Consequently, it might be unlikely that laboratory conditions have enough complexity to reconstruct or re-create the emergence of life. For those reasons, hydrothermal systems are considered as a possible important source of biomolecules on the primitive Earth. Black smokers and alkaline vents could both provide the ideal environment for supporting the emergence of the earliest metabolisms (Corliss et al. 1981; Corliss 1986; Martin et al. 2008; Dalai et al. 2016; Russell and Nitschke 2017). Hydrothermal vents are often disqualified as the reactors for the synthesis of bioorganic molecules, because of the high temperature (Brack 2010) and high pressure. In addition to that, it was recently discussed that hydrothermal vents are not appropriate for the synthesis of bioorganic molecules because inorganic membranes lying between the ocean and fluid issuing from hydrothermal alkali vents comprising the outermost compartments of the hydrothermal mound, do not form sharp gradients (Jackson 2016). Inorganic membranes consist mainly of precipitated iron in multiple distinct and chemically reactive forms, many of them of mixed valence (Russell and Hall 2009; Russell et al. 2013). There is also a hypothesis of an extraterrestrial origin of organic molecules, such as comets, meteorites and interplanetary dust, which are rich in carbonaceous material. However, it is certain that living organisms tend to keep their internal processes far from a thermodynamic equilibrium and under relatively (meta)stable conditions. In a good approximation, living organisms can be viewed as open systems in a stationary non-equilibrium state (Murphy and O'Neill 1997; Russell et al. 2013; Vitas and Dobovišek 2014). We can view life as an interconnection of thermodynamic stabilization and kinetic factors that accelerate the dissipation of free energy – a dynamic balance that is far from a thermodynamic equilibrium (Vitas 2011). Thus the hypothesis of the extraterrestrial origin of organic molecules suffers with the same drawback as the “primordial soup in warm pond” hypothesis - the need for a spontaneous emergence of non-equilibrium conditions when the molecules are precipitated on the Earth, as the non-equilibrium conditions were required at the emergence of life (Spitzer et al. 2015). Nevertheless, a recently postulated hypothesis overcomes this problem by defining first organic pigments as the fundamental molecules of life which probably absorbed, and dissipated the solar photon potential at photochemically active wavelengths (Michaelian and Simeonov 2015). Such pigments composed of pyrroles and porphirin rings could be provided abiotically (Hodgson and Ponnamperuma 1968; Simionescu et al. 1978; Fox and Strasdeit 2013). The proliferation of these pigments can be understood as an autocatalytic photochemical process obeying non-equilibrium thermodynamic directives related to an increasing solar photon dissipation rate. Under these directives, organic pigments would have evolved over time to increase the global photon dissipation rate (Michaelian and Simeonov 2015).
The origin of nucleotides, amino acids and other organic compounds is of great importance for the development of chemical evolution. In our case the importance is emphasised on the prebiotic formation of nucleo-polymers and polypeptides which will be discussed later on. Which came first, polynucleotide or polypeptide? The primacy of nucleo-polymers or polypeptides in prebiotic evolution is a frequently debated topic in the origin-of-life community (Pross 2004; Martin et al. 2008; Deamer 2009; Sharov 2009; Raffaelli 2011; Gordon-Smith 2011; Tessera 2011; Hordijk et al. 2012; Vitas and Dobovišek 2014; Adami 2015; Root-Bernstein and Root-Bernstein 2015; Sousa et al. 2015; Vitas and Dobovišek 2017). Both viewpoints face many severe problems, but the discussion on them exceeds the framework of the presented work. The issue of primacy is intuitively leading us to the next question.
The Origin of Translation – A Principal Puzzle Piece in Understanding the Origin and Evolution of Life
The ribosome represents the result of evolutionary development by growth that allowed two polymer classes, nucleic acids and proteins, to cooperatively set the central dogma of biology. The ribosome can be viewed as a digital-to-analogue converter for two distinct polymers; nucleic acids and proteins. Nucleic acids represent the digital part of the system, capable of storing large amounts of information with exact accuracy, while proteins are more analogous in their physicochemical engagement (Wächtershäuser 1998; Goodwin et al. 2012; Smith et al. 2014). One dimensional, linear, discrete information is expressed in a three dimensional analogue way (Vitas and Dobovišek 2017). The origin and evolution of translation and its corresponding genetic code are supposed to be a critical transition in the evolution of modern organisms (Szathmáry and Maynard Smith 1995; Szathmáry 2015). The canonical genetic code and translation thus represent one of the most dominant aspects of life on this planet, and its emergence is critical for understanding the evolution of life (Johnson and Wang 2010). As current scenarios are at the level of hypotheses, the origin of translation is still veiled in mystery, representing one of the principal puzzle pieces in the elucidation of the origins of life. Moreover, the problems connected to the origin of translation face the classical catch 22 paradoxical situation (Noller 2012). If the ribosome requires proteins to function, where did the proteins come from to make the first ribosome and its translation factors? It is questionable if we can explain such paradoxes solely by reductionism (Knight 2007). Nevertheless, the intention of the following discussion is to at least partially unveil the contradictions which are closely linked to the origin of translation.
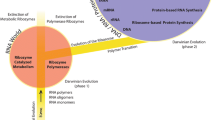
Our approach presumes that during the emergence of life, evolution had to first involve autocatalytic systems, which only subsequently acquired the capacity of genetic heredity. In the present work, we propose possible mechanisms for the emergence and subsequent molecular evolution of translation, ribosomes and enzymes as we know them today. However, since the ribosome is a ribozyme (Noller et al. 1992; Cech 2000; Ban et al. 2000; Moore and Steitz 2010; Noller 2012), it would be imprudent to overlook or to ignore the RNA World Hypothesis, which is based on the assumption that RNA polymers have primacy over other biopolymers in the origin of life. The central principle of the RNA World Hypothesis is commonly connected with the presumption that RNA could have made a copy of itself, thereby ‘setting the stage’ for evolution and hence biology (Higgs and Lehman 2014). The appeal of the RNA World Hypothesis is that it solves the “chicken or the-egg” problem; it shows that in an earlier, simplified biota the genotype/replicator and phenotype/catalyst could have been one and the same molecule. An exciting aspect of the current notion is that the genotype and phenotype could have been joined more simply, and earlier (Yarus 2010) in a ribozyme that was able to copy itself – a replicator capable of processive polymerase activity and information storage capacity. In other words, the RNA World hypothesis could be clearly categorized as a ‘template replication first’ hypothesis, vis-à-vis a ‘metabolism first’ hypothesis. Nevertheless, there are many problems and drawbacks that an RNA world scenario creates in terms of an “impossible” chemistry (Shapiro 2000, 2007). Let us, for the purpose of this discussion, expose only the most severe ones. For instance RNA’s building blocks are highly complex substances and the probability of their spontaneous synthesis is very low. Therefore, they require unrealistic conditions such as monomers (e.g., nucleotides) being readily available as resources for template-based synthesis (Sharov 2009; Kurland 2010), as presented in the The Nucleoside Problem. However, some progress has recently been reported (Pressman et al. 2015; Cafferty et al. 2016). Besides the so-called ‘error catastrophe’ and the difficulty of separating strands of moderate length, the hypothesized dual roles of RNA as both a digital information carrier and biocatalyst poses the following paradox: well-folded sequences are poor templates for copying, but poorly folded sequences are unlikely to be good ribozymes (Ivica et al. 2013 and references therein). One of the key problems of the RNA based scenario is also the high instability of RNA oligomers to hydrolysis at high temperatures or extreme pH values (Kaddour and Sahai 2014). Actually, even if the abiotic synthesis of genetic polymers was possible, the question would not be solved, as the issue is not to synthesize specific molecules – but rather to explain how non-equilibrium self-organizing systems with evolvable capacity emerged spontaneously (Tessera 2011). In this regard, there have been some attempts to describe and to connect the origins of RNA with mutually autocatalytic networks (Vaidya et al. 2012; Vasas et al. 2012; Higgs and Lehman 2014; Yeates and Lehman 2016). It is therefore worth hypothesizing that it was the chemistry that did the job in the first place, and continued to do so until protein enzymes took over (de Duve 1998). In the framework of our discussion on the origins of translation we are thus assuming, that RNA molecules were first of all, non-coding catalytic molecules involved in autocatalytic reaction networks without their role as repositories for digitalized information in template directed self-replication.
In this work, we use a previously proposed accretion model of cytochromes P450 (Vitas and Dobovišek 2017) as a tenet, which enables us to discuss the possible evolutionary development of biocatalysts. The previous model (Vitas and Dobovišek 2017) is extended and supplemented by the inclusion of the RNA-peptide development step. In this sense, ribosome is simultaneously considered as the observable remaining relict from early molecular evolution and the last survivor from the ancient RNA-peptide world. Thus, our discussion considers the co-evolution of RNA and proteins, which was already proposed as a possible and important situation for the chemical evolution of life (Brandman et al. 2012; Carter Jr and Wolfenden 2015).
Discussion
Is the Ribosome the Only Remaining Relict from Early Molecular Evolution, the RNA-Peptide World?
It is possible to define translation and the origins of translation via the irreducible Peircean semiotic triadic relation. The defining sign as the sequence of nucleotides in mRNA, object as a sequence of amino acids in the polypeptide (El-Hani et al. 2006) and ribosomes and post-translational machinery as an interpretant. The empirical testable definition hypothesis of interpretation might be of importance for solving the Origins of Life question and the interpretative role of ribosomes as ribozymes in a proto-biotic RNA world (Robinson and Southgate 2010; Lehman et al. 2014) where RNA has been bearing combined signalling, a regulating and catalytic role since the beginning (Orgel 2004; Deamer 2009; Cech and Steitz 2014; Villarreal and Witzany 2015; Witzany 2017). Moreover, the ribosome can also be viewed as a digital-to-analogue converter (Wächtershäuser 1998; Goodwin et al. 2014; Smith et al. 2014), where discrete information which is expanded in one dimension are translated and expressed into three-dimensional structures. The information in those three-dimensional structures is realized in an analogous way in the intra- and inter-molecular interactions. The converter, in our case the ribosome, and post-translational modification enzymes and epigenetic factors, should have well defined three-dimensional structures from where additional information is gained to enter the process of sequence conveyance. Therefore, this represents an aggravating circumstance for solving the origins of translation by reduction. Since the ribosome is a ribozyme (Noller et al. 1992; Cech 2000; Ban et al. 2000; Moore and Steitz 2010; Noller 2012), the origins of translation represent a challenging issue for origin of life research based on the RNA World Hypothesis (Bowman et al. 2015). In the following discussion, we will attempt to tackle this intriguing puzzle.
It is well known that nature benefits from the catalytic properties of transition metals such as Fe, Ni, Cu, Zn, Co, Mo, W, V and Mn, which are copiously used in various kinds of metal enzymes. Indeed, a vast number of exergonic but kinetically hindered (due to the high activation barriers) chemical reactions are accelerated by the presence of metal catalysts and many industrial processes take advantage of the same phenomenon. Many of the active centres of the metalloenzymes are affine with the structures of minerals, presumed to contribute to precipitate membranes produced in the mixing of hydrothermal solutions with the Hadean Ocean ~4 billion years ago (Baymann et al. 2003; Nitschke et al. 2013 and references therein). For instance, such hydrothermally catalytic iron sulphide membranes precipitated on the deep sea floor at the interface between sulfidic, alkaline, and highly reduced hot spring waters and the acidic, mildly oxidized, iron-bearing Hadean ocean. They supposedly formed the first reproducing “probotryoids” (iron sulphide bubbles) (Russell et al. 1994). Some authors thus give preference to the primordial role of metal catalysts such as iron, nickel, cobalt, molybdenum and tungsten, when explaining the origin of life (Yarus 2010). Intriguingly, the extremely evolutionary conserved ribosomal peptydil transferase centre (PTC) is supported by a framework of magnesium microclusters and it appears that Mg2+- microclusters are a primeval motif with pivotal roles in RNA folding, function and evolution (Hsiao and Williams 2009). On the other hand, coenzymes likely represent the oldest metabolic fossils within a cell, as suggested by their presence and necessity in all realms of life and the autocatalytic nature of their biosynthetic pathways. Coenzymes are often considered as remnants of the primordial metabolism. The autocatalytic nature of the coenzymes also speaks in favour of an ancient metabolic history (Jadhav and Yarus 2002; Copley et al. 2007; Sharov 2009, 2016; Raffaelli 2011). According to the review by Yarus (2010) minerals of iron, nickel, copper, manganese and molybdenum as well as nicotinamide containing cofactors were early participants in biochemistry, before the rise of complex RNA catalysts. The relation of RNA catalysts whose AMP-containing reaction centres were appropriated by protein enzymes and early peptides, which were putatively generated abiotically, will be discussed later on.
As we have already mentioned, RNA’s building blocks are highly complex substances and the probability of their spontaneous synthesis is very low (Sharov 2009). Nevertheless, via recent Miller-Urey experiments, RNA nucleobases synthesis has been achieved (Ferus et al. 2017). Alternatively, an extraterrestrial origin of nucleobases is also possible (Callahan et al. 2011). The more complex the molecule is, potentially the more likely it will interact with other molecules. If we compare nucleobases with amino acids, the latter are generally simpler molecules than the nucleobases. It was found that Ser-His dipeptide acts as a peptidyl transferase and catalyses the formation of peptide bonds, nucleotides-condensation and the synthesis of peptide nucleid acid (PNA) oligomers and also acts as protease and phosphoesterase (Gorlero et al. 2009; Adamala et al. 2014; Piast and Wieczorek 2017; Wieczorek et al. 2013, 2017). A specificity of reactions, however, would require more than just two amino acids, whereby specificity is promoted by larger interaction surfaces. Active sites of protein enzymes generally function as part of a larger structure that holds them in a proper alignment (Lehman et al. 2011; Lupas and Alva 2017; Piast and Wieczorek 2017). On the other hand, the smallest ribozyme ever described, comprising only five nucleotides acts to aminoacylate 4-nt RNA (Turk et al. 2010; Yarus 2011). Nevertheless, substrate specificity is an essential requirement for catalysis, which requires a specific structural fold (Wachowius et al. 2017). Both nucleic acids and proteins must assume a defined three-dimensional structure for their specificity and biological activity, which are prerequisites for the emergence and maintenance of complex life. This also requires a higher number of condensed monomers. How then, should the folding of RNA and polypeptides with respect to the primacy of the earlier appearance in the molecular evolution and in the context of the emergence of translation be compared? Which of them fold more readily? According to De Duve (De Duve 1995), reactions which generate life, should have been spontaneous and quick. This of course also applies to protein and RNA folding. The shortest logically possible stable secondary structure for RNA could be of length of 2, where at least one base pair incurs a stabilizing contribution to the base pair stacking. However, experimental studies have shown conclusively that the free nucleobases of RNA do not form Watson–Crick pairs in water; instead they form columnar co-planar stacks that represent a certain challenge for the current theories of RNA origins (Ts’o 1974; Hud 2016). This will be discussed further in the next section. In reality, RNA molecules start to form secondary structures in oligomers consisting of around 8 to 10 nucleotides at least (Cupal et al. 2000; Turner and Mathews 2010). Thus we may ask, what is the minimum number of amino acid types required to encode complex protein folds? Peptides start to fold in preferable stable structures at a similar number of monomers as do RNAs. In other words, the minimum number of amino acid residues in oligopeptide is in some cases between 8 and 10 (Fan and Wang 2003; Ho and Dill 2006). The minimum numbers for folding both sorts of polymers are similar. However, in general it is estimated that 20–30 amino acids are required in the shortest chains that fold (Ho and Dill 2006; Hwang 2012; Oda and Fukuyoshi 2015). As it was proposed that abiotically synthesised peptides would soon stabilize and begin to optimize metal-cofactor-based catalysts as well as introduce substrate-specificity (Brack 2007; Milner-White and Russell 2008, 2011; Nitschke et al. 2013; Vitas and Dobovišek 2014), the problem of short polypeptides folding became important in the light of early molecular evolution and the origin of translation. Multiple condensation pathways leading to the formation of short peptides have been established and recently reported amino acid polymerization assisted by volcanic gas (carbonyl sulphide) can lead to the formation of short amyloid-forming peptides (Taran et al. 2017). It seems highly unlikely that the earliest peptides could consist of large domains of tightly folded polypeptide chains as in present day proteins. Instead they would have been small, simple and heterochiral in nature. Without a genetic code as we know it, different polypeptide molecules probably had a variety of compositions and sequences and thus lacked defined large-scale three-dimensional structures. Early peptides were more exposed to solvent water and were more variable and motile in their 3D structure than present-day evolved proteins. This does not mean they lacked any structure at all, as, especially on the scale of a few Ángstrems, recurring features do occur (Milner-White and Russell 2011). Even for short homopeptide with the same monomer units might be expected to occupy certain structures or assembly with the capability to perform some biological activities (Shi et al. 2002; Friedmann et al. 2015). While nucleic acids fold spontaneously and robustly, and can in general be denatured and renatured reversibly, protein structure, in contrast, is altogether more complex. In addition, proteins tend to aggregate and can either not be renatured, or only with large loss of material, making denaturation a substantially irreversible process. Natural proteins nevertheless represent a best-case set, because in their overwhelming majority polypeptides do not appear to have a well folded structure at all. It is very difficult to estimate the actual proportion of folding polypeptides with any degree of accuracy. Given the difficulty polypeptides encounter to reach and maintain a folded state, and the exceedingly low likelihood of newly emerged polypeptides even having such a state, it is entirely non-trivial to explain how life came to rely so extensively on folded proteins (Oda and Fukuyoshi 2015; Lupas and Alva 2017). Making the problem of polypeptide folding even more acute, it is reasonable to assume that the first prebiotic amino acids were the most robust ones e.g. glycine, alanine, valine and aspartic acid, while less robust amino acids e.g. tryptophan, histidine, arginine, lysine entered biological systems by biotransformation (Miller 1998; Higgs and Pudritz 2009). A polypeptide which includes predominately simple amino acids will have more difficulty folding than the polypeptide which contains more complex amino acid residues. Moreover, simple poly amino acids consecutive (Gly - Ser) repeats are used in experiments as a flexible linker that are predicted to retain a rather unordered structure in order not to interfere with the folded specificity-determining variable domains of the engineered antibody peptide (Raag and Whitlow 1995; Škrlj et al. 2010; Škrlj and Dolinar 2014). As it is very difficult to explain how life came to rely so extensively on folded proteins, the interesting question of how early peptides circumvent the folding problem throughout early molecular evolution still remains.
Some authors are of the opinion that a solely RNA world may never have become self-sustainable (Fox 2016; Piast and Wieczorek 2017), due to the lack of structure space and therefore any functional capabilities. In this sense, it was proposed that translation originally arose not to synthesize functional proteins, but rather to provide simple (perhaps random) peptides that bound to RNA (Noller 2012; Fox 2016). De Duve (2003) proposed that specific interactions between amino acids and RNA molecules probably initiated the development of RNA-dependent protein synthesis and that these interactions may have played a key role in the selection of the biogenic amino acids and in that of the carrier RNA molecules. Moreover, it is regarded that the ability of RNA to self-aminoacylate was a key event at the transition from the RNA World to the DNA/protein based life theory, and it has also been suggested that modern protein synthesis may have evolved from a set of aminoacyl transfer reactions catalyzed by ribozymes. Later, those aminoacyl–RNAs may have played the role of starting material for the synthesis of peptides, thus constituting the pre-cursors of modern tRNA molecules as aminoacyl carriers. Modern tRNAs as key molecules for efficient and accurate protein translation are heavily modified post-transcriptionally, which is very important for a tRNA structure, function and stability. Some other RNAs were also found to catalyze peptide synthesis in addition to aminoacyl transfer (reviewed by Balke et al. 2016 and references therein). A tiny ribozyme as small as five-nucleotides was found to perform RNA acylation in trans, thus behaving like a true enzyme, and moreover, to catalyze the formation of peptides up to a length of three amino acids (Turk et al. 2010). Those “early” peptides may in turn have assisted the RNA folding by acting as chaperones (Poole et al. 1998). It has also been proposed that the rProtein and rRNA are co-chaperones (Lanier et al. 2017b). Conversely, regarding the intriguing question of early polypeptide folding, Lupas and Alva (2017) are seeking for a solution whereby the first folded domains did not arise from random processes, but from the increased complexity of the peptides that had evolved in the RNA world. This scenario emerged from the assumption that one of the properties in the starting selection must have been the ability of peptides and RNA to interact specifically, an evolutionary pressure resulting as much from a competition of primordial RNAs for a limited pool of peptides, as from the greater functional effectiveness of specific interactors. Specificity is promoted by larger interaction surfaces, a geometric fit of complementary groups and the exclusion of water from the binding sites. On the RNA side, this rewarded the emergence of ligases capable of enlarging the available peptide pool, of producing peptides of greater length than obtainable through abiotic processes, and of using an emergent code to increase the yield of peptides with useful sequences. On the peptide side, this specific interaction led to the selection of amino acids favouring nucleic-acid interaction and of sequences able to assume a defined structure on an RNA scaffold. Over geological time-scales, the increasing organizational and functional complexity of the RNA-peptide networks led to increasingly complex peptides, which structure progressed from the local formation of a secondary structure on the RNA scaffold, to the arrangement of these secondary structures into super-secondary elements and eventually, to compact defined tertiary structures (Lupas and Alva 2017) with a defined specificity of catalysis. The question therefore arises: is the ribosome the only observable remaining relict from early molecular evolution and the last survivor from the ancient RNA-peptide world?
In the sense of the origins of heredity it is also worth considering that the differentiation of plasma cells, i.e. those cells that produce antibodies, which involves the translocation of genes with intrachromosomal recombination as a response to the environmental stimulus (see Stryer 1988) as a faraway echo of the early evolutionary past and of the origins of heredity next to the formation of the genetic code. Additionally, cells also have biochemical natural genetic engineering (NGE) tools needed to make all types of changes to genome DNA. The read–write genome idea predicts that mobile DNA elements will act in evolution to generate adaptive changes in organismal DNA, where Non-coding RNAs (ncRNAs) are engaged (Shapiro 2014, 2016a, 2016b). It is worth considering the engagement of ncRNAs as they supposed to have occurred before coding RNA in early chemical evolution, which is in agreement with our basic idea that during the emergence of life evolution had to first involve autocatalytic systems which only subsequently acquired the capacity of genetic heredity. Assuming the “Metabolism first” hypothesis, a reverse translation at a certain point of the origins of genetic code should be considered (Knight 2007; Annila and Baverstock 2014). In a quest of reverse translation, it is perhaps reasonable to consider the transfer factors from immunology which represent an archaic dialect in the language of cells (Lawrence and Borkowsky 1983). Although the structure and mechanism of action at the molecular level remains so far hypothetical (Viza et al. 2013; Myles et al. 2016), it is sensible to hypothesize that translation might operate at some point in both directions.
Cytochromes P450 as a Possible Model for Enzyme Evolution
Accretion occurs pervasively in nature. Galaxies evolve by accretion, gravitational interactions, harassment - high speed galaxy encounters and dry and wet mergers of stars, gas and dust clouds (Moore et al. 1996; Fraix-Burnet et al. 2012). Stars are formed by gravitational collapse within giant molecular clouds and accrete circumstellar disks of orbiting matter that spiral inwards towards the growing central bodies. Planets arise from the proto-planetary disks of gas and solids by a process of accretion and N-body interactions (Beckwith and Sargent 1996; Öberg et al. 2015), where gravitational accretion is an example of dissipation. It is a non-equilibrium process driven by entropy production. Unsurprisingly, at the other end of the scale, macromolecules in the biological world also arise and evolve by a kind of accretion (Caetano-Anollés and Caetano-Anollés 2015). It was also proposed that the accretion model of ribosomal evolution (Petrov et al. 2014, 2015; Kovacs et al. 2017), in which the ancestral ribosome grew by the recursive accumulation of oligomers of peptide and RNA onto subunit surfaces, presume encasing and freezing the previously acquired components. The addition of new fragments onto subunit surfaces left the previous core unperturbed. During accretion, the folding of short rRNAs and polypeptides into secondary and tertiary structures were emergent phenomena dependent on the rRNA-rPeptide interactions (Kovacs et al. 2017; Lanier et al. 2017b; Lupas and Alva 2017). Separately, an alternative model of ribosome origins that putatively relies primarily on the phylogenetic methods applied to the ribosomal RNA structure, has been proposed (Harish and Caetano-Anolles 2012). This model and Petrov’s accretion model (Petrov et al. 2014, 2015; Kovacs et al. 2017) have generated recent controversy (Fox 2016; Caetano-Anollés and Caetano-Anollés 2017).
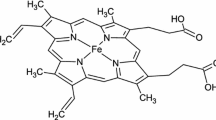
If the co-evolution of RNA and protein were accomplished within the context of the ribosome, which was therefore the cradle of early evolution (Kovacs et al. 2017), could we treat this sort of RNA-peptide networks co-evolution as a general state in the early stages of molecular evolution? It has been already illustrated how the possible complexity of the primordial catalysers’ structure, in the example of haemoproteins, cytochromes P450, has increased. It was proposed that the complexity, specificity and efficacy of the catalysers were achieved via the accretion to cope with the increasing demand for a sophisticated catalysis of complex compounds throughout evolutionary history (Vitas and Dobovišek 2017). Cytochrome P450s constitute one of the most diverse gene families, with a dizzying complexity within and between species. Cytochrome P450s are haem-thiolate enzymes that use molecular oxygen to modify substrate structure, critical in a huge number of physiological, ecological and toxicological processes. Cytochromes P450 have supposedly been present on the Earth for at least 2.7 billion years (Kelly and Kelly 2013; Nelson et al. 2013). The observed reactions that cytochrome P450s can undertake and that do not require oxygen, obviously point to the possible existence and function of ancestral cytochrome P450s being present before atmospheric molecular oxygen appeared on the Earth about 2.4 billion years ago (Kelly and Kelly 2013), e.g., cytochrome P450nor - nitric oxide reductase, involved in denitrification under anaerobic conditions in lower eukaryote, filamentous fungus Fusarium oxysporum (Su et al. 2004). Fe-S is at the core of the catalytic activity of cytochromes P450. As the iron sulphide is considered as a major component of the hatcheries of pre-cellular life, Fe-S bond was proposed as a remnant or relict from the Iron Sulphur World (de Duve 1998; Wächtershäuser 1998, 2006; Vitas and Dobovišek 2017). Some authors suggest that haemes superseded metal cations as catalysts (Milner-White and Russell 2008). We can name the acquisition of the protoporphyrin IX prosthetic group around an Fe atom as the next step in the accretion like evolution of cytochromes P450. There are also proposals that abiotically synthesised peptides would soon stabilize and begin to optimize the metal-cofactor-based catalysts as well as introduce substrate-specificity (Brack 2007; Milner-White and Russell 2008, 2011; Nitschke et al. 2013; Vitas and Dobovišek 2014), which might present the next step in the accretion like the evolutionary history of cytochromes P450 and other protein catalysers.
The idea of collectively autocatalytic sets was introduced as a “metabolism-first” scenario in the context of the origin of life (Kauffman 1971). In this scenario, life supposedly started as a functionally closed, self-sustaining reaction network in which several molecules collectively support each other’s production from basic nutrients through mutually catalyzed chemical reactions (Hordijk and Steel 2014). Real cells are non-equilibrium chemical reaction networks and the biosphere is built of such interacting networks (Kauffman 2007). The complete set of molecular components needed to build a cell must be synthesized by the reaction network in the cell, starting with some basic food set available in the environment, with the second law of thermodynamics requiring that the food has higher free energy than the waste products that are ultimately exported back into the surroundings. Cellular biochemistry thus seems to be rife with networked autocatalytic sets (Sousa et al. 2015; Wills and Carter 2018). Moreover, it was also proposed that the use of one self-assembling polymer surface to template the synthesis of another polymer may represent the next level of self-organizing feedback required to move from these pre-animate systems along a pathway to the complex chemistries of living matter (Taran et al. 2017). In this discussion, as in the “Metabolism first” hypothesis which implies that life started from an assembly of complex autocatalytic networks from the mixtures of interacting organic molecules, which only subsequently acquired the capacity of genetic heredity, it is assumed that reverse translation at a certain point of the origins of genetic code should be considered (Knight 2007; Annila and Baverstock 2014). We therefore propose, firstly, that the acquisition of the primary protein structure as a first step in the digitalization of the information contained in a biological system was achieved by accretion and secondly, that non-coding RNAs have been involved in reverse translation and at the origin of translation and the genetic code, prior to the acquiring of the ability of information storage (Vitas and Dobovišek 2017). In this sense, it should not be overlooked that some coenzymes, including NAD, FAD, coenzyme A and adenosyl-cobalamin, share a ribonucleotidyl group, even though they have completely different biochemical roles. Indeed, in none of them does the ribonucleotidyl moiety participate directly in the co-enzymatic function (Raffaelli 2011). However, they were initially connected to the nucleic acid enzymatic activity as was proposed by White (1976). Thus, this ribonucleotidyl portion might serve as a clue for looking to RNA as the main survivor of a coenzyme world. It may be assumed that the catalytic activity and specificity of sole coenzymes and mineral ions were enhanced by binding to the RNA oligomer portion, which in turn means that by harnessing a functional repertoire of cofactors, metal ions, prosthetic groups, organometallic compounds such as the various vitamins and haeme, a broader repertoire of chemical functionalities of RNA ensued, otherwise the functionality would have been rather poor compared to the one of proteins (Poon et al. 2011; Sen and Poon 2011). It is the analogue story, as discussed previously in this article that was aimed at the rRNA and rPeptides co-evolution. Indeed, it was found that Fe2+ can confer on some RNAs, a previously uncharacterized ability to catalyse single-electron transfer (Hsiao et al. 2013). RNA and Fe2+ could, in principle, support an array of RNA structures and catalytic functions more diverse than RNA with Mg2+ alone (Athavale et al. 2012). In fact, such mineral cofactors probably predate the RNA world (Yarus 2010), surviving even beyond the gigayears since the emergence of life on Earth. Enzymes containing iron in the protoporphyrin IX prosthetic group, like haemoglobins, cytochromes and cytochromes P450 deserve special attention (Eschenmoser 1988; Melton et al. 2014; Kappler et al. 2015; Okafor et al. 2017), since life presumably originated in anoxic Fe2+ rich early Archean oceans, where Iron was the most predominant catalyst (Halevy et al. 2017). This idea was supported by the simulation study of reaction sequences of small molecules in the early Archean ocean (Keller et al. 2014). Moreover, Sen and Poon (2011) showed that RNA can enhance the redox activity of iron protoporphyrin IX, which is also a constituent of modern cytochrome P450 enzymes. The proposed accretion model of cytochromes P450 (see Fig. 1) can therefore be supplemented with the RNA–peptide co-evolutionary step placed in between the acquisition of protoporphyrin IX around Fe ion and obtaining an apoprotein protein portion around haem, whereupon, in evolutionary time, proteins would entirely displace the obsolete RNA portion, while in the case of cytochrome P450, the catalytic core of the haeme would be conserved (see Fig. 2). Might we be able to generalize from this particular molecular evolutionary path, to other enzymes? What are the implications for the origins of translation and the genetic code if we consider the ribosome as the only observable remaining relict from early molecular evolution and the last survivor from the ancient RNA-peptide world?
Schematic presentation of the proposed accretion model of Cytochromes P450 molecular evolution. (1) Fe-S bond is proposed as a remnant or relict from the Iron Sulphur World at the core of the proposed model. (2) Subsequent acquisition of the Protoporphyrin IX prosthetic group around Fe atom. (3) Acquisition of RNA-peptide moiety (4) Finally, acquisition of the apoprotein portion
Conclusions and Perspectives
Conclusions
The Origins of life is a transdisciplinary field of investigation. We have discussed and proposed new aspects in the early molecular evolution of translation and more generally biocatalysts such as Co-factors, ribozymes, ribosomes and enzymes as we know them today. Assuming the hypothesis that at the emergence of life, evolution had to first involve autocatalytic systems, which only subsequently acquired the capacity of genetic heredity, reverse translation should be considered. In this sense, it was hypothesised that primordial RNAs were devoid of any repository role for discrete digital information. The inferred accretion model of the biocatalysts evolutionary course was thus appended and supplemented with the RNA-peptide development step. This step was outlined in the case of cytochromes P450 as a possible model for the evolutionary development of protein catalysers. The primacy of RNA molecules or polypeptides in the case of the origins of translation was discussed in the light of their abiotical availability. Co-evolution of both types of polymers is supported. The hypothesis that early polypeptides were folding on the RNA scaffold was also reviewed and again mutualism in the early molecular evolution of RNA and peptides is favoured. This fits into the general picture, as mutualism is common in living nature (Lanier et al. 2017a).
Perspectives
In exobiological (as well as astrobiological) terms, it has been proposed that life’s fundamental evolutionary nature might have extended beyond its origin and might be rooted in the abiotic cosmochemical evolution of the biogenic elements. It is then easy to see why the discourse about the origins of life has been multidisciplinary, broad based, and has fostered many theories, all of which, with the notable exception of the panspermia hypothesis, accept the fundamental emergent nature of life from simple molecules (Pizzarello and Shock 2010). However, the largest fraction of carbon in the universe is incorporated into solid aromatic macromolecular matter. It was proposed that assemblies based on aromatic hydrocarbons may have been the most abundant flexible and stable organic materials on the primitive Earth, with a possible integration into a minimal life-form (Ehrenfreund et al. 2006). For example, the Murchison meteorite contained about 2% by weight organic matter, most of which is insoluble macromolecular material polyaromatic hydrocarbons (PAHs) (Sephton 2002; Schönheit et al. 2016). It is interesting that Benz[a]pyren which belongs to this sort of chemical compounds can be hydroxlated by the lower eukaryotes that is in filamentous fungi by cytochrome P450 dependant monooxygenases (Dutta et al. 1983; Ghosh et al. 1983; Venkateswarlu et al. 1996). Moreover, it is also likely that the PAH derivatives contributed to the stability of the membranes, just as the polycyclic aliphatic molecule cholesterol does in current biological membranes (Deamer et al. 2002). Since biological membranes are one of the key attributes of life, consequently their origins should be carefully deliberated. Enzymes from the cytochromes P450 superfamily are involved in the metabolism and in the biosynthesis of steroids and sterols (Nelson et al. 2013). Furthermore, considering that the haeme is most likely an ancient compound, (Sen and Poon 2011) and a likely player in the supposed RNA World Theory, the abiotic synthesis and the possible role of conjugated planar systems in the reaction processes at the emergence of life should be reflected upon. Taking into account that Hud (2016) hypothesized that tetrads and intercalators, for instance tetramethylated tetra-2,3-pyrazino-porphyrazine, a protoporphirine-like molecule, could have organized the nucleobases before they were incorporated into proto-RNA polymers, the possible role of such molecules in the supposed RNA World is worth to be explored. Moreover, it should be not overlooked that glutamyl-tRNA participates in the biosynthesis of tetrapyroles, such as haemes (Jahn et al. 2006). The same applies to cholesterol and steroids, which belong to isoprenoids. Since they are potent hormones, general regulators of metabolism, and stabilizers and adjustors of the permeability of the cell membranes (Pulido et al. 2012), their appearance in early molecular evolution cannot be excluded.
References
Adamala K, Anella F, Wieczorek R, Stano P, Chiarabelli C, Luisi PL (2014) Open questions in origin of life: experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach. Comput Struct Biotechnol J 9:1–10. https://doi.org/10.5936/csbj.201402004
Adami C (2015) Information-theoretic considerations concerning the origin of life. Orig Life Evol Biosph 45:309–317. https://doi.org/10.1007/s11084-015-9439-0
Annila A, Baverstock K (2014) Genes without prominence: a reappraisal of the foundations of biology. J R Soc Interface 11:20131017. https://doi.org/10.1098/rsif.2013.1017
Athavale SS, Petrov AS, Hsiao C, Watkins D, Prickett CD, Gossett JJ, Lie L, Bowman JC, O’Neill E, Bernier CR, Hud NV, Wartell RM, Harvey SC, Williams LD (2012) RNA folding and catalysis mediated by iron (II). PLoS One 7:e38024. https://doi.org/10.1371/journal.pone.0038024
Balke D, Kuss A, Müller S (2016) Landmarks in the evolution of (t)-RNAs from the origin of life up to their present role in human cognition. Life 6:4–13. https://doi.org/10.3390/life6010001
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Ǻ resolution. Science 289:905–920. https://doi.org/10.1126/science.289.5481.905
Baymann F, Lebrun E, Brugna M, Schoepp-Cothenet B, Guidici-Oritconi MT, Nitschke W (2003) The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Phil Tran R Soc B 358:267–274. https://doi.org/10.1098/rstb.2002.1184
Beckwith SVW, Sargent AI (1996) Circumstellar disks and the search for neighbouring planetary systems. Nature 383:139–144. https://doi.org/10.1038/383139a0
Bowman JC, Hud NV, Williams LD (2015) The ribosome challenge to the RNA world. J Mol Evol 80:143–161. https://doi.org/10.1007/s0023
Brack A (2007) From interstellar amino acids to prebiotic catalytic peptides: a review. Chem Biodivers 4:665–679. https://doi.org/10.1002/cbdv.200790057
Brack A (2010) Origin of life. In: Encyclopedia of life sciences. John Wiley & Sons, Ltd. (eds), Wiley, Ltd. Chichester. https://doi.org/10.1002/9780470015902.a0001639.pub2
Brandman R, Brandman Y, Pande VS (2012) Sequence coevolution between RNA and protein characterized by mutual information between residue triplets. PLoS One 7:e30022. https://doi.org/10.1371/journal.pone.0030022
Caetano-Anollés G, Caetano-Anollés D (2015) Computing the origin and evolution of the ribosome from its structure - uncovering processes of macromolecular accretion benefiting synthetic biology. Comput Struct Biotechnol J 13:427–447. https://doi.org/10.1016/j.csbj.2015.07.003
Caetano-Anollés D, Caetano-Anollés G (2017) Commentary: history of the ribosome and the origin of translation. Front Mol Biosci 3:1–3. https://doi.org/10.3389/fmolb.2016.00087
Cafferty BJ, Fialho DM, Khanam J, Krishnamurthy R, Hud NV (2016) Spontaneous formation and base pairing of plausible prebiotic nucleotides in water. Nat Commun 7:11328. https://doi.org/10.1038/ncomms11328
Callahan MP, Smith KE, Cleaves HJ II, Ruzicka J, Stern JC, Glavin DP, House CH, Dworkin JP (2011) Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc Natl Acad Sci U S A 108:13995–13998. https://doi.org/10.1073/pnas.1106493108
Carter CW Jr, Wolfenden R (2015) tRNA acceptor stem and anticodon bases form independent codes related to protein folding. Proc Natl Acad Sci U S A 112:7489–7494. https://doi.org/10.1073/pnas.1507569112
Cech TR (2000) The ribosome is a ribozyme. Science 289:878–879. https://doi.org/10.1126/science.289.5481.878
Cech TR, Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–93. https://doi.org/10.1016/j.cell.2014.03.008
Cleaves HJ, Chalmers JH, Lazcano A, Miller SL, Bada JL (2008) A reassessment of prebiotic organic synthesis in neutral planetary atmospheres. Orig Life Evol Biosph 38:105–115. https://doi.org/10.1007/s11084-007-9120-3
Copley SD, Smith E, Morowitz HJ (2007) The origin of the RNA world: co-evolution of genes and metabolism. Bioorg Chem 35:430–443. https://doi.org/10.1016/j.bioorg.2007.08.001
Corliss JB (1986) On the creation of living cells in submarine hot spring flow reactors: attractors and bifurcations in the natural hierarchy dissipative systems. Orig Life Evol Biosph 19:381–382. https://doi.org/10.1007/BF02422084
Corliss JB, Baross JA, Hoffman SE (1981) An hypothesis concerning the relationships between submarine hot springs and the origin of life on earth. Oceanol Acta, Proccedings of 26th international geological congress, Paris, July 7-17, 1980, pp 59–69
Cupal J, Kopp S, Stadler PF (2000) RNA shape space topology. Artif Lif 6:3–23. https://doi.org/10.1162/106454600568294
Dalai P, Kaddour H, Sahai N (2016) Incubating life: prebiotic sources of organics for the origin of life. Elements 12:401–406
Deamer D (2009) First life, and next life. Synthetic biology is a new field, but it’s centered on an old question: how did life begin? Technol Rev 112:66–73
Deamer D, Dworkin JP, Sandford SA, Bernstein MP, Allamandola LJ (2002) The first cell membranes. Astrobiology 2:371–381. https://doi.org/10.1089/153110702762470482
Dixon B (1994) Power unseen, how microbes rule the world. W.H. Freeman and company, New York
Dutta D, Ghosh DK, Mishra AK, Samanta TB (1983) Induction of benz[a]pyren hydroxylase in Aspergillus ochraceus TS: evidences of multiple forms of cytochrome P-450. Biochem Biophys Res Commun 115:692–699. https://doi.org/10.1016/S0006-291X(83)80200-9
de Duve C (1995) The beginnings of life on earth. Am Sci 83:428–437
de Duve C (1998) Clues from present - day biology: the tioester world. In: Brack A (ed) The molecular origins of life. Cambridge University Press, Cambridge, pp 219–236
de Duve C (2003) A research proposal on the origin of life. Orig Life Evol Biosph 33:559–574. https://doi.org/10.1023/A:1025760311436
Ehrenfreund P, Rasmussen S, Cleaves J, Chen L (2006) Experimentally tracing the key steps in the origin of life: the aromatic world. Astrobiology 6:490–520. https://doi.org/10.1089/ast.2006.6.490
El-Hani NC, Queiroz J, Emmeche C (2006) A semiotic analysis of the genetic information system. Semiotica 160:1–68. https://doi.org/10.1515/SEM.2006.039
Eschenmoser A (1988) Vitamin B12: experiments concerning the origin of its molecular structure. Angew Chem Int Ed 27:5–39. https://doi.org/10.1002/anie.198800051
Fan K, Wang W (2003) What is the minimum number of letters required to fold a protein? J Mol Biol 328:921–926. https://doi.org/10.1016/S0022-2836(03)00324-3
Ferus M, Pietrucci F, Saitta AM, Knížek A, Kubelík P, Ivaneka O, Shestivska V, Civiša S (2017) Formation of nucleobases in a miller–Urey reducing atmosphere. Proc Natl Acad Sci U S A 114:4306–4311. https://doi.org/10.1073/pnas.1700010114
Fox GE (2016) Origins and early evolution of the ribosome. In: Hernández G, Jagus R (eds) Evolution of the protein synthesis machinery and its regulation, springer international publishing Switzerland, pp 31–60
Fox S, Strasdeit H (2013) A possible prebiotic origin on volcanic islands of oligopyrrole-type photopigments and electron transfer cofactors. Astrobiology 13:578–595. https://doi.org/10.1089/ast.2012.0934
Fraix-Burnet D, Chattopadhyay T, Chattopadhyay AK, Davoust E, Thuillard M (2012) A six-parameter space to describe galaxy diversification. A & A 545:1–24. https://doi.org/10.1051/0004-6361/201218769
Friedmann MP, Torbeev V, Zelenay V, Sobol A, Greenwald J, Riek R (2015) Towards prebiotic catalytic amyloids using high throughput screening. PLoS One 10:e0143948. https://doi.org/10.1371/journal.pone.0143948
Ghosh DK, Dutta D, Samanta TB, Mishra AK (1983) Microsomal benz[a]pyren hydrooxylase in Aspergillus ochraceus TS: assay and characterisation of the enzyme system. Biochem Biophys Res Commun 113:497–505. https://doi.org/10.1016/0006-291X(83)91753-9
Goodwin JT, Mehta AK, Lynn DG (2012) Digital and analog chemical evolution. Acc Chem Res 45:2189–2199. https://doi.org/10.1021/ar300214w
Goodwin JT, Lynn DG, Burrows C, Walker S, Amin S, Armbrust EV (2014) Alternative chemistries of life, Empirical Approaches, A Report from a workshop on alternative chemistries of life: empirical approaches. http://alternativechemistries.emory.edu/report_summary/index.html. Accessed 24 October 2014
Gordon-Smith C (2011) Non-template molecules designed for open-ended evolution. In: Lenaerts T, Giacobini M, Bersini H, Bourgin P, Dorigo M, Doursat R (eds) Advances in artificial life ECAL, proceedings of the eleventh euorpean conference on the synthesis and simulation of living systems. Massachusetts Institute of Technology, USA, pp 268–275
Gorlero M, Wieczorek R, Adamala K, Giorgi A, Schininá ME, Stano P, Luisi PL (2009) Ser-his catalyses the formation of peptides and PNAs. FEBS Lett 583:153–156. https://doi.org/10.1016/j.febslet.2008.11.052
Halevy I, Alesker M, Schuster EM, Popovitz-Biro R, Feldman Y (2017) A key role for green rust in the precambrian oceans and the genesis of iron formations. Nat Geosci 10:135–139. https://doi.org/10.1038/ngeo2878
Harish A, Caetano-Anolles G (2012) Ribosomal history reveals origins of modern protein synthesis. PLoS One 7:e32776. https://doi.org/10.1371/journal.pone.0032776
Higgs PG, Lehman N (2014) The RNA world: molecular cooperation at the origins of life. Nat Rev Genet 16:7–17. https://doi.org/10.1038/nrg3841
Higgs PG, Pudritz RE (2009) A thermodynamic basis for prebiotic amino acid synthesis and the nature of the first genetic code. Astrobiology 9:483–490. https://doi.org/10.1089/ast.2008.0280
Ho BK, Dill KA (2006) Folding very short peptides using molecular dynamics. PLoS Comput Biol 2:e27. https://doi.org/10.1371/journal.pcbi.0020060
Hodgson GW, Ponnamperuma C (1968) Prebiotic porphyrin genesis: porphyrins from electric discharge in methane, ammonia and water vapour. Proc Natl Acad Sci U S A 59:22–28
Hordijk W, Steel M (2014) Conditions for evolvability of autocatalytic sets: a formal example and analysis. Orig Life Evol Biosph 44:111–124. https://doi.org/10.1007/s11084-014-9374-5
Hordijk W, Steel M, Kauffman S (2012) The structure of autocatalytic sets: evolvability, enablement and emergence. Acta Biotheor 60:379–392. https://doi.org/10.1007/s10441-012-9165-1
Hsiao C, Williams LD (2009) A recurrent magnesium-binding motif provides a framework for ribosomal peptidyl transferase center. Nucleid Acids Res 37:3134–3142. https://doi.org/10.1093/nar/gkp119
Hsiao C, Chou IC, Okafor CD, Bowman JC, O’Neill EB, Athavale SS, Petrov AS, Hud NV, Wartell RM, Harvey SC, Williams LD (2013) RNA with iron(II) as a cofactor catalyses electron transfer. Nat Chem 5:525–528. https://doi.org/10.1038/nchem.1649
Hud NV (2016) Our odyssey to find a plausible prebiotic path to RNA: the first twenty years. Synlett 28:36–55. https://doi.org/10.1055/s-0036-1588646
Hwang S (2012) Investigation of peptide folding by nuclear magnetic resonance spectroscopy. Dissertation, Texas A&M University
Ivica NA, Obermayer B, Campbell GW, Rajamani S, Gerland U, Chen IA (2013) The paradox of dual roles in the RNA world: resolving the conflict between stable folding and templating ability. J Mol Evol 77:55–63. https://doi.org/10.1007/s00239-013-9584-x
Jackson JB (2016) Natural pH gradients in hydrothermal alkali vents were unlikely to have played a role in the origin of life. J Mol Evol 83:1–11. https://doi.org/10.1007/s00239-016-9756-6
Jadhav VR, Yarus M (2002) Coenzymes as co-ribozymes. Biochimie 84:877–888. https://doi.org/10.1016/S0300-9084(02)01404-9
Jahn D, Moser J, Schubert WD, Heinz DW (2006) Transfer RNA-dependent aminolevulinic acid formation: structure and function of glutamyl-tRNA synthetase, reductase and glutamate-1-semialdehyde-2,1-aminomutase. In: Grimm B, Porra RJ, Rüdiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications. Springer, Netherlands, pp 159–171
Johnson DBF, Wang L (2010) Imprints of the genetic code in the ribosome. Proc Natl Acad Sci U S A 107:8298–8303. https://doi.org/10.1073/pnas.1000704107
Kaddour H, Sahai N (2014) Synergism and mutualism in non-enzymatic RNA polymerization. Life (Basel) 4:598–620. https://doi.org/10.3390/life4040598
Kappler A, Emerson D, Gralnick JA, Roden EE, Muehe EM (2015) Geomicrobiology of iron, In: Ehrlich HL, Newman DK, Kappler A (eds) Geomicrobiology, 6th edition, CRC Press, pp 343–399
Kasting JF (1993) Earth’s early atmosphere. Science 259:920–926. https://doi.org/10.1126/science.11536547
Kasting JF, Catling D (2003) Evolution of a habitable planet. Annu Rev Astron Astrophys 41:429–463. https://doi.org/10.1146/annurev.astro.41.071601.170049
Kasting JF, Howard MT (2006) Atmospheric composition and climate on the early earth. Phil Trans R Soc B 361:1733–1742. https://doi.org/10.1098/rstb.2006.1902
Kauffman SA (1971) Cellular homeostasis, epigenesis and replication in randomly aggregated macromolecular systems. J Cybern 1:71–96. https://doi.org/10.1080/01969727108545830
Kauffman S (2007) Question 1: origin of life and the living state. Orig Life Evol Biosph 37:315–322. https://doi.org/10.1007/s11084-007-9093-2
Keller MA, Turchyn AV, Ralser M (2014) Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean Ocean. Mol Syst Biol 10:725. https://doi.org/10.1002/msb.20145228
Kelly SL, Kelly DE (2013) Microbial cytochromes P450: biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Phil Trans R Soc B 368:20120476. https://doi.org/10.1098/rstb.2012.0476
Knight R (2007) Reviewers’ comments: wolf YI, Koonin EV: on the origin of the translation system and the genetic code in the RNA world by means of natural selection, and sub functionalization. Biol Direct 2:1–25. https://doi.org/10.1186/1745-6150-2-14
Kovacs NA, Petrov AS, Lanier KA, Williams LD (2017) Frozen in time: the history of proteins. Mol Biol Evol 34:1252–1260. https://doi.org/10.1093/molbev/msx086
Kurland CG (2010) The RNA dreamtime. Bioassays 32:866–871. https://doi.org/10.1002/bies.201000058
Lanier KA, Petrov AS, Williams LD (2017a) The central symbiosis of molecular biology: molecules in mutualism. J Mol Evol 85:8–13. https://doi.org/10.1007/s00239-017-9804-x
Lanier KA, Roy P, Schneider DM, Williams LD (2017b) Ancestral interactions of ribosomal RNA and ribosomal proteins. Biophys J 113:268–276. https://doi.org/10.1016/j.bpj.2017.04.007
Lawrence HS, Borkowsky W (1983) A new basis for the immunoregulatory activities of transfer factor-an arcane dialect in the language of cells. Cell Immunol 82:102–116
Lehman N, Díaz Arenas C, White WA, Schmidt FJ (2011) Complexity through recombination: from chemistry to biology. Entropy 13:17–37. https://doi.org/10.3390/e13010017
Lehman N, Bernhard T, Larson BC, Robinson AJN, Southgate CCB (2014) Empirical demonstration of environmental sensing in catalytic RNA: evolution of interpretive behaviour at the origins of life. BMC Evol Biol 14:1–11. https://doi.org/10.1186/s12862-014-0248-2
Luisi PL (2006) The emergence of life, from chemical origins to synthetic biology. Cambridge University Press, Cambridge
Lupas AN, Alva V (2017) Ribosomal proteins as documents of the transition from unstructured (poly)peptides to folded proteins. J Struct Biol 198:74–81. https://doi.org/10.1016/j.jsb.2017.04.007
Martin W, Baross J, Kelley D, Russell MJ (2008) Hydrothermal vents and the origin of life. Nature 6:805–814. https://doi.org/10.1038/nrmicro1991
Maynard Smith J, Szathmáry E (1995) The major evolutionary transitions. W.H. Freeman Spectrum, Oxford
Melton DE, Swanner ED, Behrens S, Schmidt C, Kappler A (2014) The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle. Nat Rev Microbiol 12:797–808. https://doi.org/10.1038/nrmicro3347
Michaelian K, Simeonov A (2015) Fundamental molecules of life are pigments which arose and co-evolved as a response to the thermodynamic imperative of dissipating the prevailing solar spectrum. Biogeosciences 12:4913–4937. https://doi.org/10.5194/bg-12-4913-2015
Miller SL (1953) The production of amino acids under possible primitive earth conditions. Science 117:528–529. https://doi.org/10.1126/science.117.3046.528
Miller SL (1998) The endogenous synthesis of organic compounds. In: Brack A (ed) The molecular origins of life. Cambridge University Press, Cambridge, pp 59–85
Milner-White EJ, Russell MJ (2008) Predicting peptide and protein conformations in early evolution. Biol Direct 3:1–9. https://doi.org/10.1186/1745-6150-3-3
Milner-White EJ, Russell MJ (2011) Functional capabilities of the earliest peptides and the emergence of life. Genes 2:671–688. https://doi.org/10.3390/genes2040671
Moore PB, Steitz TA (2010) The roles of RNA in the synthesis of protein. Cold Spring Harb Perspect Biol 3:1–17. https://doi.org/10.1101/cshperspect.a003780
Moore B, Katz N, Lake G, Dressler A, Oemler A (1996) Galaxy harassment and the evolution of clusters of galaxies. Nature 379:613–616. https://doi.org/10.1038/379613a0
Murphy M P, O'Neill LAJ (1997) What is life? The next fifty years. An introduction. In: Murphy MP, O' Neill LAJ (eds) What is life? The next fifty years, Cambridge University Press, Cambridge, pp 1–4
Myles IA, Zhao M, Nardone G, Olano LR, Reckhow JD, Saleem D, Break TJ, Lionakis MS, Myers TG, Gardina PJ, Kirkpatrick CH, Holland SM, Datta SK (2016) CD8 + T cells produce a dialyzable antigen-specific activator of dendritic cells. J Leukoc Biol 100:1–14. https://doi.org/10.1002/eji.200526193
Nelson DR, Goldstone JV, Stegeman JJ (2013) The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Phil Trans R Soc B 368:20120474. https://doi.org/10.1098/rstb.2012.0474
Nitschke W, McGlynn SE, Milner-White EJ, Russell MJ (2013) On the antiquity of metalloenzymes and their substrates in bioenergetics. Biochim Biophys Acta 1827:871–881. https://doi.org/10.1016/j.bbabio.2013.02.008
Noller HF (2012) Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol 4:1–14. https://doi.org/10.1101/cshperspect.a003681
Noller HF, Hoffarth V, Zimniak L (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256:1416–1419. https://doi.org/10.1126/science.1604315
Öberg KI, Guzmán VV, Furuya K, Qi C, Aikawa Y, Andrews SM, Loomis R, Wilner DJ (2015) The comet-like composition of a protoplanetary disk as revealed by complex cyanides. Nature 520:198–201. https://doi.org/10.1038/nature14276
Oda A, Fukuyoshi S (2015) Predicting three-dimensional conformations of peptides constructed of only glycine, alanine, aspartic acid, and valine. Orig Life Evol Biosph 45:183–193. https://doi.org/10.1007/s11084-015-9418-5
Okafor CD, Lanier KA, Petrov AS, Athavale S, Bowman JC, Hud NV, Williams LD (2017) Iron mediates catalysis of nucleic acid processing enzymes: support for Fe(II) as a cofactor before the great oxidation event. Nucleic Acids Res 45:3634–3642. https://doi.org/10.1093/nar/gkx171
Orgel LE (2004) Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39:99–123. https://doi.org/10.1080/10409230490460765
Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, Hud NV, Williams LD (2014) Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci U S A 111:10251–10256. https://doi.org/10.1073/pnas.1407205111
Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD (2015) History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A 112:15396–15401. https://doi.org/10.1073/pnas.1509761112
Piast RW, Wieczorek R (2017) Origin of life and the phosphate transfer catalyst. Astrobiology 17:277–285. https://doi.org/10.1089/ast.2015.1463
Pizzarello S, Shock E (2010) The organic composition of carbonaceous meteorites: the evolutionary story ahead of biochemistry. Cold Spring Harb Perspect Biol 2:1–20. https://doi.org/10.1101/cshperspect.a002105
Poole AM, Jeffares DC, Penny D (1998) The path from the RNA world. J Mol Evol 46:1–17. https://doi.org/10.1007/PL00006275
Poon LCH, Methot SP, Morabi-Pazooki W, Pio F, Bennet AJ, Sen D (2011) Guanine-rich RNAs and DNAs that bind heme robustly catalyze oxygen transfer reactions. J Am Chem Soc 133:1877–1884. https://doi.org/10.1021/ja108571a
Pressman A, Blanco C, Chen IA (2015) The RNA world as a model system to study the origin of life. Curr Biol 25:953–963. https://doi.org/10.1016/j.cub.2015.06.016
Pross A (2004) Causation and the origin of life. Metabolism or replication first? Origin Life Evol Biosph 34:307–321. https://doi.org/10.1023/B:ORIG.0000016446.51012.bc
Pulido P, Perello C, Rodriguez-Concepcion M (2012) New insights into plant isoprenoid metabolism. Mol Plant 5:964–967. https://doi.org/10.1093/mp/sss088
Raag R, Whitlow M (1995) Single-chain Fvs. FASEB J 9:73–80
Raffaelli N (2011) Nicotinamide coenzyme synthesis: a case of ribonucleotide emergence or a byproduct of the RNA world? In: Egel R (ed) Origins of life: the primal self-organization. Springer-Verlag, Berlin, pp 185–208
Robinson A, Southgate C (2010) A general definition of interpretation and its application to origin of life research. Biol Philos 25:163–181. https://doi.org/10.1007/s10539-009-9188-4
Root-Bernstein M, Root-Bernstein R (2015) The ribosome as a missing link in the evolution of life. J Theor Biol 367:130–158. https://doi.org/10.1016/j.jtbi.2014.11.025
Russell MJ, Hall AJ (2009) The hydrothermal source of energy and materials at the origin of life. In: Zaikowski L, Friedrich JM, Seidel SR (eds) Chemical evolution II: from the origins of life to modern society, ACS symposium series; American chemical society, Washington, pp 45–62
Russell MJ, Nitschke W (2017) Methane: fFuel or exhaust at the emergence of life? Astrobiology 17:1053–1066. https://doi.org/10.1089/ast.2016.1599
Russell MJ, Daniel RM, Hall AJ, Sherringham JA (1994) A hydrothermally precipitated catalytic iron sulphide membrane as a first step toward life. J Mol Evol 39:231–243. https://doi.org/10.1007/BF00160147
Russell MJ, Nitschke W, Branscomb E (2013) The inevitable journey to being. Phil Trans R Soc B 368:20120254. https://doi.org/10.1098/rstb.2012.0254
Schönheit P, Buckel W, Martin WF (2016) On the origin of heterotrophy. Trends Microbiol 24:12–25. https://doi.org/10.1016/j.tim.2015.10.003
Sen D, Poon LCH (2011) RNA and DNA complexes with hemin [Fe(III) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit Rev Biochem Mol Biol 46:478–492. https://doi.org/10.3109/10409238.2011.618220
Sephton MA (2002) Organic compounds in carbonaceous meteorites. Nat Prod Rep 19:292–311
Shapiro R (2000) A replicator was not involved in the origin of life. IUBMB Life 49:173–176. https://doi.org/10.1080/713803621
Shapiro R (2007) A simpler origin for life. Sci Am 296:46–53
Shapiro JA (2014) Physiology of the read–write genome. J Physiol 592:2319–2341. https://doi.org/10.1113/jphysiol.2014.271130
Shapiro JA (2016a) Exploring the read-write genome: mobile DNA and mammalian adaptation. Crit Rev Biochem Mol Biol 52:1–17. https://doi.org/10.1080/10409238.2016.1226748
Shapiro JA (2016b) The basic concept of the read-write genome: mini-review on cell-mediated DNA modification. Biosystems 140:35–37. https://doi.org/10.1016/j.biosystems.2015.11.003
Sharov AA (2009) Coenzyme autocatalytic network on the surface of oil microspheres as a model for the origin of life. Int J Mol Sci 10:1838–1852. https://doi.org/10.3390/ijms10041838
Sharov AA (2016) Coenzyme world model of the origin of life. Biosystems 144:8–17. https://doi.org/10.1016/j.biosystems.2016.03.003
Shaw GH (2008) Earth’s atmosphere – hadean to early proterozoic. Chem Erde 68:235–264. https://doi.org/10.1016/j.chemer.2008.05.001
Shi Z, C Olson CA, Rose GD, Baldwin RL, Kallenbach NL (2002) Polyproline II structure in a sequence of seven alanine residues. Proc Natl Acad Sci U S A 99:9190–9195. https://doi.org/10.1073/pnas.112193999
Simionescu CI, Simionescu BC, Mora R, Leancâ M (1978) Porphyrin-like compounds genesis under simulated abiotic conditions. Orig Life Evol Biosph 9:103–114. https://doi.org/10.1007/978-94-009-8420-2_25
Škrlj N, Dolinar M (2014) New engineered antibodies against prions. Bioengineered 5(1):10–14. https://doi.org/10.4161/bioe.26069
Škrlj N, Čurin Šerbec V, Dolinar M (2010) Single-chain Fv antibody fragments retain binding properties of the monoclonal antibody raised against peptide P1 of the human prion protein. Appl Biochem Biotechnol 160:1808–1821. https://doi.org/10.1007/s12010-009-8699-4
Smith JE, Mowles AK, Mehta AK, Lynn DG (2014) Looked at life from both sides now. Life 4:887–902. https://doi.org/10.3390/life4040887
Sousa FL, Hordijk W, Steel M, Martin WF (2015) Autocatalytic sets in E. coli metabolism. J Syst Chem 6:1–21. https://doi.org/10.1186/s13322-015-0009-7
Spitzer J, Pielak GJ, Poolman B (2015) Emergence of life: physical chemistry changes the paradigm. Biol Direct 10:1–15. https://doi.org/10.1186/s13062-015-0060-y
Stryer L (1988) Biochemistry (3rd edition). W.H. Freeman and company, New York, pp 906–910
Su F, Takaya N, Shoun H (2004) Nitrous oxide-forming codenitrification catalyzed by cytochrome P450nor. Biosci Biotechnol Biochem 68:473–475. https://doi.org/10.1271/bbb.68.473
Szathmáry E (2015) Toward major evolutionary transitions theory 2.0. Proc Natl Acad Sci U.S.A. 112:10104–10111. https://doi.org/10.1073/pnas.1421398112
Szathmáry E, Maynard Smith J (1995) The major evolutionary transitions. Nature 374:227–232. https://doi.org/10.1038/374227a0
Taran O, Chen C, Omosun TO, Hsieh M-C, Rha A, Goodwin JT, Mehta AK, Grover MA, Lynn DG (2017) Expanding the informational chemistries of life: peptide/RNA networks. Philos Trans A Math Phys Eng Sci 28:20160356. https://doi.org/10.1098/rsta.2016.0356
Tashiro T, Ishida A, Hori M, Igisu M, Koike M, Méjean P, Takahata N, Sano Y, Komiya T (2017) Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 549:516–518. https://doi.org/10.1038/nature24019
Tessera M (2011) Origin of evolution versus origin of life: a shift of paradigm. Int J Mol Sci 12:3445–3458. https://doi.org/10.3390/ijms12063445
Ts’o POP (1974) Bases, nucleosides, and nucleotides. In: Ts’o POP (ed) Basic principles in nucleic acid chemistry. Academic Press, New York, pp 453–584
Turk RM, Chumachenko NV, Yarus M (2010) Multiple translational products from a five-nucleotide ribozyme. Proc Natl Acad Sci U S A 107:4585–4589. https://doi.org/10.1073/pnas.0912895107
Turner DH, Mathews DH (2010) NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38:280–282. https://doi.org/10.1093/nar/gkp892
Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N (2012) Spontaneous network formation among cooperative RNA replicators. Nature 491:72–77. https://doi.org/10.1038/nature11549
Vasas V, Fernando C, Santos M, Kauffman S, Szathmáry E (2012) Evolution before genes. Biol Direct 7:1–14. https://doi.org/10.1186/1745-6150-7-1
Venkateswarlu K, Marsh RM, Faber B, Kelly SL (1996) Investigation of cytochrome P450 mediated benzo[a]pirene hydroxylation in Aspergillus fumigatus. J Chem Tech Biotechnol 66:139–144. https://doi.org/10.1002/(SICI)1097-4660(199606)66:2%3C139::AID-JCTB480%3E3.0.CO;2-D
Villarreal LP, Witzany G (2015) When competing viruses unify: evolution, conservation, and plasticity of genetic identities. J Mol Evol 80:305–318. https://doi.org/10.1007/s00239-015-9683-y
Vitas M (2011) On the theory of species evolution through natural selection. Original title: O teoriji razvoja vrst s pomočjo naravne selekcije. Apokalipsa: revija za preboj v živo kulturo 152:113–122
Vitas M, Dobovišek A (2014) Evolution, transposition, transformation and flow of information. Anali Pazu 4:66–74
Vitas M, Dobovišek A (2017) On a quest of reverse translation. Found Chem 19:139–155. https://doi.org/10.1007/s10698-016-9260-5
Viza D, Fudenberg HH, Palareti A, Ablashi D, De Vinci C, Pizza G (2013) Transfer factor: an overlooked potential for the prevention and treatment of infectious diseases. Folia Biol (Praha) 59:53–67
Wachowius F, Attwater J, Holliger P (2017) Nucleic acids: function and potential for abiogenesis. Q Rev Biophys 50:e4. https://doi.org/10.1017/S0033583517000038
Wächtershäuser G (1998) Origin of life in an iron – sulfur world. In: Brack A (ed) The molecular origins of life. Cambridge University Press, Cambridge, pp 206–218
Wächtershäuser G (2006) From volcanic origins of chemoautotrophic life to bacteria, archaea and eukarya. Phil Trans R Soc B 361:1787–1808. https://doi.org/10.1098/rstb.2006.1904
White HB 3rd (1976) Coenzymes as fossils of an earlier metabolic state. J Mol Evol 7:101–104
Wieczorek R, Dörr M, Chotera A, Luisi PL, Monnard PA (2013) Formation of RNA phosphodiester bond by histidine-containing dipeptides. Chembiochem 14:217–223. https://doi.org/10.1002/cbic.201200643
Wieczorek R, Adamala K, Gasperi T, Polticelli F, Stano P (2017) Small and random peptides: an unexplored reservoir of potentially functional primitive organocatalysts. The case of seryl-histidine. Life 7:1–24. https://doi.org/10.3390/life7020019
Wills PR, Carter CW Jr (2018) Insuperable problems of the genetic code initially emerging in an RNA world. Biosystems 164:155–166. https://doi.org/10.1016/j.biosystems.2017.09.006
Witzany G (2017) Two genetic codes: repetitive syntax for active non-coding RNAs; non-repetitive syntax for the DNA archives. Commun Integr Biol 10:e1297352. https://doi.org/10.1080/19420889.2017.1297352
Yarus M (2010) Getting past the RNA world: the initial Darwinian ancestor. Cold Spring Harb Perspect Biol 1:1–8. https://doi.org/10.1101/cshperspect.a003590
Yarus M (2011) The meaning of a minuscule ribozyme. Phil Trans R Soc B 366:2902–2909. https://doi.org/10.1098/rstb.2011.0139
Yeates JAM, Lehman N (2016) RNA networks at the origins of life. Biochemist 38:8–12
Acknowledgements
We would like to sincerely thank everyone who directly or indirectly contributed to the creation of this work. In particular, we would like to thank Michael J. Russell and André Brack for their critical reading of the manuscript and their useful suggestions. We would also like to thank Marko Dolinar, David H. Mathews, Peter F. Stadler, Nita Sahai, Alexei A. Sharov, Aaron Burton, Karo Michaelian, Aleksandar Simeonov, Günther Witzany and Andrew Robinson for the productive discussions and for providing us with valuable clues on references. A special thanks is acknowledged also to Robert Root-Bernstein for a clue regarding transfer factors. We also appreciate Aljaž Bolta’s help in the form of assistance in the preparation of the figures. Andrej Dobovišek acknowledges the financial support from the Slovenian Research Agency (research core funding No. P1-0055).
Author information
Authors and Affiliations
Ethics declarations
Conflict of Interest
The authors declare no competing financial interest.
Rights and permissions
About this article
Cite this article
Vitas, M., Dobovišek, A. In the Beginning was a Mutualism - On the Origin of Translation. Orig Life Evol Biosph 48, 223–243 (2018). https://doi.org/10.1007/s11084-018-9557-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-018-9557-6